
Evaluating the Effects of a Priori Deep Learning Image Synthesis on
Multi-Modal MR-to-CT Image Registration Performance
Nils Frohwitter
1,3 a
, Alessa Hering
2 b
, Ralf M
¨
oller
1,3 c
and Mattis Hartwig
1,4 d
1
German Research Center for Artificial Intelligence, 23562 L
¨
ubeck, Germany
2
Fraunhofer MEVIS, Institute for Digital Medicine, 23562 L
¨
ubeck, Germany
3
Institute of Information Systems, University of L
¨
ubeck, 23562 L
¨
ubeck, Germany
4
singularIT GmbH, 04109 Leipzig, Germany
Keywords:
Image-to-Image Translation, Image Synthesis, Image Registration.
Abstract:
Radiation therapy often requires a computed tomography (CT) for treatment planning and an additional mag-
netic resonance (MR) imaging prior to the treatment for adaptation. With two different images from the same
scene, multi-modal image registration is needed to align areas of interest in both images. One idea to improve
the registration process is to perform an image synthesis that converts one image mode into another mode
prior to the registration. In this paper, we address the research needed to perform a thorough evaluation of
the synthesis step on overall registration performance using different well-known registration methods of the
Advanced Normalization Tools (ANTs) framework. Given abdominal images, we use CycleGAN for synthe-
sis and compare the registration performance to the one without synthesis by using four different well-known
registration methods. We show that good image synthesizing results lead to an average improvement in all reg-
istration methods, biggest improvement being achieved for the ‘Symmetric Normalization’ method with 8%
(measured with Dice-score). The overall best registration method with prior synthesis is ‘Symmetric Normal-
ization and Rigid’. Furthermore, we show that the images with bad synthetic results lead to worse registration,
thus suggesting the correlation between synthesizing quality and registration performance.
1 INTRODUCTION
Having accurate registration algorithms for multi-
modal images is highly needed in the field of im-
age guided radiation therapy. The imaging in radia-
tion therapy is often done in two steps. In the plan-
ning phase, an image based on computed tomography
(CT) is created to localise the target area and to calcu-
late a dose distribution. Right before the treatment
and/or during the treatment phase, further images
based on magnetic resonance imaging (MR) or cone-
beam computed tomography (CBCT) are taken (Qi,
2017) to deal with global displacements such as the
positioning of the patient as well as local changes
such as fill levels of organs or reductions in size of tu-
mor tissue. Multi-modal image registration is needed
to align areas of interest in the images taken with dif-
a
https://orcid.org/0000-0001-8267-281X
b
https://orcid.org/0000-0002-7602-803X
c
https://orcid.org/0000-0002-1174-3323
d
https://orcid.org/0000-0002-1507-7647
ferent imaging techniques and presumably at different
times. Especially MR-CT registration is widely used
in radiation therapy because of the high soft-tissue
contrast and post-treatment analysis with the MR im-
ages (Chandarana et al., 2018).
In the past, several authors have worked on deep
learning image synthesis to improve the multi-modal
image registration task (Wei et al., 2019; McKenzie
et al., 2019; Tanner et al., 2018). The idea is that
the registration algorithms can focus on the easier
task of mono-modal image registration if one image
type (e.g. MR) is transformed into the other type
(e.g. CT) before the actual registration takes place.
In multi-modal registration, it is generally difficult to
identify these correspondences or similarity measures
because of the often highly different physical proper-
ties (Saiti and Theoharis, 2020). Current work lacks
the clear evaluation of registration performance using
a standardised dataset and multiple registration algo-
rithms. In this paper, we focus on evaluating the in-
fluence of MR-to-CT synthesis on the multi-modal
image registration performance. Given the abdom-
322
Frohwitter, N., Hering, A., Möller, R. and Hartwig, M.
Evaluating the Effects of a Priori Deep Learning Image Synthesis on Multi-Modal MR-to-CT Image Registration Performance.
DOI: 10.5220/0011669000003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 322-329
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

inal MR and CT images from the Learn2Reg2021-
challenge (Dalca et al., 2021), we perform image-
to-image synthesis via the CycleGAN framework to
create synthetic CT images and compare the four dif-
ferent registration methods ‘Rigid’, ‘Symmetric Nor-
malization’, ‘Symmetric Normalization and Rigid’ as
well as ‘Symmetric Normalization with Elastic Regu-
larization’ of the ANTs framework with and without
the synthetic CT image instead of the MR image. We
show that good synthesizing results lead to an aver-
age improvement in all registration methods. Further-
more, with synthesis the method ‘Symmetric Normal-
ization’ achieves the biggest improvement compared
to the non-synthetic registration and ‘Symmetric Nor-
malization and Rigid’ achieves the overall best regis-
tration result.
The remainder of the paper is structured as fol-
lows. Section 2 covers the preliminaries on the Cy-
cleGAN architecture and the process of image reg-
istration in general. Section 3 describes the current
state-of-the-art of image synthesis and image registra-
tion as well as how our work fits into other researches.
Section 4 introduces the used dataset with its prepro-
cessing steps. Section 5 describes the experimental
setup consisting of image synthesis and image reg-
istration. In Section 6, the results are presented and
discussed. Lastly, Section 7 gives a short conclusion
of the paper.
2 PRELIMINARIES
In this section, we give a short introduction to the im-
portant deep learning architectures used in this paper
as well as the overall goal and functionality of image
registration.
2.1 Deep Learning Architecture
The term deep learning (DL) covers techniques that
allow for computational models that consist of multi-
ple layers to learn multiple levels of representations.
These layers and their weights are not designed by
human engineers but are learned from data given a
specific task (LeCun et al., 2015). A Generative Ad-
versarial Network (GAN) is a DL concept consisting
of two neural networks: a generator G and a discrimi-
nator D (Goodfellow et al., 2014). The generator aims
to create synthetic images that are indistinguishable
from real data by the discriminator. The discrimina-
tor on the other hand, aims to identify real and fake
data correctly which leads to the so called adversar-
ial training between generator and discriminator. In
the training process, the generator synthesizes fake
data and receives feedback of its synthesizing quality
through the evaluation of the discriminator. If the dis-
criminator is able to differentiate the fake image cor-
rectly, the generator is penalized, otherwise the dis-
criminator is penalized. If the training between those
two neural networks is done in a well balanced way,
the generator learns to imitate the distribution of the
training data. The learning problem was originally
formulated as a minmax-problem
min
G
max
D
E
x∼p
real
[log(D(x))]
+ E
z∼p
z
[log(1 − D(G(z)))], (1)
where p
real
is the distribution of the training data and
p
z
of the input noise (Goodfellow et al., 2014).
A problem that arises with GANs is its training
instability which can lead to a mode collapse. The
discriminator alone cannot assure that the generator
creates synthetic data which is either divers or clearly
shows correspondences with the input data. Espe-
cially in image-to-image synthesis, it would be prob-
lematic if the generator just learns the mapping onto a
single image to satisfy the discriminator. To solve the
mode-collapse problem in data synthesis, the Cycle-
GAN architecture was developed (Zhu et al., 2017).
The CycleGAN consists of two generators and dis-
criminators for synthesizing from both domains onto
one another in a cycle consistent way. By using a sec-
ond generator to synthesize back into the original do-
main, the recreated image can be compared pixelwise
with the original image. For the sake of completeness,
DualGAN (Yi et al., 2017) is a related architecture
to CycleGAN that has been developed in parallel and
has been used to solve similar issues. In this paper,
we focus on the CycleGAN architecture.
2.2 Image Registration
Image registration is a mathematical process that in-
volves the determination of a reasonable transforma-
tion between one or many target images and one refer-
ence image. Image registration is often used to com-
bine information of different image modalities like
the high soft-tissue contrast of the MR and the high
bone contrast of the CT or to handle images of the
same scene taken in different times (Qi, 2017). For
more details on medical image registration, see the
work of J. Modersitzki (Modersitzki, 2009).
In this paper, we use implemented registration
methods from the Advanced Normalization Tools
(ANTs) (Avants et al., 2011), more precisely from the
python version ANTsPy
12
. We choose the transform
1
https://antspy.readthedocs.io/en/latest/index.html
2
https://pypi.org/project/antspyx/0.3.3/
Evaluating the Effects of a Priori Deep Learning Image Synthesis on Multi-Modal MR-to-CT Image Registration Performance
323

types ‘Rigid’, ‘Symmetric Normalization’ (SyN),
‘Symmetric Normalization and Rigid’ (SyNRA) and
‘Symmetric Normalization with Elastic Regulariza-
tion’ (ElasticSyN) for registration. To avoid confu-
sion for the reader, we use ‘SyN’ as an abbreviation
for ‘Symmetric Normalization’ and ‘syn’ for ‘syn-
thetic’. Rigid transformation only consists of rotation
and translation but SyN (Avants et al., 2011) is used as
a symmetric diffeomorphic registration method with
mutual information as optimization metric, combined
with affine transformation as for the ANTs implemen-
tation. ElasticSyN has an additional elastic regular-
ization term and SyNRA has an additional rigid trans-
formation within.
A possible measure for the usage or evaluation of
image registration is given by a priori defined seg-
mentation masks that mark the pixelwise regions of
corresponding organs/segments in all images. Given
the segmentations of a reference image R
s
and of a
template image T
s
as well as a displacement u, the
Dice-score measures the overlap of the deformed seg-
mentation T
s
[u] and R
s
and is given by
Dice(T
s
, R
s
, u) = 2 ·
|T
s
[u] ∩ R
s
|
|T
s
[u]| + |R
s
|
, (2)
where |·| denotes the amount and ∩ the intersection of
non-zero pixels. To clarify, one can represent the seg-
mentations as sets, consisting of tuples that represent
the pixels of the segments. Totally overlapping corre-
sponding segments get the highest possible score of 1,
disjoint segments score with 0 (Dice, 1945; Sørensen,
1948).
3 RELATED WORK
At the current state of research, deep learning meth-
ods are used for a variety of applications e.g. image
recognition, image segmentation, data synthesis, im-
age restoration, natural language processing, image
registration etc. In the context of image synthesis,
derivations of GANs (Goodfellow et al., 2014) were
developed to enable text-to-image synthesis, high-
resolution synthesis of human image and even regis-
tration of medical images (Reed et al., 2016; Karras
et al., 2018; Mahapatra et al., 2018).
In 2017, Zhu and Park et al. introduced the frame-
work CycleGAN, enabling a wide usage of unpaired
image-to-image translation tasks (Zhu et al., 2017).
Some publications focused on the improvement of the
CycleGAN image synthesis in general. By enforcing
mutual information (MI) and structural consistency
on the synthesis (Ge et al., 2019), adding frequency-
supervised architecture between generator and dis-
criminator (Shi et al., 2021), studying the effects of
gradient consistency and training data size (Hiasa
et al., 2018a) or using a variety of loss functions to in-
fluence the synthesis (Kida et al., 2020), they showed
improvements on their individual datasets.
Non-registration use cases for multi-modal image
synthesis are widely investigated, especially by using
the CycleGAN framework. Wolterink et al. used the
CycleGAN to convert brain MR images into synthetic
CT images and determined an improvement of the use
of unpaired compared to paired training (Wolterink
et al., 2017). Others used CycleGAN or similar struc-
tures for MR/CT images to investigate the synthesis
using paired and unpaired data (Jin et al., 2019), to
create missing CT data (Hiasa et al., 2018b) or to im-
prove data segmentation (Jiang et al., 2018; Chart-
sias et al., 2017). In order to substitute/improve the
dose calculations of radiation treatment with CBCT
images, some researchers used CycleGAN to synthe-
size CBCT images to synthetic CT images (Kuckertz
et al., 2020; Liang et al., 2019; Gao et al., 2021; Kurz
et al., 2019).
For the specific case of image registration with
prior image synthesis, there are a few research stud-
ies investigating the influence of MR-CT image syn-
thesis on multi-modal image registration. These re-
searches use different datasets and evaluation meth-
ods, thereby coming to different results. Wei et al.
used CycleGAN synthesis and showed an improve-
ment in image registration performance by evaluat-
ing with the rigid and deformable ANTs method (Wei
et al., 2019). McKenzie et al. applied the synthesis
on images of the head-and-neck area and evaluated
the registration performance of a B-spline deformable
method, resulting in lower registration errors than
without synthesis as a prior (McKenzie et al., 2019).
On the other hand, Tanner et al. couldn’t show an
overall improvement by synthesizing whole-body MR
and CT scans and registering them with deformable
registration methods (Tanner et al., 2018). Addition-
ally, Jiahao Lu et al. evaluated the influence of syn-
thesis by CycleGAN and other synthesizing frame-
works with the registration performance on the open
source datasets ‘Zurich’, ‘Cytological’ and ‘Histolog-
ical’ and concluded that synthesis is only applicable
on easy multi-modal problems (Lu et al., 2021). Apart
from using the deep learning synthesis method Cy-
cleGAN, some researches utilized synthesis methods
from probabilistic frameworks or Patch-wise Random
Forests. In these papers, the influence of synthe-
sis was also evaluated using ‘SyN’ as a registration
method, among others (Cao et al., 2017; Roy et al.,
2014).
Because of the mixed results for the improvement
of registration performance with synthetic data, there
HEALTHINF 2023 - 16th International Conference on Health Informatics
324

Figure 1: The axial slices of the MR image on the left and
the CT image on the right are shown with 3 of their repre-
sented segments. [Light blue] as the liver, [orange] as the
left kidney and [red] as the right kidney.
is a need to further investigate this application by
performing a well-structured analysis of different use
cases with different registration methods. We con-
tribute to this analysis of deep learning MR-CT image
synthesis on multi-modal image registration by eval-
uating the performance of four registration methods
of the ANTs framework (more precisely ANTsPy).
The image synthesis prior to the registration will be
performed with a CycleGAN implementation. The
MR-CT dataset used in this study consists of ab-
dominal images coming from the Learn2Reg2021-
challenge (Dalca et al., 2021).
4 DATASET
In this study, we use the MR and CT abdomen im-
ages used in and prepared from the Learn2Reg2021
challenge (Dalca et al., 2021). The 16 paired im-
ages from the TCIA dataset (Clark et al., 2013) are
split into eight training and eight testing samples.
The Learn2Reg organizers additionally have provided
manual segmentations for all eight training MR and
CT images for the organs liver, spleen, right kidney
and left kidney. Furthermore, 40 MR images from the
CHAOS dataset (Kavur et al., 2019) as well as 49 CT
images from the BCV dataset where additionally pro-
vided for training purposes. All images are standard-
ized to the image size 192x160x192 with a voxel size
of 2x2x2mm. The challenges of registration in com-
bination of the dataset are given by its multi-modal
scans, few/noisy annotations, large deformations and
missing correspondences. As an example, Figure 1
shows an MR image and its corresponding CT image
with three different colored annotated segments. To
further preprocess the data for 2D synthesis, we sliced
all volumes into 192x160 axial 2D images and indi-
vidually removed empty or incomplete slices from the
top or bottom of the volumes. Then we replaced the
intensities of body surrounding structures to the corre-
sponding background intensity which is -1000 for CT
images and 0 for MR images. The CT images were
also clipped into the intensity interval of [−400, 600]
to artificially concentrate the relevant intensities. For
the follow-up registration process, we prepared the
segmentations of the training images to cover up the
same volume as the training images after slice reduc-
tion. After reduction, there are in total 5,448 MR and
9,189 CT 2D axial image slices available for the syn-
thesis training.
5 EXPERIMENTAL SETUP
The experiment consists of two separate image pro-
cessing methods executed consecutively. In the image
synthesis-stage, we learn the image-to-image transla-
tion between unpaired 2D MR and CT images. After-
wards, we use the synthetic CT images (synCT) to
investigate the differences in 3D image registration
performance between MR/CT and synCT/CT. The
source code is provided on github
3
.
5.1 Image Synthesis
The synthesizing framework used for the training of
MR-to-CT synthesis originates from the CycleGAN
implementation in PyTorch (Zhu et al., 2017; Isola
et al., 2017). In this implementation, the genera-
tors correspond to a ResNet structure, consisting of
mainly two down- and up-convolution blocks as well
as nine residual blocks in the lowest level in between.
The discriminators are named as 70PatchGAN and
consist of (down-)convolution blocks resulting in a
18x22 output with a 70x70 receptive field. We in-
tegrated an additional mutual information loss for the
CT-synthesis direction (MI
A
) as well as a gradient loss
into the training process for further regulation moti-
vated by the research from Ge and Wei et al. and
Hiasa et al. (Ge et al., 2019; Hiasa et al., 2018a).
The adversarial losses as well as the cycle loss and
the identity loss are originally used in the introduced
CycleGAN.
The resulting total loss function is then defined as
L
total
= L
adv
A
+ L
adv
B
+ λ
cycle
L
cycle
+ λ
idt
L
idt
+ λ
grad
L
grad
+ λ
MI
L
MI
A
. (3)
While training, the images are loaded in size
175x210, randomly cropped back to 160x192 and
scaled to the intensity range [−1, 1]. The generators
are updated prior to the discriminators (Zhu et al.,
2017).
For the purpose of hyperparameter tuning, three
of the eight paired training images described in the
3
https://github.com/nilsFrohwitter/I2I-Synthesis
Evaluating the Effects of a Priori Deep Learning Image Synthesis on Multi-Modal MR-to-CT Image Registration Performance
325
dataset Section 4 were extracted and used as val-
idation data. Following the suggestions from the
Learn2Reg-team, we chose the images with the num-
bers 12, 14 and 16 as the validation set. The hy-
pererparameter tuning resulted in the loss weights
λ
cycle
= 5, λ
idt
= 0.5, λ
grad
= 1 and λ
MI
= 0.5 with 50
epochs of Adam-optimization (Kingma and Ba, 2015)
with beta = (0.5, 0.999) and a constant learning rate
of lr = 0.0002, followed by 50 epochs with a linear
to zero decaying learning rate. With the best hyper-
parameters found for the training process, we trained
eight new models, each one by leaving out one of the
eight TCIA a priori defined training images for later
testing, performing a leave-one-out evaluation. Af-
ter training, the resulting eight individual testing im-
ages (in their original and synthetic form) are used
for the following registration evaluation. The quality
assessment of the synthesis is performed by visually
comparing the synthetic results with each other and
with its corresponding CT and MR images as well as
by controlling the synthesis continuity across all axial
slices in the 3D images. Thereby, we categorised the
images into three quality states: good, medium and
bad. In the evaluation of the following image registra-
tion, we differentiate the quality of the synthesis and
therefore analyze poorly synthesized data separately.
In our case, we have badly synthesized test images 06
and 10. Figure 2 shows a synthetic CT image slice
of the test images 14 as an example of good synthesis
and 10 as a bad one. The synthetic CT image slice
of Image 14 shows good intensity adaption while pre-
serving the organ structures properly. On the other
hand, the resulting synthetic CT image slice of Image
10 clearly shows major distortions of organ structures
after synthesis, which we rate to be an example of bad
synthesis. Image 06 also has major distortions after
synthesis, resulting especially in a high variation of
spine localisation along all image slices.
5.2 Image Registration
Image registration is performed in 3D with regis-
tration methods from the ANTsPy framework. We
use the methods ‘Rigid’, ‘SyN’, ‘SyNRA’ as well as
‘ElasticSyN’ in its predefined states. Because we aim
to evaluate the performance gain by using an addi-
tional synthesis step, we calculate two registrations
(with each method) for the eight images, one using
the original MR data and one using the prior synthe-
sized data (synCT). The determined deformations are
then applied to the segmentations of their respective
MR images.
All registration results are compared by calculat-
ing the Dice-score between the deformed segmenta-
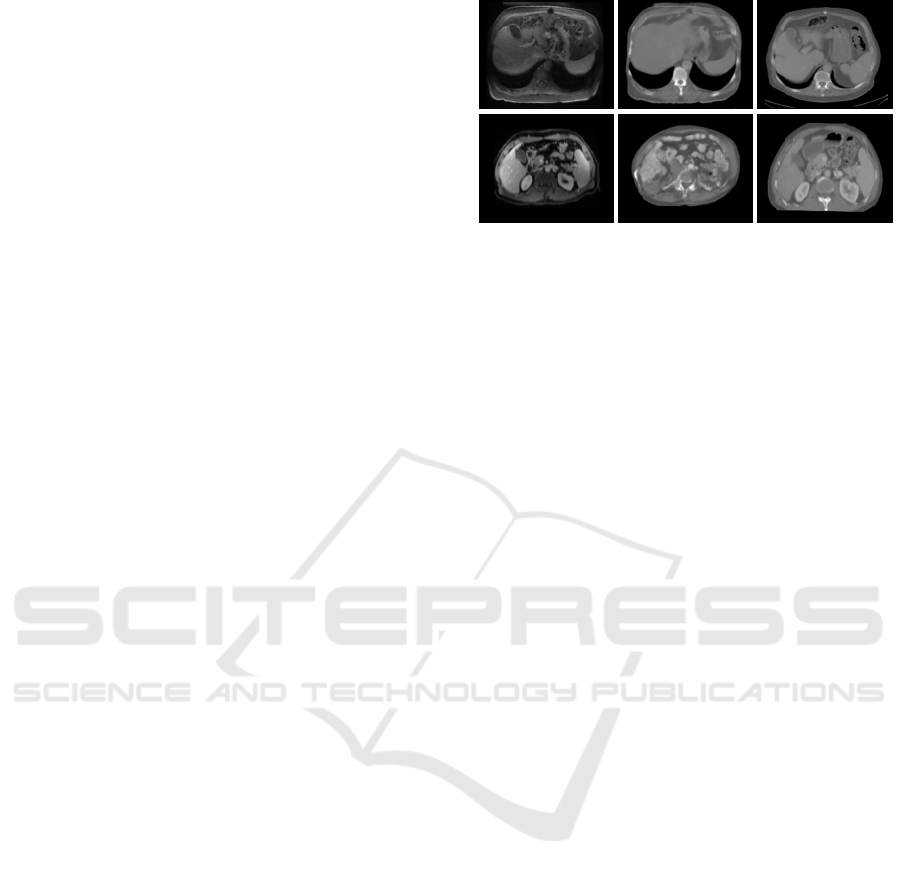
Figure 2: From left to right: MR, synCT, corresponding CT.
Note, that the corresponding CT is not properly registered to
the MR. The top row shows test image 14 as an example of a
good synthesis where intensities are adopted and structures
are mainly preserved. The bottom row shows the test image
10 as a bad synthesis example where the synthetic CT is
greatly distorted.
tions of the MR images and the segmentations of the
CT images. We reduce the influence of randomness in
the registration process by performing the whole pro-
cess of learning, synthesis and registration five times
and using the average of all runs.
5.3 Additional Image Meta Data
To further understand and interpret the results of reg-
istration in relation to the synthesis quality, we inves-
tigate some meta data of the original and synthetic
images. We calculate and compare the entropy of the
MR and CT images as well as of their synthetic cor-
respondences synCT and synMR. Furthermore, be-
sides visually interpreting, we use the Fr
´
echet Incep-
tion Distance (FID) to measure the quality of synthe-
sis in a quantitative way. The FID score compares
the mean and standard derivation of the deepest layer
of the pretrained Inception v3 model, which highly
correlates with human perception of similarity in im-
ages (Heusel et al., 2017). The lower the FID score,
the better an image represents the target dataset.
6 RESULTS AND DISCUSSION
The results in Table 1 of the four registration meth-
ods with and without a prior synthesis step show, that
when the synthesis results in realistic synthetic CT
images, this synthesis step also improves the average
image registration performance. For every ANTsPy
method, the average Dice-score of good synthesis
cases is higher with the use of the synthetic CT than
with the original MR image. The biggest improve-
ment with synthesis in registration performance is
achieved for the ‘SyN’ method with 8% Dice-score
improvement. The overall best registration method
HEALTHINF 2023 - 16th International Conference on Health Informatics
326
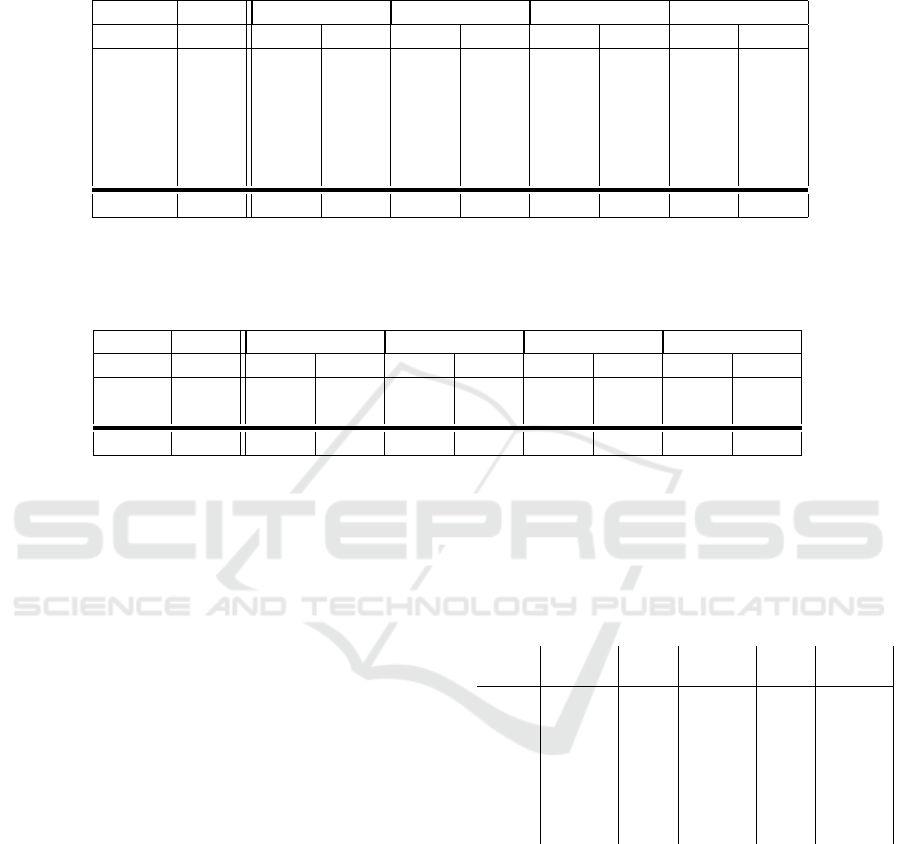
Table 1: Averaged Dice scores from 5 registration runs of the 6 good synthesized test images between the CT and MR/synthetic
CT (sCT) as initial values and after rigid, SyN, SyNRA and ElasticSyN registration. The maximum dice scores per image and
the averages over the images are highlighted as bold numbers. The highest Dice-score averaged over the 6 models is acquired
for SyNRA with the synthetic CT.
Rigid SyN SyNRA ElasticSyN
Vol init MR sCT MR sCT MR sCT MR sCT
02 0.505 0.562 0.559 0.657 0.665 0.648 0.675 0.657 0.659
04 0.095 0.254 0.382 0.497 0.405 0.538 0.526 0.598 0.381
08 0.547 0.786 0.736 0.892 0.853 0.890 0.852 0.892 0.851
12 0.257 0.472 0.538 0.626 0.653 0.620 0.649 0.625 0.654
14 0.196 0.585 0.600 0.121 0.528 0.659 0.730 0.119 0.504
16 0.493 0.693 0.685 0.847 0.826 0.843 0.827 0.856 0.822
avg
good
0.349 0.559 0.583 0.606 0.655 0.700 0.710 0.623 0.645
Table 2: Averaged Dice scores from 5 registration runs of the 2 bad synthesizing models between the CT and MR/synthetic
CT (sCT) as initial values and after rigid, SyN, SyNRA and ElasticSyN registration. The maximum dice scores per image and
the averages over the images are highlighted as bold numbers. The highest Dice-score averaged over the 2 models is acquired
for Rigid with the synthetic CT.
Rigid SyN SyNRA ElasticSyN
Vol init MR sCT MR sCT MR sCT MR sCT
06 0.465 0.505 0.599 0.401 0.252 0.326 0.272 0.345 0.248
10 0.380 0.502 0.522 0.636 0.321 0.642 0.321 0.637 0.323
avg
bad
0.423 0.504 0.561 0.519 0.287 0.484 0.297 0.491 0.286
with prior synthesis is ‘SyNRA’, which achieves a
Dice-score of 0.710. As an illustration of the differ-
ences in the registration methods used on one test im-
age, the resulting deformed MR segments on top of
the CT image with and without synthesis are given in
Figure 3 from the test image 14. The different seg-
mentation overlaps on the corresponding CT image
are clearly visible, especially for the different regis-
tration methods of the MR-CT registration.
Besides, the Dice-scores of the registrations of the
badly synthesized images 06 and 10 show a distinct
degradation of registration performance in the SyN-
methods when using the synthetic CT instead of the
original MR image (s. Table 2). Interestingly, Rigid
is the only method where the synthesis results in an
averaged better Dice-score. For the good synthesized
images, it is, on average, the exact other way around.
The results clearly show the difference on the registra-
tion performance between goodly and badly synthe-
sized images, which leads to the idea of registration
improvement by simply further enhancing the synthe-
sizing quality.
Since the synthesis quality plays an important
role, we investigated if the change in entropy or the
FID score can explain/determine the synthesis quality.
Table 3 shows that neither score gives an precise in-
dividual explanation to the synthesis quality and reg-
istration performance. The FID score of synthetic CT
Image 10 (173) is a lot higher than the scores of the
other images, considering FID has a range of [88, 173]
Table 3: After the training of the 8 models, the respective
resulting synthetic test images are visually assessed to be of
good, medium or bad quality. In order to reason this cate-
gorization, the entropy (H) as well as the fr
´
echet inception
distance (FID) is calculated before and after the MR-to-CT
synthesis. The FID score is here defined to be lower if the
image better represents the joint dataset of all the 8 corre-
sponding CT images.
image quality H H FID FID
ID (synCT) (MR) (synCT) (MR) (synCT)
02 good 14.39 5.7 239 88
04 medium 14.59 5.79 219 118
06 bad 14.84 5.72 200 129
08 medium 12.31 5.77 237 123
10 bad 9.93 5.64 226 173
12 good 13.27 5.76 153 122
14 good 13.90 5.76 213 132
16 good 14.18 5.61 186 97
for this data. Since the badly synthesized CT 06 has
a FID score of 129 and the goodly synthesized Image
14 has a comparatively high FID score of 132, the re-
liability of the FID score in terms of synthesis quality
and additionally in this research of registration per-
formance is highly questionable, which argues for a
necessary manual check of synthesis quality.
7 CONCLUSION
Given the abdominal MR and CT images of the
Learn2Reg2021-challenge, we investigated the ef-
Evaluating the Effects of a Priori Deep Learning Image Synthesis on Multi-Modal MR-to-CT Image Registration Performance
327
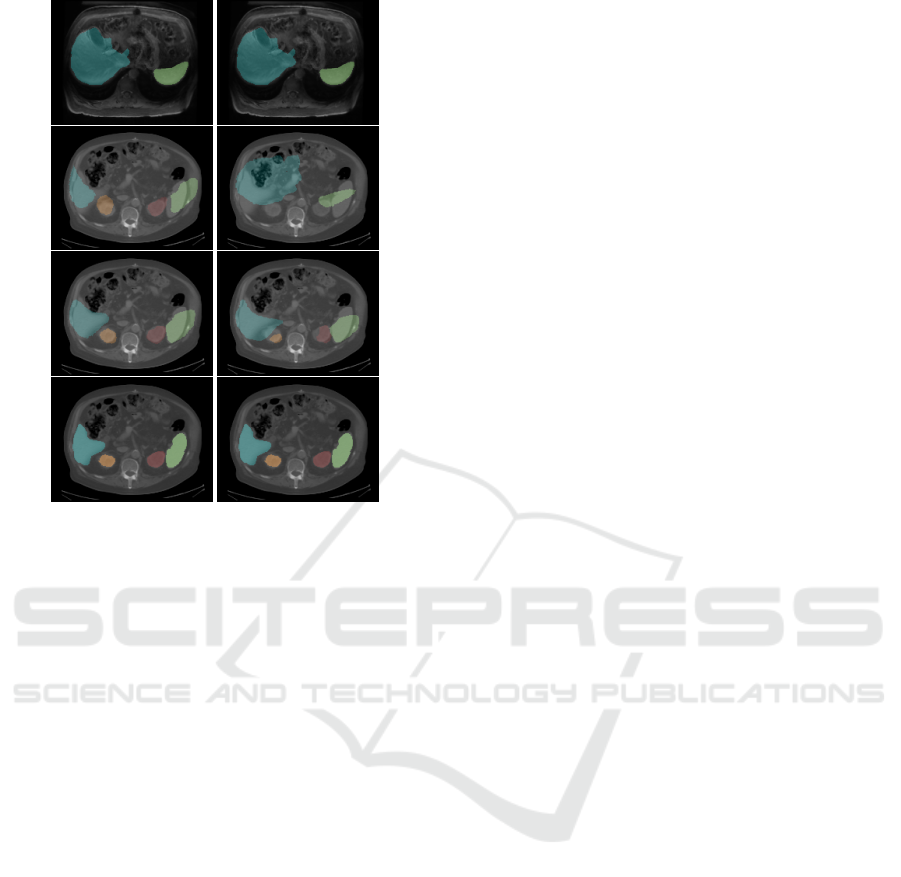
Figure 3: Both columns show the original MR on the top
and the original CT on the bottom with its corresponding
segmentations (blue: liver, green: spleen, orange: left kid-
ney, red: right kidney). The first column shows the resulting
deformed MR-segmentation in the second and third image
after registration with ‘SyNRA’, where the second column
shows it after ‘ElasticSyN’ registration. The results of the
MR-CT registration is visualized in each second image and
of the synCT-CT registration in each third image.
fects of prior image synthesis on multi-modal MR-
to-CT image registration. Precisely, we trained eight
CycleGAN-models individually by leaving out one
of the eight TCIA a priori defined training images
for later testing and compared the image registration
results of four ANTs registration methods (‘Rigid’,
‘SyN’, ‘SyNRA’ and ‘ElasticSyN’) with and without
the use of the synthetic CT instead of the original MR
image. Additionally, we investigated two metrics for
determining synthesis quality. Overall, We show that
good synthesizing results lead to an average improve-
ment in all of the four registration methods. Further-
more, the biggest improvement in registration perfor-
mance is achieved for the ‘SyN’ method with 8% in
Dice-score and the overall best registration method
with prior synthesis is ‘SyNRA’. These results are an
important contribution to the discussion on the use-
fulness of synthesis for the registration task and show
promising positive effects. Additionally, the experi-
ments indicated that further improvements in image
synthesis will also benefit the registration task. Fu-
ture research should aim to make further validation on
the synthesis benefits using other datasets and other
multi-modal registration tasks. Also, methods for au-
tomatically evaluating the synthesis performance are
needed for an automated application of synthesis in
registration tasks.
REFERENCES
Avants, B. B., Tustison, N. J., Song, G., Cook, P. A., Klein,
A., and Gee, J. C. (2011). A reproducible evaluation
of ants similarity metric performance in brain image
registration. NeuroImage, 54(3):2033–2044.
Cao, X., Yang, J., Gao, Y., Guo, Y., Wu, G., and Shen, D.
(2017). Dual-core steered non-rigid registration for
multi-modal images via bi-directional image synthe-
sis. Medical image analysis, 41:18–31.
Chandarana, H., Wang, H., Tijssen, R., and Das, I. J. (2018).
Emerging role of mri in radiation therapy. Journal of
Magnetic Resonance Imaging, 48(6):1468–1478.
Chartsias, A., Joyce, T., Dharmakumar, R., and Tsaftaris,
S. A. (2017). Adversarial image synthesis for unpaired
multi-modal cardiac data. In International workshop
on simulation and synthesis in medical imaging, pages
3–13. Springer.
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J.,
Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle,
M., et al. (2013). The cancer imaging archive (tcia):
maintaining and operating a public information repos-
itory. Journal of digital imaging, 26(6):1045–1057.
Dalca, A., Hering, A., Hansen, L., and Heinrich, M. P.
(2021). Grand challang: Learn2reg2021. Accessed:
2022-07-20.
Dice, L. R. (1945). Measures of the amount of ecologic
association between species. Ecology, 26(3):297–302.
Gao, L., Xie, K., Wu, X., Lu, Z., Li, C., Sun, J., Lin, T., Sui,
J., and Ni, X. (2021). Generating synthetic ct from
low-dose cone-beam ct by using generative adversar-
ial networks for adaptive radiotherapy. Radiation On-
cology, 16(1):1–16.
Ge, Y., Wei, D., Xue, Z., Wang, Q., Zhou, X., Zhan, Y.,
and Liao, S. (2019). Unpaired mr to ct synthesis with
explicit structural constrained adversarial learning. In
2019 IEEE 16th International Symposium on Biomed-
ical Imaging (ISBI 2019), pages 1096–1099.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. pages
2672–2680.
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., and
Hochreiter, S. (2017). Gans trained by a two time-
scale update rule converge to a local nash equilibrium.
In Guyon, I., Luxburg, U. V., Bengio, S., Wallach, H.,
Fergus, R., Vishwanathan, S., and Garnett, R., editors,
Advances in Neural Information Processing Systems,
volume 30. Curran Associates, Inc.
Hiasa, Y., Otake, Y., Takao, M., Matsuoka, T., Takashima,
K., Carass, A., Prince, J. L., Sugano, N., and Sato,
Y. (2018a). Cross-modality image synthesis from un-
paired data using cyclegan. In International workshop
HEALTHINF 2023 - 16th International Conference on Health Informatics
328

on simulation and synthesis in medical imaging, pages
31–41. Springer.
Hiasa, Y., Otake, Y., Takao, M., Matsuoka, T., Takashima,
K., Carass, A., Prince, J. L., Sugano, N., and Sato,
Y. (2018b). Cross-modality image synthesis from un-
paired data using cyclegan. In Gooya, A., Goksel,
O., Oguz, I., and Burgos, N., editors, Simulation and
Synthesis in Medical Imaging, pages 31–41, Cham.
Springer International Publishing.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017).
Image-to-image translation with conditional adversar-
ial networks. In Computer Vision and Pattern Recog-
nition (CVPR), 2017 IEEE Conference on.
Jiang, J., Hu, Y.-C., Tyagi, N., Zhang, P., Rimner, A.,
Mageras, G. S., Deasy, J. O., and Veeraraghavan, H.
(2018). Tumor-aware, adversarial domain adaptation
from ct to mri for lung cancer segmentation. In In-
ternational conference on medical image computing
and computer-assisted intervention, pages 777–785.
Springer.
Jin, C.-B., Kim, H., Liu, M., Jung, W., Joo, S., Park, E.,
Ahn, Y. S., Han, I. H., Lee, J. I., and Cui, X. (2019).
Deep ct to mr synthesis using paired and unpaired
data. Sensors, 19(10):2361.
Karras, T., Laine, S., and Aila, T. (2018). A style-based
generator architecture for generative adversarial net-
works. CoRR, abs/1812.04948.
Kavur, A. E., Selver, M. A., Dicle, O., Barıs¸, M., and Gezer,
N. S. (2019). CHAOS - Combined (CT-MR) Healthy
Abdominal Organ Segmentation Challenge Data.
Kida, S., Kaji, S., Nawa, K., Imae, T., Nakamoto, T., Ozaki,
S., Ohta, T., Nozawa, Y., and Nakagawa, K. (2020).
Visual enhancement of cone-beam ct by use of cycle-
gan. Medical physics, 47(3):998–1010.
Kingma, D. P. and Ba, J. (2015). Adam: A method for
stochastic optimization. In Bengio, Y. and LeCun,
Y., editors, 3rd International Conference on Learn-
ing Representations, ICLR 2015, San Diego, CA, USA,
May 7-9, 2015, Conference Track Proceedings.
Kuckertz, S., Papenberg, N., Honegger, J., Morgas, T.,
Haas, B., and Heldmann, S. (2020). Learning de-
formable image registration with structure guidance
constraints for adaptive radiotherapy. In
ˇ
Spiclin,
ˇ
Z.,
McClelland, J., Kybic, J., and Goksel, O., editors,
Biomedical Image Registration, pages 44–53, Cham.
Springer International Publishing.
Kurz, C., Maspero, M., Savenije, M. H., Landry, G., Kamp,
F., Pinto, M., Li, M., Parodi, K., Belka, C., and
Van den Berg, C. A. (2019). Cbct correction us-
ing a cycle-consistent generative adversarial network
and unpaired training to enable photon and proton
dose calculation. Physics in Medicine & Biology,
64(22):225004.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. nature, 521(7553):436–444.
Liang, X., Chen, L., Nguyen, D., Zhou, Z., Gu, X., Yang,
M., Wang, J., and Jiang, S. (2019). Generating syn-
thesized computed tomography (ct) from cone-beam
computed tomography (cbct) using cyclegan for adap-
tive radiation therapy. Physics in Medicine & Biology,
64(12):125002.
Lu, J.,
¨
Ofverstedt, J., Lindblad, J., and Sladoje, N. (2021).
Is image-to-image translation the panacea for multi-
modal image registration? a comparative study. arXiv
preprint arXiv:2103.16262.
Mahapatra, D., Antony, B., Sedai, S., and Garnavi, R.
(2018). Deformable medical image registration us-
ing generative adversarial networks. In 2018 IEEE
15th International Symposium on Biomedical Imaging
(ISBI 2018), pages 1449–1453. IEEE.
McKenzie, E., Santhanam, A., Ruan, D., O’Connor, D.,
Cao, M., and Sheng, K. (2019). Multimodality image
registration in the head-and-neck using a deep learn-
ing derived synthetic ct as a bridge. Medical Physics,
47.
Modersitzki, J. (2009). Fair: Flexible Algorithms for Im-
age Registration. Society for Industrial and Applied
Mathematics, USA.
Qi, X. S. (2017). Image-Guided Radiation Therapy, pages
131–173. Springer International Publishing, Cham.
Reed, S., Akata, Z., Yan, X., Logeswaran, L., Schiele, B.,
and Lee, H. (2016). Generative adversarial text to
image synthesis. In Balcan, M. F. and Weinberger,
K. Q., editors, Proceedings of The 33rd International
Conference on Machine Learning, volume 48 of Pro-
ceedings of Machine Learning Research, pages 1060–
1069, New York, New York, USA. PMLR.
Roy, S., Carass, A., Jog, A., Prince, J., and Lee, J. (2014).
Mr to ct registration of brains using image synthesis.
Proceedings of SPIE, 9034.
Saiti, E. and Theoharis, T. (2020). An application inde-
pendent review of multimodal 3d registration meth-
ods. Computers & Graphics, 91:153–178.
Shi, Z., Mettes, P., Zheng, G., and Snoek, C. (2021).
Frequency-supervised mr-to-ct image synthesis.
Sørensen, T. J. (1948). A method of establishing groups of
equal amplitude in plant sociology based on similarity
of species content and its application to analyses of
the vegetation on Danish commons, volume 5.
Tanner, C., Ozdemir, F., Profanter, R., Vishnevsky, V.,
Konukoglu, E., and Goksel, O. (2018). Generative
adversarial networks for mr-ct deformable image reg-
istration.
Wei, D., Ahmad, S., Huo, J., Peng, W., Ge, Y., Xue, Z.,
Yap, P.-T., Li, W., Shen, D., and Wang, Q. (2019).
Synthesis and inpainting-based mr-ct registration for
image-guided thermal ablation of liver tumors. In In-
ternational Conference on Medical Image Computing
and Computer-Assisted Intervention, pages 512–520.
Springer.
Wolterink, J. M., Dinkla, A. M., Savenije, M. H. F.,
Seevinck, P. R., van den Berg, C. A. T., and Isgum,
I. (2017). Deep MR to CT synthesis using unpaired
data. CoRR, abs/1708.01155.
Yi, Z., Zhang, H., Tan, P., and Gong, M. (2017). Dualgan:
Unsupervised dual learning for image-to-image trans-
lation.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Computer Vision
(ICCV), 2017 IEEE International Conference on.
Evaluating the Effects of a Priori Deep Learning Image Synthesis on Multi-Modal MR-to-CT Image Registration Performance
329
