
Hybrid Improved Physarum Learner for Structure Causal Discovery
Joao Paulo Soares
1 a
, Vitor Barth
1 b
, Alan Eckeli
2 c
and Carlos Dias Maciel
1 d
1
Department of Electrical and Computing Engineering, University of Sao Paulo, Sao Carlos, Brazil
2
Hospital das Cl
´
ınicas da Faculdade de Medicina, Ribeir
˜
ao Preto, Brazil
Keywords:
Bayesian Networks, Physarum Learner, PC Algorithm, Structure Learning.
Abstract:
Causal discovery is the problem of estimating a joint distribution from observational data. In recent years,
hybrid algorithms have been proposed to overcome computational problems that lead to better results. This
work presents a hybrid approach that combines PC algorithm independence tests with a bio-inspired Improved
Physarum Learner algorithm. The combination indicates improvement in computational time spent and yet
consistent structural results.
1 INTRODUCTION
Causal questions are present in many research fields
nowadays, enabling us to deal with everyday ques-
tions such as ”what”, ”why”, and ”what if”. Despite
the fact that causal questions are popular and instigat-
ing, the answers to this type of question are not simple
to acquire (Squires and Uhler, 2022).
The ability to answer these types of question
was the key ingredient, intrinsic to our humans, that
allowed constant evolution in decision making and
technology growth (Guo et al., 2020). If machines
were able not only to act as perceiving tools but also
to develop causal questions, it would characterize the
next generation of artificial intelligence development
(Pearl, 2018).
In the last few decades, the advancement in graph-
ical models frameworks emerged as the mathemati-
cal language for causal knowledge management, and
Bayesian networks are one of those most important
frameworks (Pearl, 2018). They are compact yet pow-
erful graphical models that efficiently encode their
probabilistic relationships among a large number of
variables (Neapolitan et al., 2004). In a Bayesian net-
work, variables are presented as nodes in a directed
acyclic graph (DAG), and the edges between nodes
represent its probabilistic dependencies.
If all edges of a Bayesian network entail a di-
rect causal relationship between two variables, then
a
https://orcid.org/0000-0002-9974-4995
b
https://orcid.org/0000-0003-2285-3314
c
https://orcid.org/0000-0001-5691-7158
d
https://orcid.org/0000-0003-0137-6678
the graph is called causal (Spirtes et al., 2000), and
the process of learning such a graph from observa-
tional data is called causal discovery (Squires and Uh-
ler, 2022; Tank et al., 2021). Finding a causal graph
that best represents a joint probability distribution has
proven to be a challenging task (Kuipers et al., 2022).
The difficulty lies in the superexponential growth of
the search space of graphs (Guo et al., 2020). Fur-
thermore, the acyclicity constraint represents a time-
consuming task, especially for large and dense graphs
(Kuipers et al., 2022).
To address the problem of learning Bayesian net-
works, different techniques were developed. They
are generally organized as a) constraints-based algo-
rithms, that use statistical tests to determine which
edges exist and then determine their orientation
(Spirtes and Glymour, 1991; Meek, 2013), b) score-
based algorithms, in which a score criterion evaluates
the quality of DAG candidates and selects the best fit
(Chickering, 2002), and c) hybrid approaches, which
combine both of the previous strategies to reduce the
number of DAG candidates and accelerate the search
(Tsamardinos et al., 2006; Gasse et al., 2014; Kuipers
et al., 2022; Huang and Zhou, 2022).
In fact, the acceleration is archive by a consid-
erable restriction in DAG search space normally en-
coded by a completed partially directed acyclic graph.
A similar structure is obtained as intermediate re-
sult in PC algorithm what makes it a popular choice
for hybrid causal discovery solutions. (Nandy et al.,
2018) proved that hybrid methods like Greedy Equiv-
alence Search (GES) and Adaptively Restricte Greedy
Equivalence Search (ARGES) leads to consistent re-
234
Soares, J., Barth, V., Eckeli, A. and Maciel, C.
Hybrid Improved Physarum Learner for Structure Causal Discovery.
DOI: 10.5220/0011671000003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 234-240
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

sults for several sparse high-dimensional settings.
Also, to efficient navigate through DAG candidates in
Markov Equivalence Class, (Kuipers et al., 2022) pro-
posed a hybrid method based on PC Algorithm and
a Markov Chain Monte Carlo (MCMC) sampler that
reduces computational complexity for large and dense
graphs.
Compelled by the development of bioinspired al-
gorithms based on the slime mold Physarum poly-
cephalum, (Sch
¨
on et al., 2014) combined a Bayesian
score with a bioinspired algorithm, creating the
Physarum Learner algorithm. This algorithm uses the
Physarum solver to find the shortest path between two
nodes inside a Physarum maze and uses this informa-
tion to determine whether or not an edge exists in a
Bayesian network.
The modified version Improved Physarum
Learner was proposed in which the difficulties
in learning the edge orientation for the Physarum
Learner were addressed as well as optimization
changes to improve computational time (Ribeiro
et al., 2022).
In this work, we are looking forward to improv-
ing Improved Physarum Learner computational ef-
ficiency by combining it with the well-known PC
Algorithm to learn causal structures from observa-
tional data. First, we perform the PC algorithm
to acquire an initial structure based on conditional
independence tests, which are used to initiate the
Physarum maze. It is then possible to check the capa-
bility of the proposed method to learn a known causal
structure verifying the consistency of the discovered
graph compared with the ground-truth graph. We
also expect that with a better initial guess for the Im-
proved Physarum Learner, the hybrid approach may
encounter the best score structure with lower compu-
tational time, therefore, being feasible for large data.
In Section 2, we present the theoretical back-
ground of Bayesian networks and some state-of-the-
art causal learning strategies. Section 3 describes the
computational environment, data analysis with graph
structures and probabilities, and the hybrid method-
ology of this work. The structures obtained are pre-
sented in Section 4 with a further discussion presented
in Section 5.
2 THEORY
In this section, we will introduce the notation, main
equations and cite relevant references in each topic.
2.1 Bayesian Networks
Bayesian networks are a class of Graphical Models
(GM), and as in all other GMs the BNs objective is to
represent a joint distribution by making assumptions
of Conditional Independencies (CIs). Structurally, the
graph nodes represent random variables and the pres-
ence or absence of edges indicates the statistical re-
lations between variables. What separates Bayesian
networks from all other GMs is the usage of directed
acyclic graphs (DAGs) to comply with the Markov
assumption (Koller and Friedman, 2009; Neapolitan
et al., 2004).
The main characteristic of DAGs is that, when or-
dered, all nodes will always be placed after their par-
ents. This characteristic, called the Markov condi-
tion, can be seen as a generalization of the first-order
Markov condition from chains to DAGs. If a graph
satisfies the Markov condition, each node in the graph
will only depend on its immediate parents, being in-
dependent of all other predecessors. Given a DAG
G = (V, E) and a set of conditional probability dis-
tribution Θ, we say (G, Θ) satisfies the Markov con-
dition if for each random variable x ∈ V , x is condi-
tionally independent of the sets of its non-descendants
(ND(x)) given the set of its parents (Pa(x)) (Koller
and Friedman, 2009),
I
p
(x, ND(x)|Pa(x)) (1)
The structure formed by (G, Θ) configures a joint
probability distribution over Θ, which can be obtained
by
P(θ
1
, θ
2
, θ
3
, ..., θ
n
) =
n
∏
i=1
P(θ
i
|par(θ
i
)) (2)
where par(θ
i
) denote the parents of θ
i
(Koller and
Friedman, 2009).
Given a data set D and a structure G, estimating
the set of conditional probability distributions Θ is
generally straightforward. However, in most practi-
cal applications, finding the structure G that best en-
tails the dependencies between the variables is a really
hard task, especially for large D.
The Equation 2 represents the Chain Rule for
Bayesian networks in which the Markov condition is
essential. That means that each variable is statistically
independent of its non-descendants once its parents
are known (Koller and Friedman, 2009).
2.2 Structure Learning Methods
A same distribution might factorize in different ways.
The Markov Class is a group that contain all DAGs
Hybrid Improved Physarum Learner for Structure Causal Discovery
235

A
B
C
(a)
A
B
C
(b)
A
B
C
(c)
Figure 1: Possible three node DAGs. The structures 1b and
1a share the same set of independences therefore they be-
long to the same Markov class. The structure 1c are in a
different Markov class.
that share the same independence set (Glymour et al.,
2019; Spirtes, 2013). Figure 1 shows some possible
distributions with three variables. 1a entailed inde-
pendence between A and C given B (I(A, C|B)) and
1b also entailed I(A, C|B) that place it in the same
Markov Class. On the other hand, 1c entailed I(A, C)
and thus constitutes a different Markov Class.
Conventional approaches for discovering causal
structures rely on conditional independence proper-
ties, but there is another class of algorithms that com-
mits in search for a DAG best fit the joint distribu-
tion. These methods may not use any of those inde-
pendence proprieties and yet lead to good results.
2.2.1 Constraint-based algorithms
These algorithms discover DAGs by testing the inde-
pendence between variables and adding (or removing)
edges based on these results. The PC (Spirtes et al.,
2000; Glymour et al., 2019) is the most known algo-
rithm of this class and represents a Bayesian network
as a set of independences. At first, given the data,
the algorithm first creates a complete graph in which
each node is a variable, which means an empty set of
independencies. For each round of the algorithm, all
combinations of nodes are tested for conditional inde-
pendence in the form I(X, Y |Z), with conditional set
Z starting from |Z| = 0 and adding 1 for the round.
For positive results in an independent test, the edge
between X and Y is removed and the set Z is saved in
association with the edge removed. When |Z| is less
than the DAG maximum degree, the independent test-
ing process stops and the sets Z are used to orient the
edges.
Since conditional independence relationships
presents symmetric aspects, the orientation round can
only obtain the Markov equivalence class of DAGs
(Kuipers et al., 2022).
The Conditional independence test adopted by the
algorithm has major impact in quality of obtained
structure and it may vary if the random variable is
continuous or discrete. Some commonly employed
conditional independence tests are Pearson’s correla-
tion (Baba et al., 2004) for continuous data, χ
2
test for
categorical data (Spirtes et al., 2000) and yet some
likelihood-based tests for all types of data (Tsagris
et al., 2018).
The estimating process of a conditional distribu-
tion Z for higher-order conditional independence tests
tends to deteriorate test results as long as |Z| in-
creases, especially for discrete variables with several
possible values. In fact, the number of sample sizes
needed to efficiently estimate the distribution grows
rapidly, leading to empty or nearly empty variable
cells (Spirtes et al., 2000).
2.2.2 Score-based Algorithms and Hybrid
Approaches
Unlike Constraint-based methods that rely on statisti-
cal proprieties and independence tests to achieve and
DAG on which to build the Bayesian network, score-
based algorithms settle the causal discovery problem
by using an evaluation method as a criterion to judge
whether a DAG candidate is good or not (Squires and
Uhler, 2022). Every DAG in the search space is a
possible solution; therefore, it becomes an optimiza-
tion problem based on a specific score method and a
sampler strategy for searching the DAG space (Koller
and Friedman, 2009).
2.2.3 Improved Physarum Learner
Inspired by the maze-solving ability of the slime mold
Physarum polycephalum, the Physarum Learner al-
gorithm was proposed adapting the Physarum Solver
capability of finding the optimal path in a maze
to the Bayesian network causal discovery problem
(Miyaji and Ohnishi, 2008). The Physarum-Maze is
formed by an initial fully connected graph with ran-
dom weights. In each Physarum Solver iteration, the
Source and Sink nodes are changed randomly, and the
weights are updated. Edges with weights above a cer-
tain threshold are marked as Bayesian network pos-
tulate edges, and then a score criterion defines if the
edge is kept or not in the final network (Sch
¨
on et al.,
2014).
An improved version of the Physarum Learner al-
gorithm was proposed in (Ribeiro et al., 2022). The
proposed implementation adds a search step, once a
new edge is inserted in the graph, for a configuration
of reoriented edges that maximizes the score inside
a Markov class. Also, an extra procedure checks for
score stagnation, and if detected, the current iteration
is finished, minimizing time spent.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
236

The evidence supports that Improved Physarum
Learner shows better computational performance
converging faster than the original method.
3 MATERIAL AND METHODS
In this section, descriptions are made to highlight
how the Pc Algorithm is combined with the Improved
Physarum Learner for solving the causal structure
learning problem, with the specifications of each step
and the validation methodology. In addition, the data
set and the ground truth structure are described. The
experiments were performed on a computer with the
following characteristics:
• Processor: Intel Core i5-10300H
• RAM: 16GB
• Operating System: Pop! OS 22.04 LTS
• Python: 3.8
• NumPy: 1.19.2
In its initial steps, Physarum Learner creates a
fully connected undirected graph called Physarum-
Maze. Data variables are nodes in the maze and each
edge has a weight randomly sampled from a uniform
distribution weight ∼ uniform[0.78, 0.79] that repre-
sents the impact of that edge on a Bayesian scoring
function. The Physarum Learner proposes to trans-
form the Physarum-Maze (a fully-undirected graph)
into a Bayesian Network (a directed acyclic graph) by
removing edges with an impact lower than a thresh-
old. Each edge weight is gradually updated using the
Physarum Solver output until it reaches stagnation,
which can be very time-consuming for large graphs.
The total number of Physarum Solver iterations is ap-
proximately n
2
, where n is the number of nodes in the
maze.
We believe that the process of updating edge
weights, already optimized in Improved Physarum
Learner, can take advantage of the use of a constraint
algorithm like the PC to further improve its perfor-
mance, especially in sparse graphs, which is where
the PC algorithm demonstrates its best results.
Both algorithms start with a similar structure and
then evaluate the effectiveness of each edge using dif-
ferent strategies. The independence tests in the PC
algorithm run faster than the estimation used in Im-
proved Physarum Learner, but it also leads to less pre-
cise results.
The idea is to modify the edge sampling distribu-
tion in the Physarum-Maze accordingly to the exis-
tence or not of that edge in the PC algorithm output,
expecting to accelerate the convergence process of
the Physarum-Maze edge weights. The base code for
the implementation of the PC algorithm was adapted
from (Callan, 2018) coupled with the χ
2
indepen-
dence test.
First, the PC algorithm is performed with the max-
imum order for the independence test equal to 1.
Then, Physarum-Maze structure are initialized, and
the edge weights are sampled as follows:
W (e) ∼
(
uniform[0.68 , 0.79], if e ∃ in PC output
uniform[0.28 , 0.39], otherwise
(3)
Where W (e) in Equation 3 is a sampling func-
tion that attributes a weight to edge e. In this case,
all edges preserved by the PC algorithm start with a
higher probability of existence in the final structure.
A popular metric to test the performance of causal
discovery algorithms is to check the difference be-
tween the resulting graph structure and a known
ground-truth graph used to generate the dataset.
The LUng CAncer Simple set (LUCAS) (Guyon,
2022) is a popular dataset for learning causal graphs
and will be used in this work in addition to the Struc-
tural Hamming Distance (SHD) as a graph distance-
based metric (Cheng et al., 2022).
Lung Cancer
Smoking
Genetics
Anxiety
Peer Pressure
Yellow Fingers
Attention Disorder
Car Accident
Allergy
Coughing
Fatigue
Born an Even Day
Figure 2: Original LUng CAncer Simple set (LUCAS)
structure extracted from (Guyon, 2022). This structure was
artificially designed to model a Lung Cancer medical ap-
plication. It represents the statistical relationship between
behavioral and genetic variable in the likelihood of devel-
oping cancer in humans. Illustrate causes and possible con-
sequences.
LUCAS is an artificial dataset in which samples
are generated from a Bayesian network that represents
a medical application to diagnose, prevent and cure
lung cancer. All variables are listed in Table 1. Vari-
ables are divided into three main groups based on the
number of parents. Anxiety, Peer Pressure, Genetics,
Allergy and Born an Even Day are marked cyan and
do not have parents, for that reason they are not influ-
enced by any other variable. In magenta are Yellow
Fingers and Attention Disorder which have Smoking
and Genetics as nodes with edges connecting to them,
respectively. And finally, in yellow, we have Smok-
ing influenced by Peer Pressure and Anxiety, Lung
cancer with edges coming from Smoking and Genet-
ics, Coughing with edges coming from Allergy and
Lung cancer, Fatigue influenced by Lung cancer and
Hybrid Improved Physarum Learner for Structure Causal Discovery
237
Coughing, and the last variable is Car accident with
Attention disorder and Fatigue as parent variables.
The subscript letter
t
or
f
after the variable name in
Table 1 represents the assumed value for the variables
corresponding to the respective probability shown in
the last column. From the data in Table 1 is possible
to marginalize all conditional probability tables by ex-
ploiting the fact that each entry in the conditional dis-
tribution must have a sum of 1 for a fixed value of its
parents: for example, from the last line in Table 1 we
know that P(CarAccident = T |AttentionDisorder =
T, Fatigue = T ) = 0.97169 then we can com-
pute that P(CarAccident = F|AttentionDisorder =
T, Fatigue = T ) = 1 − 0.97169 = 0.02831. The joint
distribution generated a dataset with 1 million sam-
ples and was used for the causal discovery task.
Table 1: The Joint Probability Distribution for LUCAS
dataset. First column contain variable names, second col-
umn has variable parents (if exists) and the their current
state necessary for observe the conditional probability de-
scribed in column three. The Cyan rows represents vari-
ables without parents. In Magenta, the random variables
with one parent and the Yellow rows represents the nodes
with two parents.
Variable Parents Probability
Anxiety
t
0.64277
PeerPressure
t
0.32997
Genetics
t
0.15953
Allergy
t
0.32841
BornanEvenDay
t
0.5
YellowFingers
t
Smoking
f
0.23119
YellowFingers
t
Smoking
t
0.95372
AttentionDisorder
t
Genetics
f
0.28956
AttentionDisorder
t
Genetics
t
0.68706
Smoking
t
PeerPressure
f
, Anxiety
f
0.43118
Smoking
t
PeerPressure
t
, Anxiety
f
0.74591
Smoking
t
PeerPressure
f
, Anxiety
t
0.8686
Smoking
t
PeerPressure
t
, Anxiety
t
0.91576
Lungcancer
t
Genetics
f
, Smoking
f
0.23146
Lungcancer
t
Genetics
t
, Smoking
f
0.86996
Lungcancer
t
Genetics
f
, Smoking
t
0.83934
Lungcancer
t
Genetics
t
, Smoking
t
0.99351
Coughing
t
Allergy
f
, Lungcancer
f
0.1347
Coughing
t
Allergy
t
, Lungcancer
f
0.64592
Coughing
t
Allergy
f
, Lungcancer
t
0.7664
Coughing
t
Allergy
t
, Lungcancer
t
0.99947
Fatigue
t
Lungcancer
f
, Coughing
f
0.35212
Fatigue
t
Lungcancer
t
, Coughing
f
0.56514
Fatigue
t
Lungcancer
f
, Coughing
t
0.80016
Fatigue
t
Lungcancer
t
, Coughing
t
0.89589
CarAccident
t
AttentionDisorder
f
, Fatigue
F
0.2274
CarAccident
t
AttentionDisorder
t
, Fatigue
f
0.779
CarAccident
t
AttentionDisorder
f
, Fatigue
t
0.78861
CarAccident
t
AttentionDisorder
t
, Fatigue
t
0.97169
Figure 2 shows the graph structure of the 12 binary
random variables and their edges dependencies.
4 RESULTS AND DISCUSSION
In this section, relevant details for the LUCAS causal
structure are given, which include isolated variables
and their impact on the causal discovery problem.
The final structure obtained from the proposed hybrid
methodology is also presented in this section in ad-
dition to the intermediate structure learned from PC
algorithm.
One characteristic of the structure of the LUCAS
network is the variable Born an Even Day that mea-
sures the impact of the day of birth on the chances of
developing lung cancer and, as expressed in Figure 2,
that the influence is negligible once there are no con-
nections with the rest of the DAG structure and, as a
consequence, should not influence any of the other 11
variables.
Identifying isolated variables is extremely impor-
tant once they have an irrelevant impact on tasks such
as forecasting or inference. In this case, the difference
between the search space for DAGs with 12 variables,
like LUCAS, has 5.2 × 10
26
more elements than the
search space for DAGs with 11 variables. So, it is
crucial that the statistical independence verification
of the PC algorithm detects the Born an Even Day
isolated node from LUCAS and avoid the Improved
Physarum Learner from searching for irrelevant paths.
For that reason, different conditional indepen-
dence tests were performed and the χ
2
was selected
for PC algorithm execution. The experiment also
counted with Pearson Correlation (Hemmings and
Hopkins, 2006) and Fast Conditional Independence
Test (Chalupka, 2022).
Lung Cancer
Smoking
Genetics
Anxiety
Peer Pressure
Yellow Fingers
Attention Disorder
Car Accident
Allergy
Coughing
Fatigue
Born an Even Day
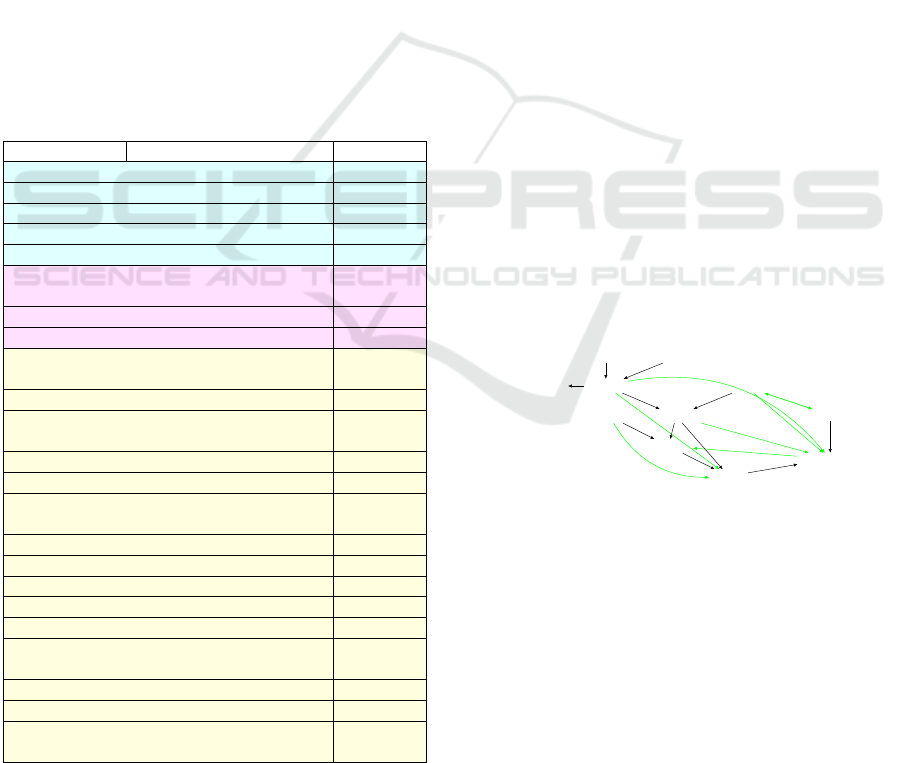
Figure 3: The PC algorithm obtained structure. In black are
the edges were preserved by the algorithm that are present in
the ground-truth structure. In green are the edges wrongly
kept by the algorithm. It has SHD = 7.
Figure 3 shows the structure obtained from the ex-
ecution of the PC Algorithm. The black edges are
the true positive edges kept by the algorithm that be-
longs to the ground truth graph, and all the edges of
the ground truth are present in the output of the PC
algorithm. The edge between Genetics and Attention
Disorder has arrowheads at each endpoint, showing
ambiguity in determining the direction of the edge
using the PC Algorithm. In addition, the algorithm
has truly isolated the variable Born an Even Day re-
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
238

ducing the chances of the improved Physarum learner
connecting it to anything else.
Lung Cancer
Smoking
Genetics
Anxiety
Peer Pressure
Yellow Fingers
Attention Disorder
Car Accident
Allergy
Coughing
Fatigue
Born an Even Day
Figure 4: The obtained structure from hybrid approach. In
black are the edges were preserved by the algorithm that are
present in the ground-truth structure. The green edge from
Attention Disorder to Genetics has reverse orientation. It
has SHD = 1.
Based on his partial result, the edge weights were
sampled as mentioned in section 3 as a starting point
for Improved Physarum Learner. Figure 4 shows the
learned structure that has SHD = 1. The only edge in-
correctly oriented from Attention Disorder to Genet-
ics is the same edge in which the PC algorithm had
difficulty determining the orientation. Despite that,
all edges kept by the Improved Physarum Learner be-
long to the ground-truth graph.
No major difference between the structure learned
by the methodology proposed in this work and the Im-
proved Physarum Learner, however, the hybrid ver-
sion presented a decrease in computational time. In
10 executions, the Improved Learner had an average
217.2 seconds to find a structure, while the hybrid
had an average of 155.3 seconds, showing a consis-
tent 28% of time savings.
5 CONCLUSIONS
In this work, we presented a hybrid alternative for
Improved Physarum Learner in which we tested the
quality of the founded causal structure proposed in
(Guyon, 2022) by counting the Structural Hamming
Distance (SHD) between the learned structure and the
ground-truth graph. We also measured the computa-
tional time saved by adding information from Condi-
tional Independence tests into the Physarum maze.
The results showed consistency in the causal dis-
covery of the true structure with almost no errors.
The SHD = 1 refers to the green edge between Ge-
netics and Attention Disorder misoriented. In our
tests, the proposed methodology outperforms Im-
proved Physarum Learner, finding the causal struc-
ture on average 28% faster.
Although promising, the proposed combination of
algorithms needs, in future works, to be compared
with strategies of learning structures, both algorithms
consolidated in the literature and new approaches, us-
ing the same hardware and the same amounts of data
for all algorithms. Also, it is important to check the
Hybrid Improved Physarum behavior in different sce-
narios such as non-binary data, networks with a large
number of nodes, or even how it behaves with scarce
samples.
Furthermore, parallel implementation strategies
can be highly beneficial for the Hybrid Improved
Physarum Learner. For the PC algorithm, the method-
ology proposed by (Le et al., 2016) seems promising
especially in high-dimensional data. But no parallel
technique was found by the authors relating causal
discovery problem and Physarum.
ACKNOWLEDGEMENTS
This work was partially supported by the follow-
ing agencies: CAPES, FAPESP 2014/50851-0, CNPq
465755/2014-3 and BPE Fapesp 2018/19150-6.
REFERENCES
Baba, K., Shibata, R., and Sibuya, M. (2004). Partial corre-
lation and conditional correlation as measures of con-
ditional independence. Australian & New Zealand
Journal of Statistics, 46(4):657–664.
Callan, J. (2018). Learning Causal Networks in Python.
Master’s thesis, University of York, England.
Chalupka, K. (2022). A fast conditional independence test
(fcit). https://github.com/kjchalup/fcit.
Cheng, L., Guo, R., Moraffah, R., Sheth, P., Candan, K. S.,
and Liu, H. (2022). Evaluation methods and measures
for causal learning algorithms. IEEE Transactions on
Artificial Intelligence.
Chickering, D. M. (2002). Optimal structure identification
with greedy search. Journal of machine learning re-
search, 3(Nov):507–554.
Gasse, M., Aussem, A., and Elghazel, H. (2014). A hy-
brid algorithm for bayesian network structure learn-
ing with application to multi-label learning. Expert
Systems with Applications, 41(15):6755–6772.
Glymour, C., Zhang, K., and Spirtes, P. (2019). Review of
causal discovery methods based on graphical models.
Frontiers in genetics, 10:524.
Guo, R., Cheng, L., Li, J., Hahn, P. R., and Liu, H. (2020).
A survey of learning causality with data: Problems
and methods. ACM Computing Surveys (CSUR),
53(4):1–37.
Guyon, I. (2022). Causality workbench project.
http://www.causality.inf.ethz.ch/data/LUCAS.html.
Hemmings, H. C. and Hopkins, P. M. (2006). Foundations
of anesthesia: basic sciences for clinical practice. El-
sevier Health Sciences.
Hybrid Improved Physarum Learner for Structure Causal Discovery
239

Huang, J. and Zhou, Q. (2022). Partitioned hybrid learning
of bayesian network structures. Machine Learning,
pages 1–44.
Koller, D. and Friedman, N. (2009). Probabilistic graphical
models: principles and techniques. MIT press.
Kuipers, J., Suter, P., and Moffa, G. (2022). Efficient
sampling and structure learning of bayesian networks.
Journal of Computational and Graphical Statistics,
pages 1–12.
Le, T. D., Hoang, T., Li, J., Liu, L., Liu, H., and Hu,
S. (2016). A fast pc algorithm for high dimensional
causal discovery with multi-core pcs. IEEE/ACM
transactions on computational biology and bioinfor-
matics, 16(5):1483–1495.
Meek, C. (2013). Causal inference and causal expla-
nation with background knowledge. arXiv preprint
arXiv:1302.4972.
Miyaji, T. and Ohnishi, I. (2008). Physarum can solve the
shortest path problem on riemannian surface mathe-
matically rigourously. International Journal of Pure
and Applied Mathematics, 47(3):353–369.
Nandy, P., Hauser, A., and Maathuis, M. H. (2018).
High-dimensional consistency in score-based and hy-
brid structure learning. The Annals of Statistics,
46(6A):3151–3183.
Neapolitan, R. E. et al. (2004). Learning bayesian networks,
volume 38. Pearson Prentice Hall Upper Saddle River.
Pearl, J. (2018). Theoretical impediments to machine learn-
ing with seven sparks from the causal revolution.
arXiv preprint arXiv:1801.04016.
Ribeiro, V. P., RP, B., Maciel, C. D., and Balestieri,
J. A. (2022). An improved bayesian network super-
structure evaluation using physarum polycephalum
bio-inspiration. In Congresso Brasileiro de Au-
tom
´
atica-CBA, volume 2.
Sch
¨
on, T., Stetter, M., Tom
´
e, A. M., Puntonet, C. G., and
Lang, E. W. (2014). Physarum learner: A bio-inspired
way of learning structure from data. Expert systems
with applications, 41(11):5353–5370.
Spirtes, P. and Glymour, C. (1991). An algorithm for fast
recovery of sparse causal graphs. Social science com-
puter review, 9(1):62–72.
Spirtes, P., Glymour, C. N., Scheines, R., and Heckerman,
D. (2000). Causation, prediction, and search. MIT
press.
Spirtes, P. L. (2013). Directed cyclic graphical rep-
resentations of feedback models. arXiv preprint
arXiv:1302.4982.
Squires, C. and Uhler, C. (2022). Causal structure learn-
ing: a combinatorial perspective. arXiv preprint
arXiv:2206.01152.
Tank, A., Covert, I., Foti, N., Shojaie, A., and Fox, E. B.
(2021). Neural granger causality. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
44(8):4267–4279.
Tsagris, M., Borboudakis, G., Lagani, V., and Tsamardi-
nos, I. (2018). Constraint-based causal discovery with
mixed data. International journal of data science and
analytics, 6(1):19–30.
Tsamardinos, I., Brown, L. E., and Aliferis, C. F. (2006).
The max-min hill-climbing bayesian network struc-
ture learning algorithm. Machine learning, 65(1):31–
78.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
240
