
SAT-Based Method for Finding Attractors in Asynchronous
Multi-Valued Networks
Takehide Soh
1 a
, Morgan Magnin
2 b
, Daniel Le Berre
3 c
, Mutsunori Banbara
4 d
and Naoyuki Tamura
1 e
1
Kobe University, Information Infrastructure and Digital Transformation Initiatives Headquarters, 1-1, Rokko-dai, Nada,
Kobe, Hyogo 657-8501 Japan
2
Nantes Université, École Centrale Nantes, CNRS, LS2N, UMR 6004, F-44000 Nantes, France
3
Univ. Artois, CNRS, Centre de Recherche en Informatique de Lens (CRIL), F-62300 Lens, France
4
Nagoya University, Graduate School of Informatics, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan
Keywords:
Automata Network, Attractor, Constraint Programming, SAT.
Abstract:
In this paper, we propose a SAT-based method for finding attractors of bounded size in asynchronous automata
networks. The automata network is a multi-valued mathematical model which has been studied for the quali-
tative modeling of biological regulatory networks. An attractor is a minimal set of states in automata networks
that cannot be escaped and thus loops indefinitely. Attractors are crucial to validate the initial design of a
biological model and predict possible asymptotic behaviors, e.g., how cells may result through maturation in
differentiated cell types. Developing an efficient computational method to find attractors is thus an important
research topic. Our contribution is a translation of the problem of finding attractors of automata networks into
a sequence of propositional satisfiability (SAT) problems. We also propose to add two optional constraints
to improve the computation time of attractors. Experiments are carried out using 30 automata networks, 8
coming from real biological case studies and 22 crafted ones with controlled attractor size. The experimental
results show that our method scales better than the state-of-the-art ASP method when the size of the attractors
increases.
1 INTRODUCTION
Background. Understanding the mechanisms in-
volved in biological regulation is a fundamental is-
sue in analyzing living systems. The formal study of
the dynamics of biological systems raises many prob-
lems, e.g., identification of attractors, bifurcations,
and reachability, that are combinatorial by essence.
Making these issues scalable requires designing ef-
ficient methods that rely on efficient programming
frameworks. Considering this viewpoint, the long-
term behavior of a regulatory network’s dynamics
is of specific interest (Wuensche, 1998). Such out-
line has been interpreted as distinct responses of the
organism, such as differentiating into distinct cell
a
https://orcid.org/0000-0001-5897-9192
b
https://orcid.org/0000-0001-5443-0506
c
https://orcid.org/0000-0003-3221-9923
d
https://orcid.org/0000-0002-5388-727X
e
https://orcid.org/0000-0002-5466-1010
types in multicellular organisms (Huang et al., 2005).
Moreover, when refining a model of a living system,
one way to remove inconsistencies or to predict miss-
ing information in biological models consists in com-
paring the attractors of the model with the experimen-
tally observed long-term behavior. This explains why
our property of interest in this paper is the computa-
tion of attractors, an attractor being a minimal set of
states that cannot be escaped and thus loops indefi-
nitely.
Automata Network. Various kinds of mathemat-
ical models have been proposed for the qualitative
modeling of biological regulatory networks (BRNs).
These models include neural networks, differential
equations, Petri Nets, Boolean Networks (BN), prob-
abilistic Boolean networks, and other multi-valued
models such as synchronous/asynchronous Automata
Networks (AN).
In this paper, we focus on a subclass of automata
networks called Asynchronous Automata Networks
Soh, T., Magnin, M., Le Berre, D., Banbara, M. and Tamura, N.
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks.
DOI: 10.5220/0011675100003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 163-174
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
163

(AAN) (Folschette et al., 2015; Paulevé, 2016a),
which is convenient to model BRNs. In Automata
Networks, biological components (e.g., genes) are
represented as abstracted in the form of automata.
The different local states (that are not restricted to
Boolean values) of each automaton correspond to the
different discrete qualitative levels of the components
represented by the automaton. Interaction between bi-
ological components is modeled in an atomic way by
local transitions on the automaton, where each one is
conditioned by a set of required local states in dif-
ferent automata and can modify the local state of a
unique automaton. In other words, AANs allow to
have multiple requirements for a local transition to oc-
cur, but not to synchronize several local transitions in
different automata. In this sense, they are considered
Asynchronous Automata Networks. Synchronous se-
mantics could also be defined on such a model, but
this is out of the scope of the current paper. While
building a model, the choice of an appropriate seman-
tics is of crucial importance with regard to dynamical
properties. This discussion is a matter of research in
itself, with many papers balancing the merits of each
semantics. For some applications, like the biologi-
cal ones, asynchronous semantics is said to capture
more realistic behaviors in the sense that, at a given
time, a single gene can change its expression level.
For a more comprehensive discussion on this aspect,
(Garg et al., 2008) considers the differences and re-
spective merits of synchronous and asynchronous se-
mantics to model biological networks and identify
attractors. Note also that, depending on the cho-
sen semantics (asynchronous or synchronous), AANs
encompass the well-known Boolean frameworks of
René Thomas (Thomas, 1973) and Stuart Kauffman
(Kauffman, 1969).
Studies on Automata Networks. Automata net-
works are a very concise and convenient framework
to model BRNs. Their connection with other for-
mal models used in the Systems Biology commu-
nity (like logical networks, which they generalize) has
been formally proven in the past (Paulevé et al., 2012;
Folschette et al., 2015). In a more practical perspec-
tive, respective translations between model frame-
works have previously been implemented and inte-
grated altogether in the CoLoMoTo initiative (Naldi
et al., 2018) through the tool implementing (Paulevé,
2017). As such, the existing literature defining log-
ical models of gene regulatory networks or signal-
ing networks, e.g., (Grieco et al., 2013; Abou-Jaoudé
et al., 2015; Sahin et al., 2009) (among many oth-
ers) can benefit from results obtained on automata
networks (Levy et al., 2018). Many approaches have
thus been developed to analyze dynamical properties
on automata networks (Paulevé et al., 2013; Paulevé,
2016b; Fitime et al., 2017; Chai et al., 2020). In
(Rougny et al., 2021), the authors offer a representa-
tive example of the benefits of automata networks to
capture the behavior of complex biological systems
like the circadian clock while recognizing that such
works would greatly benefit the analysis of long-term
behaviors, i.e. attractors, untractable when too many
nodes and interactions are involved.
Computing Attractors. In recent years, the design
of efficient approaches to identify attractors in bio-
logical regulator networks mainly relied on the ASP
framework (Gebser et al., 2012). In (Ben Abdallah
et al., 2017), the authors proposed a first ASP encod-
ing to enumerate attractors up to a given length. Their
approach is iterative and without any assumption on
the topology of the interaction graph. Contrastingly,
the authors of (Khaled and Benhamou, 2020) enumer-
ate attractors in asynchronous Boolean networks that
have the form of a circuit using ASP by introducing a
new resolution semantics that does not use the usual
negation by failure. Instead, they rely on a weak ver-
sion of the negation by failure that allows them to pro-
vide an enumeration approach preventing the simula-
tion of the underlying networks. This approach tar-
gets networks with a given structure (all nodes must
have an incoming edge and some results have been
obtained only for cyclic interaction graphs). In this
paper, we do not assume any specific structure for the
networks.
Using SAT instead of ASP. If ASP solvers be-
came very popular thanks to their versatile model-
ing capability, SAT solvers became in two decades de
facto NP-complete oracles for solving a wide range
of problems, either decision or optimization ones,
on a wide range of complexity (from NP-complete
to PSPACE-complete)(Biere et al., 2021) thanks to
the availability of numerous, robust and increasingly
more efficient solvers. The use of SAT solvers re-
quires translating the original problem into a set of
clauses, with Boolean variables. Such translation is
most often ad-hoc, but can also use an intermedi-
ate modeling language for which a generic transla-
tion to SAT exists. In this paper, we use constraints
with integer variables as an intermediate modeling
language. We propose a constraints-based method for
finding attractors of bounded size in asynchronous au-
tomata networks implemented by translation into a
sequence of SAT problems. Our contribution is the
translation of the problem of finding attractors of au-
tomata networks into the enumeration of the solutions
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
164
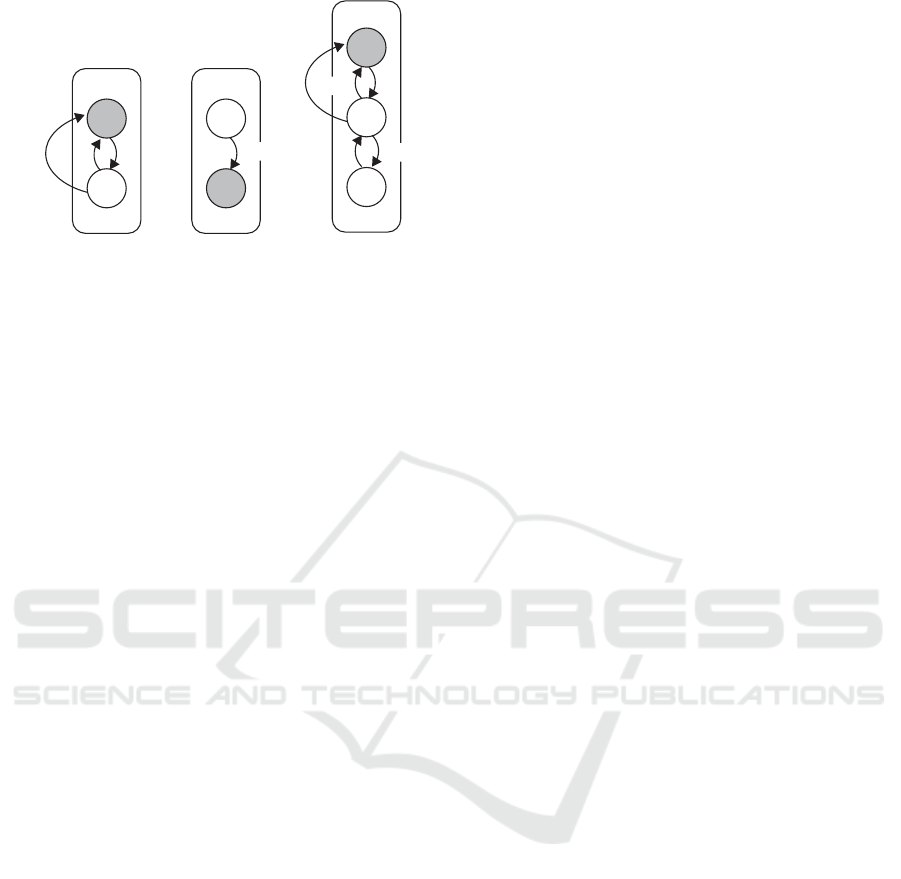
a b c
c
c
b
b
a
c
a
c
a
b
a
b
a
b
a
b
a
a
a
b
a
b
b
c
b
c
Σ = {a, b,c}
S = {a
0
,a
1
} × {b
0
,b
1
} × {c
0
,c
1
,c
2
}
T
a
= {(a
0
,a
1
,{c
0
}),(a
0
,a
1
,{b
1
,c
2
}),(a
1
,a
0
,{b
0
})}
T
b
= {(b
1
,b
0
,{a
0
,c
0
})}
T
c
= {(c
0
,c
1
,{a
0
}),(c
1
,c
0
,{a
0
,b
0
}),(c
1
,c
2
,{a
1
,b
1
}),
(c
1
,c
2
,{a
0
,b
0
}),(c
2
,c
1
,{})}
Figure 1: Automata Network Example: grey is used to represent one possible state of the model, that is (a
1
,b
0
,c
2
).
of a set of constraints. Notably, we redefine attrac-
tors by using two predicates about local playability
and global playability. Moreover, we propose to im-
prove the computation time of our approach thanks
to the addition of two optional constraints. The non-
Boolean constraints are finally translated to a Boolean
domain to complete the translation into a SAT prob-
lem, which allows using a wide variety of very effi-
cient SAT solvers. Experiments are carried out using
30 automata networks. The SAT approach is com-
pared favorably to the existing ASP approach on in-
stances containing larger-sized attractors.
2 PRELIMINARIES
2.1 Automata Networks
Definition 1. An automata network is a triple
(Σ,S, T) where Σ, S and T are defined as follows:
• Σ is a set of automata. We use a symbol α to de-
note an automaton α ∈ Σ.
• S =
∏
α∈Σ
S
α
is the finite set of global states,
where S
α
= {α
0
,α
1
,. ..} is a finite set of local
states of an automaton α ∈ Σ and α
v
denotes an
automaton α is in the state v.
• T is a finite set of transitions. A transition can
change the state of only one automaton α and is
represented in the form of (α
u
,α
v
,C). It changes
the state of the automaton α from u to v using a
condition C ⊂
S
α∈Σ
S
α
. In addition, we define
the transitions changing the state of an automaton
α as T
α
⊆ {(α
u
,α
v
,C) | α
u
,α
v
∈ S
α
,u ̸= v,C ⊂
S
α∈Σ
S
α
}.
Figure 1 shows an example of an automata net-
work. It contains three automata named a,b, c. We
focus on the automaton a to explain the example.
The automaton a can be in one of the two states 0
or 1. The automaton a has three transitions T
a
=
{(a
0
,a
1
,{c
0
}),(a
0
,a
1
,{b
1
,c
2
}),(a
1
,a
0
,{b
0
})}. The
transition (a
0
,a
1
,{b
1
,c
2
}) means that the transition
changes the state of the automaton a from 0 to 1 when
the two conditions b
1
– automaton b is in state 1 – and
c
2
– automaton c is in state 2 – hold.
2.2 Playability in Asynchronous
Automata Network
This section explains how a transition t ∈T on a sin-
gle automaton is used to define the transitions among
global states g ∈ S in an automata network (Σ, S, T ).
Asynchronous Update. The state of an automa-
ton can be updated either synchronously or asyn-
chronously. The synchronous update is the simplest
method and all automata are updated simultaneously.
The asynchronous update is a more realistic model
and all automata are not necessarily updated. A dif-
ference between the two update methods appears in
their state transition graphs (STGs). The synchronous
update is deterministic and thus the outdegree of each
node in STG is at most one. The asynchronous update
is nondeterministic and thus the outdegree of each
node in STG is not limited. In this paper, we focus
on the asynchronous update method. In particular, we
allow only one automaton to update, called general-
ized asynchronous, which is known to contain many
other update schemes as its special case (Gershenson,
2002).
In the following, we treat a global state g as a set
rather than a tuple if it is clear from the context.
Definition 2. (Local Playability of Transition). A
transition (α
u
,α
v
,C) ∈ T is locally playable in a
global state g ∈ S if {α
u
} ∪C ⊆ g holds.
Let g
i
,g
j
be two global states in S such that g
i
̸= g
j
and t = (α
u
,α
v
,C) ∈ T
α
is a transition changing the
state of α ∈ Σ.
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks
165
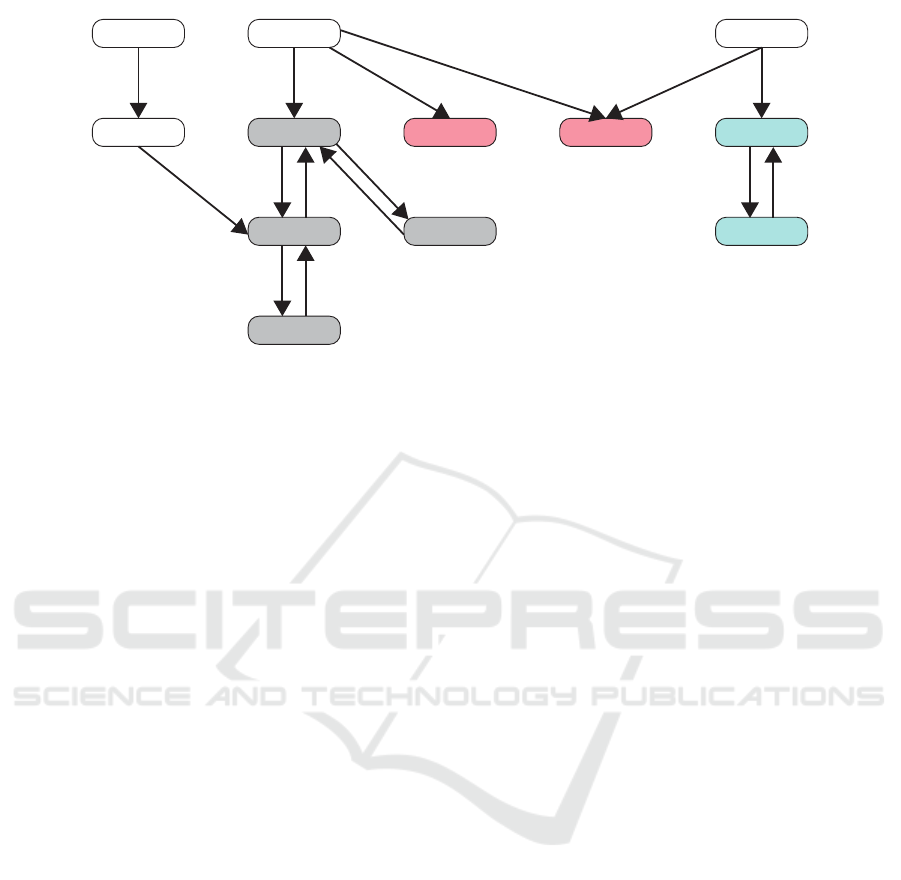
aa
b
b c
c
Figure 2: State Transition Graph Drawn from the Example Automata Network: red, blue, and grey are used to represent
attractors of size 1, 2 and 4, respectively.
Definition 3. (Global Playability of Transition). A
transition t = (α
u
,α
v
,C) ∈ T is globally playable
from a global state g
i
∈ S to a global state g
j
∈ S such
that the transition t is locally playable in the global
state g
i
, α
v
∈ g
j
, and |g
i
\ g
j
| = 1.
Note that the condition |g
i
\g
j
| = 1 guarantees the
difference between g
i
and g
j
is only one automaton,
i.e., the automaton α whose state is changed from u to
v according to the generalized asynchronous update.
2.3 Attractors
We define two predicates about the playability of a
transition t with respect to global states which facil-
itate the understanding of the definition of trap do-
mains and our proposed constraint model described
in Section 3.2.
P(t, g
i
)
def
⇐=⇒ {α
u
} ∪C ⊆ g
i
P(t, g
i
,g
j
)
def
⇐=⇒ P(t, g
i
) ∧ α
v
∈ g
j
∧ |g
i
\ g
j
| = 1
Then, trap domains and attractors are defined as
follows.
Definition 4. (Trap Domain). G ⊆ S is a trap do-
main if and only if ∀t ∈ T.∀g
i
∈ G.(P(t,g
i
) → ∃g
j
∈
G.(P(t, g
i
,g
j
))
Definition 5. (Attractor (or Minimal Trap Domain)).
G ⊆ S is an attractor if and only if there is no G
′
⊂ G
such that G
′
is a trap domain.
That is, attractors are trap domains that are
inclusion-minimal. The size of an attractor G is the
cardinality of G denoted as |G|.
Figure 2 shows the state transition graph of the
automata network of Figure 1. All attractors in this
network are:
• Size 1: {(a
0
,b
1
,c
1
)},{(a
1
,b
1
,c
0
)}
• Size 2: {(a
1
,b
1
,c
2
),(a
1
,b
1
,c
1
)}
• Size 4: {(a
0
,b
0
,c
0
),(a
0
,b
0
,c
1
),(a
0
,b
0
,c
2
),
(a
1
,b
0
,c
0
)}
In the state transition graph, attractors can be seen
as strongly connected components (SCCs).
Definition 6. Bounded Attractor Enumeration Prob-
lem (BAE). BAE is the problem of finding all attrac-
tors sized less than or equal to a given size.
• Input: An automata network (Σ,S, T) and a
bound k.
• Output: All attractors of the given automata net-
work whose sizes are less than or equal to k.
Given a BAE whose inputs are the automata net-
work of Figure 1 and k = 1, the output is 2 attractors.
In the case of k = 2, the output is 3 attractors. In the
case of k = 4, the output is 4 attractors.
3 A SAT-based MODEL
The proposed SAT-based approach is based on two
parts. The first one is our constraint model for con-
structing trap domains. The second one is the incre-
mental computation of trap domains. We ensure the
minimality of trap domains by computing them start-
ing from the smallest one.
3.1 Propositional Logic Recap
We consider propositional formulas in conjunctive
normal form (CNF). A literal is either a propositional
variable or its negation. A clause is a disjunction of
literals and a CNF formula is a conjunction of clauses.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
166
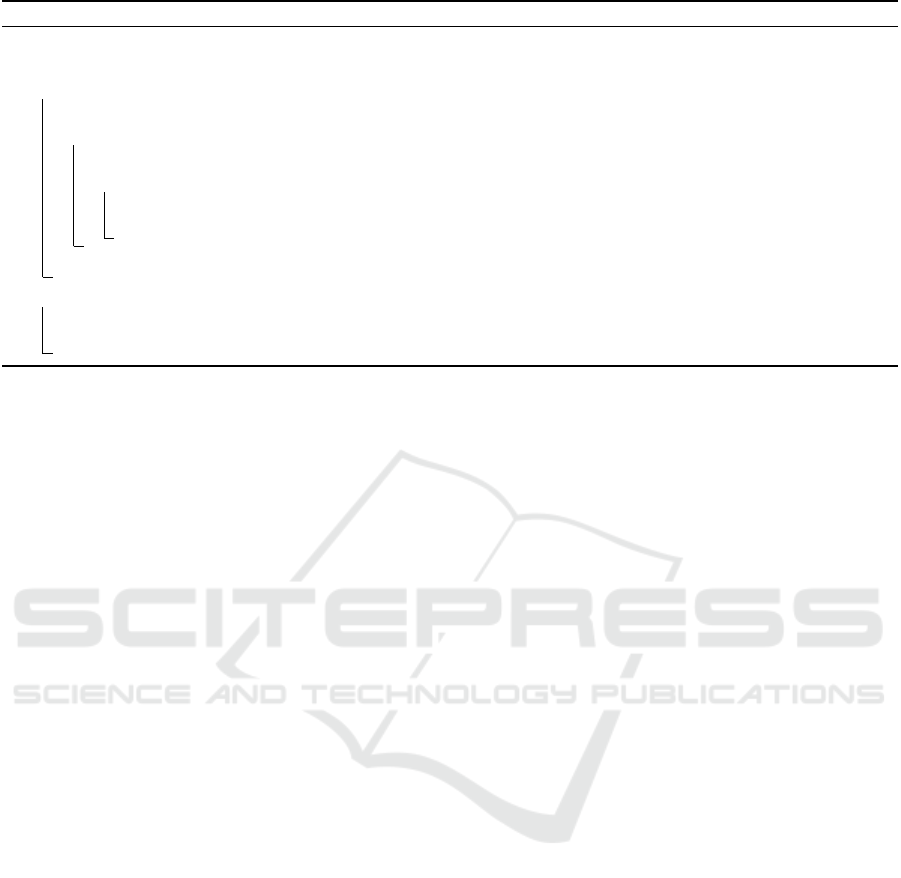
Algorithm 1: Algorithm for Computing Attractors.
Data: BAE: Automata network (Σ,S, T) and a bound k
Result: All attractors of size ≤ k
1 Function Main((Σ, S, T ) and a bound k):
2 Ω := {} ; // set of trap domains
3 for i = 1 to k do
4 Ψ
i
:= Construct Ψ
L
i
∧ Ψ
G
i
∧ Ψ
T
i
∧
V
ω∈Ω
BlockClauses(ω,i) ;
5 while Ψ
i
has a solution ω do
6 Ω := Ω ∪ { f (ω)} ; // f (ω) returns a trap domain from the solution ω.
7 Ψ
i
= Ψ
i
∧ BlockClauses(ω,i) ;
8 return Ω;
9 Function BlockClauses(Solution ω, Current Bound i):
10 generate clauses Ψ
ω
i
avoiding computation of trap domains containing a trap domain f (ω).;
11 return Ψ
ω
i
An assignment is a mapping from a set of proposi-
tional variables to the Boolean values true or false.
A propositional formula is satisfiable (SAT) if there
exists an assignment that satisfies it and is unsatisfi-
able (UNSAT) otherwise. In addition to ∨, ∧, ¬, we
use Boolean operators ↔, → which are easily trans-
lated in CNF formulas. A SAT solver is a program
that computes a solution (if any) from a CNF formula.
Note that the constraint model in the following
section is partially not propositional, i.e., we use
some constraints over integer variables for clarity to
the reader. Those non-Boolean constraints are sur-
rounded by symbols JK. They will be translated into
Boolean constraints using the encodings in Table 1 in
Section 5.
3.2 Constraint Model for Trap Domain
Variables. For each t ∈ T and 1 ≤ i ≤ k, we intro-
duce a propositional variable p
t,i
that is true when a
transition t ∈ T is locally playable from a global state
g
i
∈ S. For each t ∈ T and 1 ≤ i ̸= j ≤ k, we introduce
a propositional variable p
t,i, j
that is true when a tran-
sition t ∈ T is globally playable from a global state
g
i
∈ S to a global state g
j
∈ S. For each α ∈ Σ and
1 ≤ i ≤ k, we introduce an integer variable x
α,i
that is
x
α,i
= u iff an automaton α is in a local state u ∈ S
α
at
a global state g
i
∈ S, i.e., α
u
∈ g
i
.
Constraints. We introduce the following con-
straints to compute trap domains using the variables
explained. A symbol •t denotes the condition of
the local playability of a transition t, that is, •t =
{{α
u
} ∪C} when t = (α
u
,α
v
,C). The first constraint
is the definition of the local playability of transitions.
We denote this constraint as Ψ
L
k
.
p
t,i
↔
^
α
u
∈•t
Jx
α,i
= uK
!
(t ∈ T, 1 ≤ i ≤ k)
The second constraint is the definition of the global
playability of transitions. We denote this constraint
as Ψ
G
k
.
p
t,i, j
↔
p
t,i
∧ Jx
α, j
= vK ∧
^
α
′
∈Σ\α
Jx
α
′
,i
= x
α
′
, j
K
(t = (α
u
,α
v
,C) ∈ T, 1 ≤ i ̸= j ≤ k)
The third constraint is the definition of the trap do-
main. We denote this constraint as Ψ
T
k
.
p
t,i
→
_
j̸=i
1≤ j≤k
p
t,i, j
(t ∈ T, 1 ≤ i ≤ k)
Solutions. Let ω be any solution of Ψ
L
k
∧ Ψ
G
k
∧ Ψ
T
k
,
which are assignments to propositional or integer
variables t
i
, t
i, j
, and x
α,i
that satisfies it. In a solu-
tion ω, x
α,i
= u means α
u
∈ g
i
. Let f be a mapping
from a solution to global states. For instance, suppose
that a solution ω contains x
a,0
= 1, x
b,0
= 1, x
c,0
= 2
and x
a,1
= 1,x
b,1
= 1,x
c,1
= 1. Then, global states
f (ω) are {g
1
,g
2
}, where g
1
= (a
1
,b
1
,c
2
) and g
2
=
(a
1
,b
1
,c
1
).
Proposition 1. Global states f(ω) from solutions ω
of Ψ
L
k
∧ Ψ
G
k
∧ Ψ
T
k
are trap domains.
(Proof) The two propositional variables p
t,i
in Ψ
L
k
and p
t,i, j
in Ψ
G
k
correspond to the definition of the two
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks
167

predicates P(t, g
i
) and P(t, g
i
,g
j
), respectively. The
constraint Ψ
T
k
is the same as the definition of the trap
domain: ∀t ∈ T.∀g
i
∈ G.(P(t,g
i
) → ∃g
j
(P(t, g
i
,g
j
)).
Thus, global states f (ω) are trap domains.
Note that we do not need ← of Ψ
G
k
to solve BAE
since p
t,i, j
appears only positively in Ψ
T
k
.
3.3 Computing Attractors
Algorithm 1 shows an algorithm to compute attractors
using the constraint model from the previous section.
Input is a BAE instance, i.e., an automata network
(Σ,S, T ) and a bound k. Output is all attractors whose
sizes are less than or equal to k. Line 2 initializes a
set Ω that contains attractors. Lines 3 to 7 incremen-
tally compute attractors from size 1 to size k. Line 5
launches a SAT solver to compute an attractor of size
i. Lines 9 to 11 discard the attractors found so far.
Specifically, the blocking clauses Ψ
ω
i
are as follows.
^
1≤ j≤i
_
α∈Σ
α
u
∈g
Jx
α, j
̸= uK (g ∈ f(w))
Proposition 2. Algorithm 1 outputs all attractors
sized less than or equal to k.
(Proof) We use mathematical induction on k. i) in the
case of k = 1, Algorithm 1 outputs all trap domains
sized 1. Obviously, those trap domains are minimal.
Thus, the proposition holds at k = 1. ii) if the propo-
sition holds at k, then Algorithm 1 outputs all trap
domains sized k + 1 and those trap domains are min-
imal since all trap domains less than or equal to k are
enumerated and blocked. Therefore, by i) and ii), the
proposition holds.
4 TUNING OUR MODEL
To represent the next constraints, we need to intro-
duce new propositional variables. A propositional
variable p
t,i→
is true iff a propositional variable p
t,i, j
is true for some j. A propositional variable p
t, j←
is
true iff a propositional variable p
t,i, j
is true for some
i. A propositional variable p
t
is true iff propositional
variables p
t,i→
or p
t,i←
are true for some i.
p
t,i→
↔
_
1≤ j≤k
p
t,i, j
(t ∈ T, 1 ≤ i ≤ k)
p
t, j←
↔
_
1≤i≤k
p
t,i, j
(t ∈ T, 1 ≤ j ≤ k)
p
t
↔
_
1≤i≤k
p
t,i→
∨ p
t,i←
(t ∈ T )
Note that the propositional variable p
t,i→
is redundant
because it is the same as the propositional variable p
t,i
in Section3.2. We rewrite here for clarity to the reader.
4.1 For Attractors of Size Greater
than 1
When we search attractors sized ≥ 2, we can add the
following constraints since each global state in attrac-
tors should have at least one indegree and one outde-
gree in STGs.
_
t∈T
p
t,i→
(1 ≤ i ≤ k)
_
t∈T
p
t, j←
(1 ≤ j ≤ k)
These constraints are useful when there are a huge
number of attractors sized 1 since we do not need to
block each of them individually.
4.2 With Optional Cycle Constraints
Let T
′
α
be a set of transitions, which changes the state
of an automaton α, i.e., T
′
α
⊆ T
α
. We call T
′
α
are cy-
cles of transitions in an automaton α when T
′
α
are (not
simple, and possibly multiple) cycles.
An important property of attractors sized ≥ 2 is
that these consist of (not simple, and possibly multi-
ple) cycles of transitions in each automata T
α
. Con-
sider the attractor {(a
1
,b
1
,c
2
),(a
1
,b
1
,c
1
)} in the ex-
ample, it consists of the cycle of transitions of the
automata c, i.e., (c
1
,c
2
,{a
1
,b
1
}) and (c
2
,c
1
,{}). In
general, we can say the following proposition.
Proposition 3. (Transitions consisting attractors).
Let T
′
α
be a set of transitions such that T
′
α
⊆ T
α
.
Given an attractor G, we denote all transitions glob-
ally playable in G as
S
α∈Σ
T
′
α
. Then, for each α, T
′
α
are cycles of transitions or an empty set.
(Proof) We use a proof by contradiction. Suppose that
the set of transitions T
′
α
are not cycles of transitions.
Then, there is at least one state that cannot be reached
by using transitions of T
′
α
. Since any attractors are
strongly connected components in its state transition
graph, it is a contradiction.
For instance, simple cycles of transitions in Fig-
ure 1 are listed as follows.
γ
1
= {(a
0
,a
1
,{c
0
}),(a
1
,a
0
,{b
0
})}
γ
2
= {(a
0
,a
1
,{b
1
,c
2
}),(a
1
,a
0
,{b
0
})}
γ
3
= {(c
0
,c
1
,{a
0
}),(c
1
,c
0
,{a
0
,b
0
})}
γ
4
= {(c
1
,c
2
,{a
1
,b
1
}),(c
2
,c
1
,{})}
γ
5
= {(c
1
,c
2
,{a
0
,b
0
}),(c
2
,c
1
,{})}
Then, all sets of transitions globally playable in at-
tractors are combinations from the following list.
T
′
a
∈ {{},γ
1
,γ
2
,γ
1
∪ γ
2
}
T
′
b
∈ {{}}
T
′
c
∈ {{}, γ
3
,γ
4
,γ
5
,γ
3
∪ γ
4
,γ
3
∪ γ
5
,γ
4
∪ γ
5
,γ
3
∪ γ
4
∪ γ
5
}
For instance, all transitions globally playable in the
attractors sized ≥ 2 in the example are as follows.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
168

• {(a
1
,b
1
,c
2
),(a
1
,b
1
,c
1
)}
– T
′
a
= {} and T
′
c
= γ
4
.
• {(a
0
,b
0
,c
0
),(a
0
,b
0
,c
1
),(a
0
,b
0
,c
2
),(a
1
,b
0
,c
0
)}
– T
′
a
= γ
1
and T
′
c
= γ
3
∪ γ
4
.
Using Proposition 3, we can add the following op-
tional constraints. Let Γ be the set of simple cycles in
a given automata network. We introduce a proposi-
tional variable p
γ
that is true if a cycle γ ∈ Γ is con-
tained in a solution. We add the following constraints.
p
γ
→
^
t∈γ
p
t
(γ ∈ Γ)
p
t
→
_
γ∈Γ
t∈γ
p
γ
(t ∈ T )
The first constraint ensures that if a cycle γ is con-
tained in a solution then all its transitions also belong
to the solution. The second constraint ensures that if
a transition t is playable then one of the cycles γ that
contain the transition t belongs to the solution.
4.3 With Optional Ordering
Constraints
Solutions of the basic constraints discussed in Section
3.2 contain many symmetries. In the case of an attrac-
tor {(a
1
,b
1
,c
2
),(a
1
,b
1
,c
1
)} from the example in Fig-
ure 2, two solutions can be computed. The first one
ω
1
is:
• x
a,0
= 1,x
b,0
= 1,x
c,0
= 2 (i.e., (a
1
,b
1
,c
2
))
• x
a,1
= 1,x
b,1
= 1,x
c,1
= 1 (i.e., (a
1
,b
1
,c
1
))
The second one ω
2
is:
• x
a,0
= 1,x
b,0
= 1,x
c,0
= 1 (i.e., (a
1
,b
1
,c
1
))
• x
a,1
= 1,x
b,1
= 1,x
c,1
= 2 (i.e., (a
1
,b
1
,c
2
))
Those two solutions are identical in the sense that
both solutions represent the same attractor. The tran-
sitions are simply not in the same order.
Using the current constraints, the solver could find
any of them. The other one will be later discarded by
the blocking clauses.
It is possible to force the order of the transitions,
i.e. to represent each attractor only once. This is a
standard technique in constraints programming called
"symmetry breaking" (Crawford et al., 1996).
Given a sequence of global states g
1
,g
2
,. . . , g
k
,
we add constraints forcing g
1
≺ g
2
∧ ·· · ∧ g
k−1
≺ g
k
,
where g
i
≺ g
i+1
means the global state g
i
is lexi-
cographically strictly smaller than g
i+1
. Let Σ be
{a
1
,a
2
,. . . a
n
}. Then the constraints g
i
≺ g
i+1
are de-
fined as follows. Note that ◁ is < if j = n and ≤
otherwise.
Jx
a
j
,i
≤ x
a
j
,i+1
K ( j = 1)
^
1≤m≤ j−1
Jx
a
m
,i
= x
a
m
,i+1
K
!
→ Jx
a
j
,i
◁ x
a
j
,i+1
K (2 ≤ j ≤ n)
By this constraint, only ω
2
can be a solution. It is
known that symmetry breaking constraints are often
effective for unsatisfiable instances since it reduces
search spaces.
5 ENCODING CONSTRAINTS IN
CNF
The constraints in Section 3.2 and 4 are almost propo-
sitional constraints but we need to encode some inte-
ger constraints surrounded by JK. There have been
many encoding methods that encode integer con-
straints into propositional clauses: direct encoding (de
Kleer, 1989; Walsh, 2000), log encoding (Iwama
and Miyazaki, 1994; Gelder, 2008), order encod-
ing (Crawford and Baker, 1994; Tamura et al., 2009),
and hybrid encoding (Soh et al., 2017). The bench-
marks used in the next section contain automata with
at most 3 states, and biological systems rarely have
more than 4 states (Folschette et al., 2015). Thus, we
use the direct encoding because it is simple and effi-
cient when there are few integer or domain values to
encode.
Let D be a mapping from a variable to its domain
values. Let p
x=d
be a propositional variable that is
true when x = d holds, where d ∈ D(x). We encode in-
teger variables’ axiom, a variable is assigned exactly
one value, as follows.
_
d∈D(x)
p
x=d
(x ∈ X)
^
d,d
′
∈D(x)
d̸=d
′
¬p
x=d
∨ ¬p
x=d
′
(x ∈ X)
Using the propositional variable p
x=d
, integer
constraints are encoded as summarized in Table 1.
Note that the translation of symmetry breaking con-
straints in Section 4.3 can be done in linear size by
using Tseitin translation (Tseitin, 1968).
Table 2 shows the order of the number of con-
straints and clauses. We here assume the followings
to ease the discussion: |S
α
| and | •t| are constant val-
ues; |T| > |Γ|; |Γ| ≫ |γ|. The table shows us that the
encoding is dominated by the definition of globally
playable transitions when k becomes large.
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks
169

Table 1: Encoding Constraints.
Constraints Clauses
Jx = vK p
x=v
Jx = x
′
K
^
d∈D(x)
d
′
∈D(x
′
)
d=d
′
p
x=d
↔ p
x
′
=d
′
Jx < x
′
K
^
d∈D(x)
d
′
∈D(x
′
)
¬(d<d
′
)
¬p
x=d
∨ ¬p
x
′
=d
′
Jx ≤ x
′
K
^
d∈D(x)
d
′
∈D(x
′
)
¬(d≤d
′
)
¬p
x=d
∨ ¬p
x
′
=d
′
Table 2: Number of Constraints and Clauses.
Name #Constraints #Clauses
Ψ
L
k
O(|T||k|) O(|T||k|| •t|)
Ψ
G
k
O(|T||k|
2
|Σ|) O(|T||k|
2
|Σ||S
α
|
2
)
Ψ
T
k
O(|T||k|) O(|T||k|)
Cycle O(|T|) O(|T|)
Sym. Break. O(|k||Σ|) O(|k||Σ||S
α
|
2
)
6 EXPERIMENTS
To check the performance of the proposed SAT-based
method, we carried out an experimental compari-
son against the state-of-the-art ASP-based method
from (Ben Abdallah et al., 2017). Our approach is
implemented using Scala 2.12.4 to generate the CNF
from the automata network description. We use two
SAT solvers in our implementation. For k = 1, we use
BDD_MINISAT_ALL (Toda and Soh, 2016). This solver
is good at problems that are easy but have many solu-
tions. For k > 1, we use CaDiCaL (Biere et al., 2020).
This solver is good at problems that are hard to solve.
6.1 Experimental Conditions
Existing Benchmark. 30 automata networks are
used. The first 8 networks are all instances used
in the previous study (Ben Abdallah et al., 2017),
which are inspired from real organisms and found in
the literature: Example (Ben Abdallah et al., 2017),
Lambda phage (Thieffry and Thomas, 1995), Trp-
reg (Simão et al., 2005), Fission-yeast (Davidich and
Bornholdt, 2008), Mamm (Fauré et al., 2006), Tcr-
sig (Klamt et al., 2006), FGF (Mbodj et al., 2013),
and T-helper (Abou-Jaoudé et al., 2015). These net-
works contain relatively small-sized attractors.
Figure 3: The State Transition Graph of star03.
Crafted Benchmark. We further contribute to an
artificial benchmark set named star to check the scal-
ability w.r.t. the number of automata, the number
of transitions, and the size of cyclic attractors. This
benchmark is available on the repository
1
.
Given a parameter N > 1, starN is an automata
network (Σ,S, T) defined as follows.
• Σ = {a
i
| 1 ≤ i ≤ N}
• S = {a
1
0
,a
1
1
} × {a
2
0
,a
2
1
} × ·· · × {a
N
0
,a
N
1
}
• T = {(a
i
0
,a
i
1
,{a
j
0
| 1 ≤ j ≤ N,i ̸= j}) | 1 ≤ i ≤
N} ∪ {(a
i
1
,a
i
0
,{}) | 1 ≤ i ≤ N}.
Each starN automata network contains N automata
that have two states, 2N transitions, and only one at-
tractor of size N + 1.
star03 is given below.
• Σ = {a
1
,a
2
,a
3
}
• S = {a
1
0
,a
1
1
} × {a
2
0
,a
2
1
} × {a
3
0
,a
3
1
}
• T = T
a
1
∪ T
a
2
∪ T
a
3
– T
a
1
= {(a
1
0
,a
1
1
,{a
2
0
,a
3
0
}),(a
1
1
,a
1
0
,{})}
– T
a
2
= {(a
2
0
,a
2
1
,{a
1
0
,a
3
0
}),(a
2
1
,a
2
0
,{})}
– T
a
3
= {(a
3
0
,a
3
1
,{a
1
0
,a
2
0
}),(a
3
1
,a
3
0
,{})}
Figure 3 shows the attractor of star03 in gray.
Bound k. For all benchmarks, the attractors are
known. In this experiment, we set a bound k for
each automata network so that solvers can compute
the largest attractors.
Environment. We execute all experiments on the
machine that equips 3GHz CPU and 16GB RAM. The
time limit is 3 hours.
6.2 Results
Table 3 shows comparisons between the proposed
method and the state-of-the-art existing ASP-based
method (Ben Abdallah et al., 2017). The first col-
umn denotes the name of the automata networks. The
1
https://doi.org/10.5281/zenodo.7460387
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
170

Table 3: Comparisons between Proposed SAT-based Methods and the Existing Method.
Statictics Proposed Existing
Instance |Σ| |T| Attractors k |S
max
| Basic +C +S +C+S ASP
Example 4 12 1(3):2(1):4(1) 4 3 1.6 1.6 1.7 1.7 0.2
Lambda phage 4 46 1(1):2(1) 2 4 1.7 1.8 1.8 1.7 0.1
Trp-reg 4 14 1(2):4(1) 4 3 1.7 1.7 1.7 1.7 0.2
Fission-yeast 9 43 1(1) 1 3 1.4 1.4 1.5 1.4 <0.1
Mamm. 10 34 1(1) 1 2 1.3 1.3 1.3 1.3 <0.1
Tcrsig 40 85 1(8) 1 2 1.6 1.7 1.8 1.8 <0.1
FGF 59 102 1(1536) 1 3 1.8 1.8 1.9 1.8 <0.1
T-helper 101 316 1(5875504) 1 3 88.6 86.6 88.9 88.4 148.3
star02 2 4 3(1) 3 2 1.3 1.3 1.3 1.3 0.1
star03 3 6 4(1) 4 2 1.4 1.5 1.3 1.5 0.3
star04 4 8 5(1) 5 2 1.7 1.6 1.6 1.6 0.6
star05 5 10 6(1) 6 2 2.0 1.9 1.7 1.8 1.6
star06 6 12 7(1) 7 2 2.6 2.2 2.1 2.1 3.6
star07 7 14 8(1) 8 2 6.5 2.8 2.4 2.3 10.7
star08 8 16 9(1) 9 2 46.5 5.9 2.9 2.7 27.1
star09 9 18 10(1) 10 2 538.6 36.7 3.4 3.2 65.2
star10 10 20 11(1) 11 2 6598.8 442.7 4.1 4.0 148.0
star11 11 22 12(1) 12 2 T.O. 8787.0 4.6 4.6 1088.2
star12 12 24 13(1) 13 2 T.O. T.O. 5.9 4.8 9959.3
star13 13 26 14(1) 14 2 T.O. T.O. 7.8 5.2 T.O.
star14 14 28 15(1) 15 2 T.O. T.O. 12.1 5.9 T.O.
star15 15 30 16(1) 16 2 T.O. T.O. 19.8 7.6 T.O.
star16 16 32 17(1) 17 2 T.O. T.O. 24.5 9.7 T.O.
star17 17 34 18(1) 18 2 T.O. T.O. 38.8 11.5 T.O.
star18 18 36 19(1) 19 2 T.O. T.O. 57.7 14.5 T.O.
star19 19 38 20(1) 20 2 T.O. T.O. 87.7 19.3 T.O.
star20 20 40 21(1) 21 2 T.O. T.O. 146.3 28.5 T.O.
star30 30 60 31(1) 31 2 T.O. T.O. 6250.3 614.4 T.O.
star40 40 80 41(1) 41 2 T.O. T.O. T.O. 7296.7 T.O.
second column denotes the number of automata in-
cluded. The third column denotes the number of
transitions included. The fourth column denotes the
sizes of the attractors. The number of attractors for
each size is displayed between the parenthesis. The
fifth column denotes the bound k given to the solvers.
The sixth column denotes the maximum number of
states of automata denoted as S
max
. The seventh
to tenth columns denote the CPU time of the pro-
posed method. “Basic” denotes the basic constraint
model described in Section 3.2 with at least one con-
straint in Section 4.1. “+C” denotes the basic con-
straint model with the cycle constraint discussed in
Section 4.2. “+S” denotes the basic constraint model
with the symmetry breaking constraint discussed in
Section 4.3. “+C+S” denotes all constraints described
in Section 3.2 and Section 4. The eleventh col-
umn denotes the CPU time of the existing ASP-based
method.
In the biologically inspired benchmarks, all meth-
ods solved them within a few seconds. The main
reason is that they contain small-sized attractors and
thus k is also small. The difference between the pro-
posed methods and the ASP-based method mainly
comes from the implementation language: C++ and
Scala. The proposed method is implemented on Scala
which is running on a Java Virtual Machine (JVM) for
the modeling part and thus there is a small disadvan-
tage on this point. One exception is T-helper which
contains a large number of attractors. On this prob-
lem, our approach takes advantage of the SAT solver
BDD_MINISAT_ALL dedicated to the fast enumeration
of solutions.
On star benchmarks, the difference between each
method is more obvious. The CPU time of the ASP-
based method increases exponentially, and it cannot
solve star13 within 3 hours. On the other hand, al-
though “Basic” cannot solve star11, the two optional
constraints successfully improve “Basic”. The best
results are obtained by combining those constraints.
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks
171

7 CONCLUSIONS
This paper describes a SAT-based method for finding
attractors of bounded size in asynchronous automata
networks. Attractors are crucial to validate the ini-
tial design of a biological model and predict possi-
ble asymptotic behaviors, e.g., how cells may result
through maturation in differentiated cell types. Given
a bound k, we propose a constraint model of trap do-
mains and compute attractors by incrementally com-
puting them from 1 to k. We further propose two op-
tional kinds of constraints to improve the computation
time of our approach. The first one denotes cycle con-
straints which utilize a property of transitions in au-
tomata networks. The second one denotes symmetry
breaking constraints which reduce redundant search
spaces contained in the initial constraint model. Ex-
perimental evaluations are carried out over 30 au-
tomata networks. While the proposed SAT-based ap-
proach and the state-of-the-art ASP one could hardly
be discriminated on the few existing biologically in-
spired benchmarks, their behavior is quite different on
the crafted benchmarks we contribute. While the per-
formance of the initial SAT-based model is not so ef-
fective, adding optional constraints allows it to scale
much better than the ASP approach on benchmarks
with controlled attractor size. Future work is listed
as follows. Extending the comparisons to methods
of other networks like Boolean network (Mori and
Akutsu, 2022; Trinh et al., 2022; Rozum et al., 2021;
Inoue, 2011) is necessary. Extending the proposed
method to more complex networks like Petri net is
interesting. Supplementary materials of this paper is
available on the repository
2
.
ACKNOWLEDGEMENTS
This work was financially supported by the “PHC
Sakura” program (43009SC, JPJSBP120193213), im-
plemented by the French Ministry for Europe and
Foreign Affairs, the French Ministry of Higher Ed-
ucation, Research and Innovation and the Japan So-
ciety for Promotion of Science. This work was
also supported by JSPS KAKENHI Grant Numbers
JP20K11748 and JP20H05794.
REFERENCES
Abou-Jaoudé, W., Monteiro, P. T., Naldi, A., Grandclaudon,
M., Soumelis, V., Chaouiya, C., and Thieffry, D.
2
https://doi.org/10.5281/zenodo.7460387
(2015). Model checking to assess t-helper cell plas-
ticity. Frontiers in bioengineering and biotechnology,
2:86.
Ben Abdallah, E., Folschette, M., Roux, O., and Magnin,
M. (2017). Asp-based method for the enumeration
of attractors in non-deterministic synchronous and
asynchronous multi-valued networks. Algorithms for
Molecular Biology, 12(1):1–23.
Biere, A., Fazekas, K., Fleury, M., and Heisinger, M.
(2020). CaDiCaL, Kissat, Paracooba, Plingeling and
Treengeling entering the SAT Competition 2020. In
Balyo, T., Froleyks, N., Heule, M., Iser, M., Järvisalo,
M., and Suda, M., editors, Proc. of SAT Competition
2020 – Solver and Benchmark Descriptions, volume
B-2020-1 of Department of Computer Science Report
Series B, pages 51–53. University of Helsinki.
Biere, A., Heule, M., van Maaren, H., and Walsh, T., editors
(2021). Handbook of Satisfiability - Second Edition,
volume 336 of Frontiers in Artificial Intelligence and
Applications. IOS Press.
Chai, X., Ribeiro, T., Magnin, M., Roux, O., and Inoue,
K. (2020). Static analysis and stochastic search for
reachability problem. Electronic Notes in Theoretical
Computer Science, 350:139–158.
Crawford, J. M. and Baker, A. B. (1994). Experimental
results on the application of satisfiability algorithms
to scheduling problems. In Proceedings of the 12th
National Conference on Artificial Intelligence (AAAI
1994), pages 1092–1097.
Crawford, J. M., Ginsberg, M. L., Luks, E. M., and
Roy, A. (1996). Symmetry-breaking predicates for
search problems. In Aiello, L. C., Doyle, J., and
Shapiro, S. C., editors, Proceedings of the Fifth Inter-
national Conference on Principles of Knowledge Rep-
resentation and Reasoning (KR’96), Cambridge, Mas-
sachusetts, USA, November 5-8, 1996, pages 148–
159. Morgan Kaufmann.
Davidich, M. I. and Bornholdt, S. (2008). Boolean network
model predicts cell cycle sequence of fission yeast.
PloS one, 3(2):e1672.
de Kleer, J. (1989). A comparison of ATMS and CSP tech-
niques. In Proceedings of the 11th International Joint
Conference on Artificial Intelligence (IJCAI 1989),
pages 290–296.
Fauré, A., Naldi, A., Chaouiya, C., and Thieffry, D. (2006).
Dynamical analysis of a generic boolean model for the
control of the mammalian cell cycle. Bioinformatics,
22(14):e124–e131.
Fitime, L. F., Roux, O., Guziolowski, C., and Paulevé,
L. (2017). Identification of bifurcation transitions
in biological regulatory networks using Answer-Set
Programming. Algorithms for Molecular Biology,
12(1):19.
Folschette, M., Paulevé, L., Magnin, M., and Roux, O.
(2015). Sufficient conditions for reachability in au-
tomata networks with priorities. Theoretical Com-
puter Science, 608:66–83.
Garg, A., Di Cara, A., Xenarios, I., Mendoza, L., and
De Micheli, G. (2008). Synchronous versus asyn-
chronous modeling of gene regulatory networks.
Bioinformatics, 24(17):1917–1925.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
172

Gebser, M., Kaminski, R., Kaufmann, B., and Schaub, T.
(2012). Answer Set Solving in Practice. Synthe-
sis Lectures on Artificial Intelligence and Machine
Learning. Morgan & Claypool Publishers.
Gelder, A. V. (2008). Another look at graph coloring via
propositional satisfiability. Discrete Applied Mathe-
matics, 156(2):230–243.
Gershenson, C. (2002). Classification of random boolean
networks. In Proceedings of the Eighth International
Conference on Artificial Life, pages 1–8.
Grieco, L., Calzone, L., Bernard-Pierrot, I., Radvanyi, F.,
Kahn-Perles, B., and Thieffry, D. (2013). Integrative
modelling of the influence of mapk network on can-
cer cell fate decision. PLoS computational biology,
9(10):e1003286.
Huang, S., Eichler, G., Bar-Yam, Y., and Ingber, D. E.
(2005). Cell fates as high-dimensional attractor states
of a complex gene regulatory network. Physical re-
view letters, 94(12):128701.
Inoue, K. (2011). Logic programming for boolean net-
works. In IJCAI 2011, Proceedings of the 22nd In-
ternational Joint Conference on Artificial Intelligence,
Barcelona, Catalonia, Spain, July 16-22, 2011, pages
924–930.
Iwama, K. and Miyazaki, S. (1994). SAT-variable complex-
ity of hard combinatorial problems. In Proceedings of
the IFIP 13th World Computer Congress, pages 253–
258.
Kauffman, S. A. (1969). Metabolic stability and epigene-
sis in randomly constructed genetic nets. Journal of
Theoretical Biology, 22(3):437–467.
Khaled, T. and Benhamou, B. (2020). An asp-based ap-
proach for boolean networks representation and at-
tractor detection. In LPAR, pages 317–333.
Klamt, S., Saez-Rodriguez, J., Lindquist, J., Simeoni, L.,
and Gilles, E. (2006). A methodology for the struc-
tural and functional analysis of signaling and regula-
tory networks. BMC Bioinformatics, 7(1):56.
Levy, N., Naldi, A., Hernandez, C., Stoll, G., Thieffry, D.,
Zinovyev, A., Calzone, L., and Paulevé, L. (2018).
Prediction of Mutations to Control Pathways Enabling
Tumour Cell Invasion with the CoLoMoTo Interactive
Notebook (Tutorial) . Frontiers in Physiology, 9:787.
Mbodj, A., Junion, G., Brun, C., Furlong, E. E., and Thief-
fry, D. (2013). Logical modelling of drosophila sig-
nalling pathways. Molecular BioSystems, 9(9):2248–
2258.
Mori, T. and Akutsu, T. (2022). Mini review attractor de-
tection and enumeration algorithms for boolean net-
works. Computational and Structural Biotechnology
Journal.
Naldi, A., Hernandez, C., Levy, N., Stoll, G., Monteiro,
P. T., Chaouiya, C., Helikar, T., Zinovyev, A., Cal-
zone, L., Cohen-Boulakia, S., et al. (2018). The
colomoto interactive notebook: accessible and repro-
ducible computational analyses for qualitative biolog-
ical networks. Frontiers in physiology, 9:680.
Paulevé, L. (2016a). Goal-oriented reduction of automata
networks. In International Conference on Compu-
tational Methods in Systems Biology, volume 9859
of Lecture Notes in Bioinformatics, pages 252–272.
Springer.
Paulevé, L. (2016b). Goal-oriented reduction of automata
networks. In International Conference on Computa-
tional Methods in Systems Biology, pages 252–272.
Springer.
Paulevé, L. (2017). Pint: a static analyzer for transient
dynamics of qualitative networks with IPython inter-
face. In CMSB 2017 - 15th conference on Computa-
tional Methods for Systems Biology, volume 10545 of
Lecture Notes in Computer Science, pages 309–316.
Springer International Publishing.
Paulevé, L., Andrieux, G., and Koeppl, H. (2013). Under-
approximating cut sets for reachability in large scale
automata networks. In International Conference on
Computer Aided Verification, pages 69–84. Springer.
Paulevé, L., Magnin, M., and Roux, O. (2012). From the
Process Hitting to Petri Nets and Back. Technical Re-
port hal-00744807, ETH Zürich.
Rougny, A., Paulevé, L., Teboul, M., and Delaunay, F.
(2021). A detailed map of coupled circadian clock and
cell cycle with qualitative dynamics validation. BMC
bioinformatics, 22(1):1–24.
Rozum, J. C., Deritei, D., Park, K. H., Gómez Tejeda Za-
ñudo, J., and Albert, R. (2021). pystablemotifs:
Python library for attractor identification and control
in Boolean networks. Bioinformatics, 38(5):1465–
1466.
Sahin, Ö., Fröhlich, H., Löbke, C., Korf, U., Burmester,
S., Majety, M., Mattern, J., Schupp, I., Chaouiya, C.,
Thieffry, D., et al. (2009). Modeling erbb receptor-
regulated g1/s transition to find novel targets for de
novo trastuzumab resistance. BMC systems biology,
3(1):1–20.
Simão, E., Remy, E., Thieffry, D., and Chaouiya, C. (2005).
Qualitative modelling of regulated metabolic path-
ways: application to the tryptophan biosynthesis in e.
coli. Bioinformatics, 21(suppl 2):ii190–ii196.
Soh, T., Banbara, M., and Tamura, N. (2017). Proposal and
evaluation of hybrid encoding of CSP to SAT integrat-
ing order and log encodings. International Journal on
Artificial Intelligence Tools, 26(1):1–29.
Tamura, N., Taga, A., Kitagawa, S., and Banbara, M.
(2009). Compiling finite linear CSP into SAT. Con-
straints, 14(2):254–272.
Thieffry, D. and Thomas, R. (1995). Dynamical behaviour
of biological regulatory networks—ii. immunity con-
trol in bacteriophage lambda. Bulletin of Mathemati-
cal Biology, 57(2):277–297.
Thomas, R. (1973). Boolean formalization of genetic
control circuits. Journal of Theoretical Biology,
42(3):563 – 585.
Toda, T. and Soh, T. (2016). Implementing efficient all
solutions SAT solvers. ACM J. Exp. Algorithmics,
21(1):1.12:1–1.12:44.
Trinh, V.-G., Hiraishi, K., and Benhamou, B. (2022). Com-
puting attractors of large-scale asynchronous boolean
networks using minimal trap spaces. In Proceedings
of the 13th ACM International Conference on Bioin-
SAT-Based Method for Finding Attractors in Asynchronous Multi-Valued Networks
173

formatics, Computational Biology and Health Infor-
matics, pages 1–10.
Tseitin, G. S. (1968). On the complexity of derivations
in the propositional calculus. Studies in Mathematics
and Mathematical Logic Part II, pages 115—-125.
Walsh, T. (2000). SAT v CSP. In Proceedings of the 6th
International Conference on Principles and Practice
of Constraint Programming (CP 2000), pages 441–
456.
Wuensche, A. (1998). Genomic regulation modeled as a
network with basins of attraction. In Pacific Sympo-
sium on Biocomputing, volume 3, pages 89–102.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
174
