
(ε, k)-Randomized Anonymization: ε-Differentially Private Data Sharing
with k-Anonymity
Akito Yamamoto
1 a
, Eizen Kimura
2 b
and Tetsuo Shibuya
1 c
1
Human Genome Center, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
2
Medical School of Ehime University, Ehime, Japan
Keywords:
Differential Privacy, Randomized Response, k-Anonymity, Data Sharing.
Abstract:
As the amount of biomedical and healthcare data increases, data mining for medicine becomes more and
more important for health improvement. At the same time, privacy concerns in data utilization have also been
growing. The key concepts for privacy protection are k-anonymity and differential privacy, but k-anonymity
alone cannot protect personal presence information, and differential privacy alone would leak the identity. To
promote data sharing throughout the world, universal methods to release the entire data while satisfying both
concepts are required, but such a method does not yet exist. Therefore, we propose a novel privacy-preserving
method, (ε, k)-Randomized Anonymization. In this paper, we first present two methods that compose the
Randomized Anonymization method. They perform k-anonymization and randomized response in sequence
and have adequate randomness and high privacy guarantees, respectively. Then, we show the algorithm for
(ε, k)-Randomized Anonymization, which can provide highly accurate outputs with both k-anonymity and dif-
ferential privacy. In addition, we describe the analysis procedures for each method using an inverse matrix
and expectation-maximization (EM) algorithm. In the experiments, we used real data to evaluate our methods’
anonymity, privacy level, and accuracy. Furthermore, we show several examples of analysis results to demon-
strate high utility of the proposed methods.
1 INTRODUCTION
With the recent increase in health awareness, the vol-
ume of biomedical data has grown, and data mining
for medicine and healthcare has gained importance
(Rakesh Kumar et al., 2019; Wu et al., 2021). At
the same time, privacy concerns in releasing data have
been recognized (Hl
´
avka, 2020; Su et al., 2021), and
the development and discussion of privacy-preserving
methods of personal information contained in datasets
are now one of essential research topics. Further-
more, general data sharing methods in compliance
with European Union’s General Data Protection Reg-
ulation (GDPR) (European Commission, 2016) and
other regulations are required to promote the utiliza-
tion of medical data on a global basis in the future.
The two most important concepts to publish data
while protecting privacy are k-anonymity (Sweeney,
2002) and differential privacy (Dwork, 2006). k-
a
https://orcid.org/0000-0002-3769-3352
b
https://orcid.org/0000-0002-0690-8568
c
https://orcid.org/0000-0003-1514-5766
anonymity can prevent identity disclosure and is
widely used for healthcare data sharing (Emam and
Dankar, 2008; Lee et al., 2017). Differential privacy
is a framework to protect information on individu-
als’ participation and has increasingly been applied
to genomic data and other health research (Aziz et al.,
2019; Ficek et al., 2021). These two concepts focus
on different aspects of data privacy and need to com-
plement each other: k-anonymity alone would reveal
the presence of an individual, while differential pri-
vacy alone would leak the identity. In fact, it is re-
ported that for a data application system to be GDPR-
compliant, we need to satisfy differential privacy in
addition to anonymity (Cummings and Desai, 2018).
In this study, we propose new privacy-preserving
methods for data sharing that satisfy both k-
anonymity and ε-differential privacy. Our methods
differs from existing methods (Li et al., 2012; Holo-
han et al., 2017; Tsou et al., 2021) in that they do not
assume data sampling and can release all the infor-
mation in the original data. In particular, the contri-
butions of this study are as follows:
1. We employ the randomized response as a mecha-
Yamamoto, A., Kimura, E. and Shibuya, T.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity.
DOI: 10.5220/0011665600003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 287-297
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
287

nism to satisfy ε-differential privacy, and by com-
bining it with k-anonymization, we propose three
methods to release data while protecting both
identity and presence of individuals. The first
two methods perform k-anonymization and ran-
domized response in sequence. While these algo-
rithms are straightforward, each has its strength:
adequate randomness and high privacy guaran-
tees. Then, based on these two algorithms, we
propose a novel privacy-preserving data sharing
method with both of their advantages, (ε, k)-
Randomized Anonymization. We theoretically
guarantee that all of these three algorithms satisfy
ε-differential privacy. In addition, we describe the
analysis procedures using an inverse matrix and
an expectation-maximization (EM) algorithm.
2. In the experiments, we evaluated each method’s
anonymity, privacy level, and output accuracy
by using real data. The first method, k-
anonymization followed by randomized response,
reduces the anonymity of the output, but provides
the highest accuracy. The second method, ran-
domized response followed by k-anonymization,
can achieve stronger privacy guarantees, although
the accuracy is lower due to the high random-
ness. (ε, k)-Randomized Anonymization, which
combines the above two methods, can provide
both high privacy assurance and accuracy with-
out compromising anonymity. These results in-
dicate that (ε, k)-Randomized Anonymization is a
novel privacy-preserving method for data sharing
that can achieve high utility while satisfying both
k-anonymity and ε-differential privacy.
In Section 2, we briefly review the related work.
In Section 3, we describe the preliminary definitions
for this study. In Section 4, we propose new privacy-
preserving data sharing methods that satisfy both k-
anonymity and ε-differential privacy. In Section 5,
we evaluate our proposed methods by experiments
using real data. In Section 6, we summarize our
study with future direction. Python codes of our
methods are available at https://github.com/ay0408/
Randomized-Anonymization.
2 RELATED WORK
The most prominent concept for privacy-preserving
data sharing is k-anonymity (Sweeney, 2002). This
aims to prevent the identity disclosure of a targeted
individual in a dataset. However, there is still a risk
of identifying whether the target is in the dataset de-
pending on the adversary’s prior knowledge (Li et al.,
2012). In contrast to k-anonymity, differential privacy
(Dwork, 2006) is a framework to protect information
about the presence of the target. In the concept of
differential privacy, we can guarantee data privacy no
matter what information the adversary knows. How-
ever, should the information that the dataset contains
the target be leaked, the individual’s identity could be
revealed.
In this situation, it is desirable to develop new
data sharing methods that can protect both the iden-
tity and presence of individuals, and there have been
several studies (Li et al., 2012; Holohan et al., 2017;
Tsou et al., 2021) to connect k-anonymity and dif-
ferential privacy. First, Li et al. showed that k-
anonymization can achieve differential privacy when
preceded by random sampling (Li et al., 2012). Sub-
sequently, Tsou et al. presented an anonymization
method that satisfies k-anonymity and differential pri-
vacy by applying KD-tree in addition to random sam-
pling (Tsou et al., 2021). These studies assume that
we sample the data before releasing them. As yet, no
method can release all the information in the original
data while satisfying anonymity and privacy. Other
methods proposed by Holohan et al. consider vary-
ing the approach depending on data types and apply
k-anonymization to attribute data and differentially
private methods to numerical data (Holohan et al.,
2017), but the dataset as a whole is not completely
privacy-preserving. This paper proposes novel meth-
ods to publish all the data while achieving both k-
anonymity and differential privacy without sampling.
Although our methods do not protect against attribute
disclosure, they may achieve stronger privacy guaran-
tees by combining with the existing work on the rela-
tionship between t-closeness and differential privacy
(Domingo-Ferrer and Soria-Comas, 2015).
3 PRELIMINARIES
3.1 k-Anonymity
The concept of k-anonymity (Sweeney, 2002) was
proposed for privacy-preserving microdata sharing.
The k-anonymity requires that each tuple value of
quasi-identifier (QI) attributes appears at least k times
in a dataset, so that even if adversaries know a tu-
ple value of a particular individual’s QIs, they cannot
uniquely identify the exact record of the individual.
The following is the definition of k-anonymity.
Definition 1. (k-anonymity (Sweeney, 2002))
Let T be a table and QI
T
be the quasi-identifiers as-
sociated with it. T satisfies k-anonymity if and only if
each tuple value in T [QI
T
] appears at least k times in
T [QI
T
].
HEALTHINF 2023 - 16th International Conference on Health Informatics
288

The k-anonymity protects against identity disclo-
sure, but does not against a risk of identifying the
presence of the individual in the dataset. Therefore,
when we also aim to protect the information of the in-
dividual’s participation, a stronger privacy guarantee
is required.
3.2 ε-Differential Privacy
Differential privacy (Dwork, 2006) was developed in
the field of cryptography as a framework that allows
statistical analysis of databases while preserving per-
sonal data in the database from adversaries. Unlike k-
anonymity, differential privacy can protect the infor-
mation about whether the individual is in the dataset
or not. The idea of differential privacy is based on
the fact that it should be almost impossible to distin-
guish between two neighboring datasets differing in
just one record. The privacy level in differential pri-
vacy is evaluated by the parameter ε > 0. Smaller ε
values achieve stronger privacy guarantees but reduce
the utility of the data. The following is the definition
of ε-differential privacy.
Definition 2. (ε-Differential Privacy (Dwork, 2006))
A randomized mechanism M satisfies ε-differential
privacy if, for any S ⊂ range M and all neighboring
datasets D and D
′
, Pr[M(D) ∈ S] ≤ e
ε
·Pr[M(D
′
) ∈ S].
One main mechanism to satisfy ε-differential pri-
vacy is the Laplace mechanism (Dwork et al., 2006),
which only adds a random noise according to the sen-
sitivity of the function to the original data and outputs
private values. The definition of the sensitivity is as
follows.
Definition 3. (Sensitivity for the Laplace Mechanism
(Dwork et al., 2006))
The sensitivity of a function f : D
M
→ R
d
is
∆ f = max
D,D
′
|| f (D) − f (D
′
)||
1
,
where D, D
′
∈ D
M
are neighboring datasets.
Releasing f (D) + b satisfies ε-differential privacy
when b is random noise derived from a Laplace distri-
bution with mean 0 and scale
∆ f
ε
(Dwork et al., 2006).
The Laplace mechanism is highly practical when
the sensitivity is small, as in the case of histogram
publication (Meng et al., 2017). However, if the sen-
sitivity is large compared to the original value or the
dataset consists of discrete values, the outputs may be
less accurate. Therefore, in this study, we consider to
satisfy ε-differential privacy by the technique of ran-
domized response, which randomly perturbs each in-
dividual’s attribute values.
3.2.1 Randomized Response
Randomized response was first introduced by Warner
(Warner, 1965) to encourage survey participants to
answer sensitive questions truthfully. This mecha-
nism was shown to be differentially private (Dwork
and Roth, 2014) and has been well used for hypothe-
sis testing (Gaboardi and Rogers, 2018) and crowd-
sourcing (Erlingsson et al., 2014). In the follow-
ing, we describe the randomized response approach
in the case where all the participants in a dataset is di-
vided into m (≥ 2) mutually exclusive and exhaustive
classes.
The randomized response with m classes follows
an m × m distortion matrix:
P =
p
11
p
12
··· p
1m
p
21
p
22
··· p
2m
.
.
.
.
.
.
.
.
.
.
.
.
p
m1
p
m2
··· p
mm
,
where p
uv
= Pr[x
′
= u|x = v] (u, v ∈ {1, 2, · ·· , m}) de-
notes the probability that the randomized output is u
when the real class of the participant is v. Here, the
sum of probabilities of each column is 1. When the
following inequality holds:
ε ≥ max
u=1,2,···,m
max
v=1,2,···,m
p
uv
min
v=1,2,···,m
p
uv
,
the randomized response satisfies ε-differential pri-
vacy (Wang et al., 2016).
4 METHODS
In this study, we propose new privacy-preserving
methods for medical data sharing that satisfy both
k-anonymity and ε-differential privacy. First,
we present two algorithms that apply the k-
anonymization method and randomized response in
sequence, and discuss the advantages and disadvan-
tages of each. Then, we propose a novel method,
(ε, k)-Randomized Anonymization, based on the first
two algorithms. This method is expected to achieve
both high accuracy and strong privacy assurance of
the output. Also, we theoretically prove that each
algorithm satisfies differential privacy. Furthermore,
we describe the procedures for conducting statistical
analysis using the published data.
4.1 k-Anonymization → Randomized
Response
First, we present an algorithm that performs k-
anonymization on the original table followed by the
randomized response.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity
289

Algorithm 1: k-Anonymization → Randomized Response.
Input: A table with QIs, privacy parameters k and ε.
Output: A k-anonymized and ε-differentially private
table.
1: Group input tuples of QIs into clusters, s.t., each
cluster has at least k tuples.
2: Let m be the number of clusters and c
i
be the clus-
ter to which each tuple i belongs.
3: Construct an m × m distortion matrix P, and per-
form the randomized response for each c
i
accord-
ing to P. Let ˜c
i
be the randomized output from
c
i
.
4: For each tuple i, replace the QI values with the
representative values of ˜c
i
.
The distortion matrix P in Algorithm 1 satisfies
the following equation to maximize the sum of the
diagonal components (Wang et al., 2016):
P
uv
=
(
e
ε
e
ε
+m−1
(u = v)
1
e
ε
+m−1
(u ̸= v)
.
We provide a privacy guarantee of Algorithm 1 by
Theorem 1.
Theorem 1. Algorithm 1 Satisfies ε-differential Pri-
vacy.
Proof. Let T be the output table from Algorithm 1,
and g,g
′
be input tables differing in a tuple of one
individual. We let A be the mechanism represented
by Algorithm 1 and show
Pr[A(g) = T ] ≤ e
ε
· Pr[A (g
′
) = T ].
Here, we denote the procedure of applying the ran-
domized response to a cluster c by a function RR. Let
c
i
be the cluster containing tuple i, then the following
equation holds:
Pr[A(g) = T ]
= Pr[RR(c
g
0
) = c
T
0
] · Pr[RR(c
g
1
) = c
T
1
]
··· Pr[RR(c
g
|T |−1
) = c
T
|T |−1
] (̸= 0).
Suppose that the j-th tuple is different in g and g
′
,
then
Pr[A(g) = T ]
Pr[A(g
′
) = T ]
=
Pr[RR(c
g
j
) = c
T
j
]
Pr[RR(c
g
′
j
) = c
T
j
]
. (1)
Since the elements of the distortion matrix in Algo-
rithm 1 are
e
ε
e
ε
+m−1
or
1
e
ε
+m−1
, we can show (1) ≤
e
ε
. Therefore, Algorithm 1 satisfies ε-differential pri-
vacy.
When analyzing the data using the output from
this algorithm, we first create a vector d ∈ N
m
repre-
senting the number of elements in each cluster. Then,
we recover the original distribution by
˜
d = P
−1
d.
Here, the elements of P
−1
are as follows:
(P
−1
)
uv
=
(
e
ε
+m−2
e
ε
−1
(u = v)
−1
e
ε
−1
(u ̸= v)
.
After that, change the negative elements of
˜
d to 0, and
finally, calculate
˜
d ×
||d||
1
||
˜
d||
1
so that the sum of elements
of
˜
d becomes equal to that of d.
In addition, we can also use an expectation-
maximization (EM) algorithm to reconstruct
˜
d fol-
lowing some existing studies (Fanti et al., 2016; Ye
et al., 2019). Unlike the above procedure using P
−1
,
the EM algorithm has an advantage that it does not
output negative numbers. The detailed procedure is
as follows:
i. Initialization:
Let s be the number of individuals in the dataset.
Create x ∈ R
s×m
s.t.
x
h,i
=
(
1
∑
i−1
j=0
d
j
≤ h <
∑
i
j=0
d
j
0 (otherwise)
Set θ
0
0
= θ
0
1
= · ·· = θ
0
m−1
=
1
m
. (This is a uniform
distribution and m is the number of clusters.)
ii. e-Step:
For any individual h (0 ≤ h < s) and any cluster i
(0 ≤ i < m),
θ
k
h,i
= Pr[z
h,i
= 1|x
h,i
]
=
Pr[x
h,i
|z
h,i
= 1] · Pr[z
h,i
= 1]
∑
m−1
j=0
Pr[x
h,i
|z
h, j
= 1] · Pr[z
h, j
= 1]
=
Pr[x
h,i
|z
h,i
= 1] · θ
k−1
i
∑
m−1
j=0
Pr[x
h,i
|z
h, j
= 1] · θ
k−1
j
.
iii. m-Step:
θ
k
i
=
1
s
s−1
∑
h=0
θ
k
h,i
iv. Repeat steps ii and iii until
∑
i
θ
k
i
− θ
k−1
i
< δ for
some δ > 0, then calculate
˜
d = s · θ
k
.
Here, z
h,i
in the EM algorithm is unobserved data and
satisfies the following equations:
Pr[x
h,i
|z
h,i
] =
(
e
ε
e
ε
+m−1
(x
h,i
= 1)
m−1
e
ε
+m−1
(x
h,i
= 0)
Pr[x
h,i
|z
h, j
] =
(
1
e
ε
+m−1
(x
h,i
= 1)
e
ε
+m−2
e
ε
+m−1
(x
h,i
= 0)
(i ̸= j).
HEALTHINF 2023 - 16th International Conference on Health Informatics
290

The computational complexity of the E-Step is
O(sm
2
), so when the dataset size is large or the
anonymity parameter k is small, it could take a much
longer time than when using P
−1
.
Algorithm 1 is the first method for privacy-
preserving medical data sharing that satisfies both
anonymity and differential privacy. One drawback of
this method is that the output table does not strictly
satisfy k-anonymity. For a large k, anonymity of the
dataset is expected to be little compromised, but for
small values of k, more accurate algorithm is desired.
4.2 Randomized Response →
k-Anonymization
The next algorithm is to perform randomized re-
sponse first, then k-anonymization. Unlike Algorithm
1, the output exactly satisfies k-anonymity.
Algorithm 2: Randomized Response → k-Anonymization.
Input: A table with QIs, privacy parameters k and ε.
Output: A k-anonymized and ε-differentially private
table.
1: Let X be the set of possible tuples of QIs and n be
the size of X.
2: Let t
i
∈ X be the i-th tuple value.
3: Construct an n × n distortion matrix P, and per-
form the randomized response for each t
i
accord-
ing to P.
4: Group the randomized tuples into clusters, s.t.,
each cluster has at least k tuples.
5: Let c
i
be the cluster to which each tuple i belongs.
6: For each tuple i, replace the QI values with the
representative values of c
i
.
Similar to Algorithm 1, the distribution matrix P
satisfies the following equation:
P
uv
=
(
e
ε
e
ε
+n−1
(u = v)
1
e
ε
+n−1
(u ̸= v)
.
Here, for privacy assurance of Algorithm 2 and
data analysis using the output, we consider the prob-
ability of the i-th tuple belonging to cluster c
i
in the
output table. We let ˆc
i
be the cluster to which tuple i
should belong based on the input data, and r
j
be the
number of possible tuple values that cluster j can con-
tain. Then, the probability that ˆc
i
changes to c
i
is as
follows:
Pr[ ˆc
i
→ c
i
] =
(
e
ε
+(r
c
i
−1)
e
ε
+n−1
( ˆc
i
= c
i
)
r
c
i
e
ε
+n−1
( ˆc
i
̸= c
i
)
.
Using this probability, the privacy guarantee of Algo-
rithm 2 is shown by Theorem 2.
Theorem 2. Algorithm 2 Satisfies ε-differential Pri-
vacy.
Proof. Similar to the proof of Theorem 1, we con-
sider the following equation:
Pr[A(g) = T ]
= Pr[c
g
0
→ c
T
0
] · Pr[c
g
1
→ c
T
1
]
··· Pr[c
g
|T |−1
→ c
T
|T |−1
].
When the j-th tuple is different in g and g
′
,
Pr[A(g) = T ]
Pr[A(g
′
) = T ]
=
Pr[c
g
j
→ c
T
j
]
Pr[c
g
′
j
→ c
T
j
]
. (2)
Here, using the probability above, the following in-
equalities hold:
(2) ≤
e
ε
+ (r
c
g
j
− 1)
r
c
g
j
= 1 +
e
ε
− 1
r
c
g
j
≤ e
ε
.
Therefore, Algorithm 2 satisfies ε-differential pri-
vacy. In particular, when min
j
r
c
g
j
≥ 2, this algorithm
achieves a truly higher privacy guarantee than ε.
When analyzing the data based on the output from
Algorithm 2, we consider the following m× m matrix
Q:
Q =
1
e
ε
+ n − 1
e
ε
+ r
0
− 1 r
0
··· r
0
r
1
e
ε
+ r
1
− 1 ··· r
1
.
.
.
.
.
.
.
.
.
.
.
.
r
m−1
r
m−1
··· e
ε
+ r
m−1
− 1
,
where m is the number of clusters, and note that
∑
i
r
i
= n. Similar to the case of Algorithm 1, we first
create a vector d ∈ N
m
representing the number of el-
ements in each cluster, then reconstruct the original
distribution using Q
−1
. The elements of Q
−1
are as
follows:
(Q
−1
)
uv
=
(
e
ε
+n−r
u
−1
e
ε
−1
(u = v)
−r
u
e
ε
−1
(u ̸= v)
.
When using the EM algorithm, we can follow the
same procedure as Algorithm 1 and the unobserved
data z
h,i
satisfies the following equations:
Pr[x
h,i
|z
h,i
] =
(
e
ε
+r
i
−1
e
ε
+n−1
(x
h,i
= 1)
n−r
i
e
ε
+n−1
(x
h,i
= 0)
Pr[x
h,i
|z
h, j
] =
(
r
i
e
ε
+n−1
(x
h,i
= 1)
e
ε
+n−r
i
−1
e
ε
+n−1
(x
h,i
= 0)
(i ̸= j).
Algorithm 2 can guarantee that the output table is
k-anonymized and, moreover, achieve a truly stronger
privacy guarantee than ε. Therefore, Algorithm 2 is
superior to Algorithm 1 in terms of anonymity and
privacy protection, but when n is much larger than
the number of clusters, the accuracy is expected to
decrease because the randomness of the output in-
creases.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity
291

4.3 (ε, k)-Randomized Anonymization
Finally, we propose a novel method that combines
the moderate randomness of Algorithm 1 and high
anonymity and privacy guarantees of Algorithm 2.
In this method, we perform k
′
(< k)-anonymization
first, and then apply the randomized response to
the anonymized data. After that, we perform k-
anonymization on the randomized tuples. The de-
tailed algorithm is shown in Algorithm 3.
Algorithm 3: (ε, k)-Randomized Anonymization.
Input: A table with QIs, privacy parameters k
′
, k and
ε.
Output: A k-anonymized and ε-differentially private
table.
1: Group input tuples of QIs into clusters, s.t., each
cluster has at least k
′
tuples.
2: Let m
′
be the number of clusters and c
′
i
be the
cluster to which each tuple i belongs.
3: Construct an m
′
×m
′
distortion matrix P, and per-
form the randomized response for each c
′
i
accord-
ing to P. Let
˜
c
′
i
be the randomized output from
c
′
i
.
4: For each tuple i, replace the QI values with the
representative values of
˜
c
′
i
.
5: Group the randomized tuples into clusters, s.t.
each cluster has at least k tuples.
6: Let c
i
be the cluster to which each tuple i belongs.
7: For each tuple i, replace the QI values with the
representative values of c
i
.
The distribution matrix P in Algorithm 3 satisfies
the following equation:
P
uv
=
(
e
ε
e
ε
+m
′
−1
(u = v)
1
e
ε
+m
′
−1
(u ̸= v)
.
Similar to the case of Algorithm 2, the probabil-
ity that ˆc
i
changes to c
i
through Algorithm 3 can be
expressed as follows:
Pr[ ˆc
i
→ c
i
] =
e
ε
+(r
′
c
i
−1)
e
ε
+m
′
−1
( ˆc
i
= c
i
)
r
′
c
i
e
ε
+m
′
−1
( ˆc
i
̸= c
i
)
,
where r
′
j
is the number of possible randomized tuple
values that cluster j can contain. Using this probabil-
ity, we can show the privacy guarantee of Algorithm
3 by Theorem 3.
Theorem 3. Algorithm 3 satisfies ε-differential pri-
vacy.
Proof. Similar to the proof of Theorem 2, the follow-
ing inequalities hold:
Pr[A(g) = T ]
Pr[A(g
′
) = T ]
≤
e
ε
+ (r
′
c
g
j
− 1)
r
′
c
g
j
= 1 +
e
ε
− 1
r
′
c
g
j
≤ e
ε
.
Therefore, Algorithm 3 satisfies ε-differential privacy
and when min
j
r
′
c
g
j
≥ 2, the privacy guarantee is truly
higher than ε.
When analyzing the data based on the output from
Algorithm 3, we consider an m × m matrix Q
′
whose
elements are as follows:
Q
′
uv
=
(
e
ε
+r
′
u
−1
e
ε
+m
′
−1
(u = v)
r
′
u
e
ε
+m
′
−1
(u ̸= v)
.
Then, we can analyze the data based on the recovered
distribution
˜
d = Q
′
−1
d, like in the previous cases.
If we use the EM algorithm, consider the unobserved
data z
h,i
satisfying the following equations:
Pr[x
h,i
|z
h,i
] =
(
e
ε
+r
′
i
−1
e
ε
+m
′
−1
(x
h,i
= 1)
m
′
−r
′
i
e
ε
+m
′
−1
(x
h,i
= 0)
Pr[x
h,i
|z
h, j
] =
(
r
i
e
ε
+m
′
−1
(x
h,i
= 1)
e
ε
+m
′
−r
′
i
−1
e
ε
+m
′
−1
(x
h,i
= 0)
(i ̸= j).
5 EXPERIMENTS AND
DISCUSSION
In the experiments, we used the data provided
by Japan Medical Association Medical Information
Management Organization (Control Number: 2021-
3). Medical ethical approval was obtained from
Anonymized Medical Data Provision Review Board
of Japan Medical Association Medical Information
Management Organization.
Using the provided data, we first examined the
characteristics of our proposed methods, including
the anonymity of Algorithm 1 and the privacy level of
Algorithms 2 and 3. Then, we conducted an age dis-
tribution analysis for a disease and measured the dif-
ference between the original data and the analysis re-
sults obtained from our methods to show their utility.
Furthermore, we present several examples of analy-
sis results using our methods and demonstrate that
the results are roughly identical to the original data
while satisfying both k-anonymity and ε-differential
privacy.
5.1 Data Description
We used the data on diseases in this experiment. The
data size is 1, 512, 673. The data contains four at-
HEALTHINF 2023 - 16th International Conference on Health Informatics
292

tributes that can be regarded as QIs: Medical Institu-
tion Code, Consultation Date, Sex, and Age. Medical
Institution Code is from 1 to 38, Consultation Date is
from April 1, 2020 to April 30, 2021, Sex is M or F
and Age is from 0 to 105. We considered performing
k-anonymization based on these attributes. In the fol-
lowing, we show the k-anonymization method for the
experiment.
5.1.1 k-Anonymization
First, we represent the set of QIs of each individual as
a single integer score. Note that the score and the set
of QIs are in one-to-one correspondence. Then, we
can satisfy k-anonymity by grouping the individuals
whose scores are close to each other. Finally, by re-
placing each individual’s score with a representative
value of the group, a k-anonymized table can be out-
put. The detailed algorithm is shown in Algorithm 4.
This k-anonymization method does not mask any
of the QI attributes. Therefore, analyses for all the QI
attributes can be performed in the similar way, and the
representative values of each cluster can be calculated
easily for our proposed methods. However, the score
calculation in Algorithm 4 may result in poor accu-
racy of the QI information in the lower bits (i.e., c
i
or
d
i
) because individuals with different values tend to
be clustered in the same group. Should the analysis
purpose and the use of the data be known in detail, a
better k-anonymization method would exist, so this is
one important problem for the future work.
5.2 Results
In this subsection, we evaluate the utility of the pro-
posed methods in terms of anonymity, privacy level,
and accuracy of the analysis results.
5.2.1 Anonymity of Algorithm 1
Algorithm 1 applies randomized response after k-
anonymization, so the anonymity of the output table
becomes less than or equal to k. In this experiment,
we varied the values of k (from 10 to 100) and ε (from
1 to 20), the inputs to Algorithm 1, to measure the
output anonymity. The results are plotted in Figure 1.
The results show that the output anonymity is
roughly proportional to the input k, and the rate of de-
crease in anonymity is almost independent of k. Re-
garding the effect of ε, a larger ε preserves higher
anonymity. However, as ε value increases, the pri-
vacy guarantee under differential privacy decreases
and the information about the presence of individuals
is more likely to be revealed. Therefore, Algorithm 1
has a strong trade-off between anonymity and privacy
Algorithm 4: k-Anonymization method for the experiment.
Input: A table with QIs (Medical Institution Code,
Consultation Date, Sex, and Age) and privacy pa-
rameter k.
Output: A k-anonymized table.
1: Let C, D, S, and A be the number of possible
values of Medical Institution Code, Consultation
Date, Sex, and Age, respectively.
2: Rewrite each value of QIs as a natural number in
the range of [0,C − 1], [0, D − 1], [0, S − 1], and
[0, A − 1], respectively.
3: Denote the set of QIs of the individual i by the
following SCORE
i
:
SCORE
i
= c
i
+C · d
i
+C · D · s
i
+C · D · S · a
i
,
where c
i
, d
i
, s
i
, and a
i
are the QIs of individual i.
4: Let n
j
be the number of individuals with a score
j.
5: Partition the distribution of scores so that each
group contains k or more individuals by the fol-
lowing procedure:
m = 0, t = 0
Let p be a vector representing the maximum
score in each group.
for j in 0, 1, . . . ,C · D · S · A − 1 do
m = m + n
j
if m ≥ k do
p
t
= j; t = t +1; m = 0
if m < k do
p
t−1
= C · D · S · A − 1
6: Replace SCORE
i
of each individual i with the
representative value of the group in which the
score is included. When the score is in the group
g, the replaced SCORE
i
can be calculated as fol-
lows:
SCORE
i
= ⌊
p
g−1
+p
g
2
⌋.
Figure 1: Anonymity of the output by Algorithm 1 when
varying the input k and ε.
level, requiring a larger ε for stronger anonymity and
a smaller ε for a stronger privacy assurance.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity
293
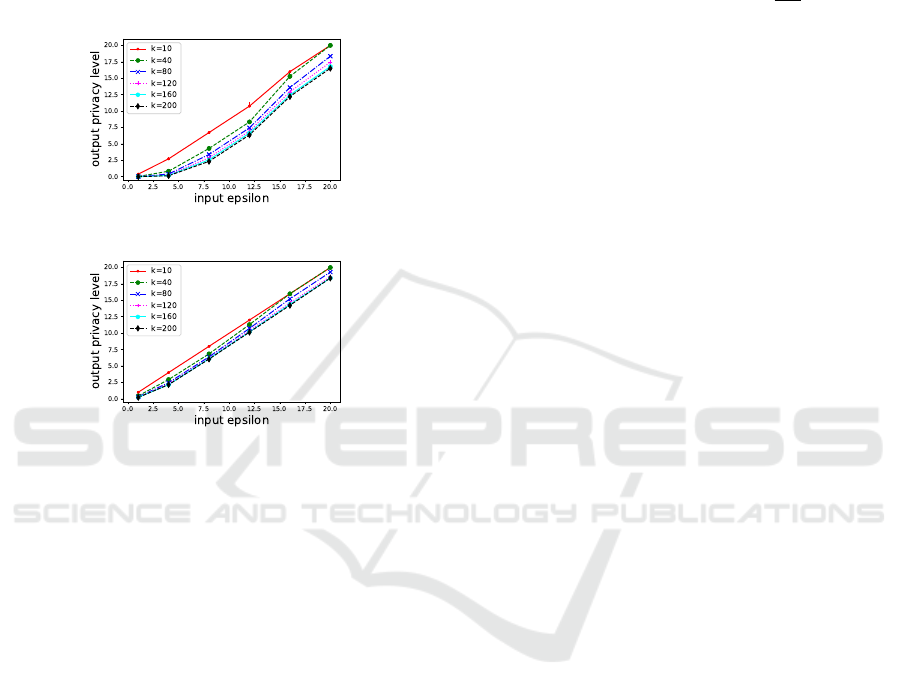
5.2.2 Privacy Level of Algorithms 2 and 3
Unlike Algorithm 1, the output tables from Algo-
rithms 2 and 3 are truly k-anonymized, and stronger
privacy guarantees than the input ε can be provided.
In this experiment, we varied the values of ε (from 1
to 20) and k (from 10 to 200) and measured the output
privacy level. The results are shown in Figure 2.
(A)
(B)
Figure 2: Privacy level of the output by Algorithms 2 (A)
and 3 (B) when varying the input ε and k.
As for Algorithm 2, a smaller value of ε makes the
privacy assurance stronger. However, the added per-
turbation also increases, which may lead to poor out-
put accuracy. As for Algorithm 3, the privacy levels
are relatively stable for any ε compared to Algorithm
2, and the accuracy of Algorithm 3 is expected to be
higher than that of Algorithm 2. Regarding the effect
of k, a higher k can provide a stronger privacy guar-
antee. Therefore, when using Algorithms 2 and 3, we
can enhance both anonymity and privacy guarantee by
increasing k.
5.2.3 Accuracy
Next, to evaluate the output accuracy from each al-
gorithm, we compared the original data and the anal-
ysis results by our methods for the age distribution
of those diagnosed with gastritis. Here, we use an
inverse matrix to analyze data for quick execution.
Because the data size used in this study was large
(1, 512, 673) and the number of QIs was small (4),
we can set relatively large values as the anonymity
parameter k. In this experiment, we considered four
cases of k = 100, 400, 800, and 1, 600. To measure the
difference between the original age distribution and
the analysis results from our methods, we use the KL
divergence (Kullback and Leibler, 1951). The defini-
tion is as follows.
Definition 4. (KL Divergence (Kullback and Leibler,
1951))
For discrete probability distributions p and q defined
on the same probability space X, the KL divergence is
defined by D
KL
(p||q) =
∑
x∈X
p(x)log
p(x)
q(x)
.
In this experiment, we let q be the original distri-
bution. A smaller KL divergence indicates that p and
q are closer together. The results are plotted in Figure
3.
The results in all the cases show a similar trend
of accuracy when varying the value of ε. When the
value of k increased, the input values were easily per-
turbed by grouping in k-anonymization, resulting in
lower accuracy.
Then, we will discuss each algorithm. Algorithm
2 (k-RR) has too strong a privacy guarantee when
ε is small as we showed in Figure 2, so the out-
put accuracy was poor. Algorithm 1 (RR-k) pro-
vided the highest accuracy because the output pri-
vacy level does not change from ε, but we should
note that anonymity is reduced. Algorithm 3 (RA)
also maintains high accuracy without compromis-
ing anonymity and with stronger privacy guarantees
than ε. These results indicate that our Algorithm 3,
(ε, k)-Randomized Anonymization, can provide high-
quality results in all aspects of anonymity, privacy
guarantee, and output accuracy.
5.3 Examples
As examples of analysis results, we examined the dis-
tributions of those diagnosed with gastritis. Here, we
performed both the analysis using an inverse matrix
and EM algorithm. The values of k and ε were set to
800 and 16, respectively. We analyzed the age dis-
tribution and consultation month, and the results are
plotted in Figures 4 and 5.
These results show that using either an inverse ma-
trix or EM algorithm, the analysis results are equiva-
lent to the original distribution, indicating high utility
of our methods.
6 CONCLUSION
In this study, we proposed new privacy-preserving
methods for data sharing that satisfy both k-
anonymity and ε-differential privacy. Our meth-
ods have the advantage that they do not assume
data sampling and can release all the information in
HEALTHINF 2023 - 16th International Conference on Health Informatics
294
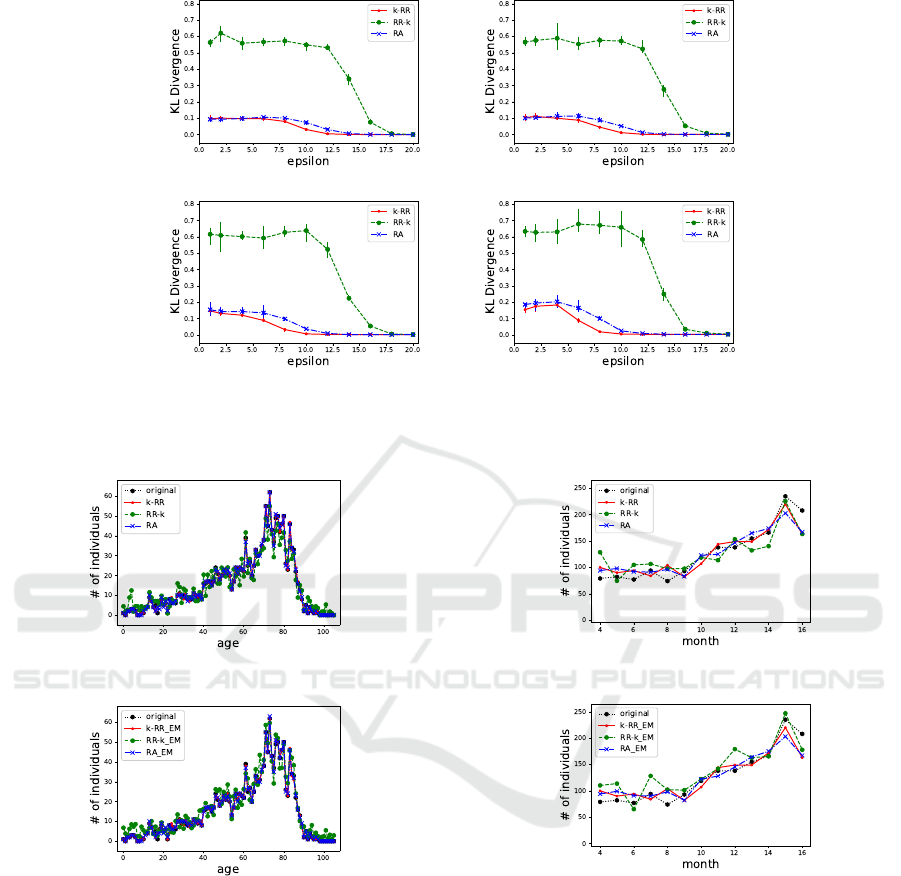
(a) (b)
(c) (d)
Figure 3: KL Divergence between the original age distribution and the analysis results from our methods when (a) k = 100,
(b) k = 400, (c) k = 800, and (d) k = 1, 600. k-RR (red, solid), RR-k (green, dashed), and RA (blue, dash-dot) represent our
Algorithms 1, 2, and 3, respectively.
(I)
(II)
Figure 4: Age distributions of those diagnosed with gastritis
when using an inverse matrix (I) and EM algorithm (II).
the original data. In particular, our third method,
(ε, k)-Randomized Anonymization, is a novel method
that can achieve a stronger privacy guarantee than ε
while truly satisfying k-anonymity. Also, the exper-
iments using real data show that (ε, k)-Randomized
Anonymization can provide highly accurate results
close to the original data. Not only data sharing meth-
ods, we also described two analysis procedures: one
using an inverse matrix and the other using an EM
algorithm.
An important future work is the development of
k-anonymization methods suited for integration with
(I)
(II)
Figure 5: Consultation month distributions (from April
2020 to April 2021) of those diagnosed with gastritis when
using an inverse matrix (I) and EM algorithm (II).
the randomized response technique. The optimized
methods may strongly depend on the data usage and
analysis purposes, so we plan to explore this prob-
lem continuously. Furthermore, combination with the
concept of t-closeness and the use of RAPPOR (Er-
lingsson et al., 2014) instead of randomized response
will also be beneficial. We hope that this study will
help in free sharing of biomedical and healthcare data
throughout the world in the future.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity
295

ACKNOWLEDGEMENTS
This research was supported by Health Labour Sci-
ences Research Grant 21AC1001.
REFERENCES
Aziz, M. M. A., Sadat, M. N., Alhadidi, D., Wang, S.,
Jiang, X., Brown, C. L., and Mohammed, N. (2019).
Privacy-preserving techniques of genomic data-a sur-
vey. Brief Bioinform., 20(3):887–895.
Cummings, R. and Desai, D. (2018). The role of differential
privacy in GDPR compliance.
Domingo-Ferrer, J. and Soria-Comas, J. (2015). From t-
closeness to differential privacy and vice versa in data
anonymization. Knowl.-Based Sys., 74:151–158.
Dwork, C. (2006). Differential privacy. In Bugliesi, M.,
Preneel, B., Sassone, V., and Wegener, I., editors, Au-
tomata, Languages and Programming, pages 1–12,
Berlin, Heidelberg. Springer Berlin Heidelberg.
Dwork, C., McSherry, F., Nissim, K., and Smith, A. (2006).
Calibrating noise to sensitivity in private data anal-
ysis. In Halevi, S. and Rabin, T., editors, Theory
of Cryptography, pages 265–284, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Dwork, C. and Roth, A. (2014). The algorithmic foun-
dations of differential privacy. Found. Trends Theor.
Comput. Sci., 9(3–4):211–407.
Emam, K. E. and Dankar, F. K. (2008). Protecting pri-
vacy using k-anonymity. J. Am. Med. Inform. Assoc.,
15(5):627–637.
Erlingsson, U., Pihur, V., and Korolova, A. (2014). RAP-
POR: Randomized aggregatable privacy-preserving
ordinal response. In Proceedings of the 2014 ACM
SIGSAC Conference on Computer and Communica-
tions Security, CCS ’14, page 1054–1067, New York,
NY, USA. Association for Computing Machinery.
European Commission (2016). Regulation (EU) 2016/679
of the European Parliament and of the Council of 27
April 2016 on the protection of natural persons with
regard to the processing of personal data and on the
free movement of such data, and repealing Directive
95/46/EC (General Data Protection Regulation), OJ
2016 L 119/1.
Fanti, G., Pihur, V., and Erlingsson, U. (2016). Build-
ing a RAPPOR with the unknown: Privacy-
preserving learning of associations and data dictionar-
ies. Proceedings on Privacy Enhancing Technologies
(PoPETS), issue 3, 2016.
Ficek, J., Wang, W., Chen, H., Dagne, G., and Daley, E.
(2021). Differential privacy in health research: A
scoping review. Journal of the American Medical In-
formatics Association, 28(10):2269–2276.
Gaboardi, M. and Rogers, R. (2018). Local private hy-
pothesis testing: Chi-square tests. In Dy, J. and
Krause, A., editors, Proceedings of the 35th Interna-
tional Conference on Machine Learning, volume 80
of Proceedings of Machine Learning Research, pages
1626–1635. PMLR.
Hl
´
avka, J. P. (2020). Chapter 10 - security, privacy, and
information-sharing aspects of healthcare artificial in-
telligence. In Bohr, A. and Memarzadeh, K., editors,
Artificial Intelligence in Healthcare, pages 235–270.
Academic Press.
Holohan, N., Antonatos, S., Braghin, S., and Aonghusa,
P. M. (2017). (k,ε)-anonymity: k-anonymity with ε-
differential privacy. arXiv: Cryptography and Secu-
rity.
Kullback, S. and Leibler, R. A. (1951). On information and
sufficiency. Ann. Math. Statist., 22(1):79–86.
Lee, H., Kim, S., Kim, J. W., and Chung, Y. D. (2017).
Utility-preserving anonymization for health data pub-
lishing. BMC Med. Inform. Decis. Mak., 17:104.
Li, N., Qardaji, W., and Su, D. (2012). On sam-
pling, anonymization, and differential privacy or, k-
anonymization meets differential privacy. In Proceed-
ings of the 7th ACM Symposium on Information, Com-
puter and Communications Security, ASIACCS ’12,
page 32–33, New York, NY, USA. Association for
Computing Machinery.
Meng, X., Li, H., and Cui, J. (2017). Different strategies for
differentially private histogram publication. J. Com-
mun. Inf. Netw., 2:68–77.
Rakesh Kumar, S., Gayathri, N., Muthuramalingam, S.,
Balamurugan, B., Ramesh, C., and Nallakaruppan, M.
(2019). Chapter 13 - medical big data mining and pro-
cessing in e-healthcare. In Balas, V. E., Son, L. H.,
Jha, S., Khari, M., and Kumar, R., editors, Internet
of Things in Biomedical Engineering, pages 323–339.
Academic Press.
Su, J., Cao, Y., Chen, Y., Liu, Y., and Song, J. (2021).
Privacy protection of medical data in social network.
BMC Med. Inform. Decis. Mak., 21:286.
Sweeney, L. (2002). K-anonymity: A model for protect-
ing privacy. Int. J. Uncertain. Fuzziness Knowl.-Based
Syst., 10(5):557–570.
Tsou, Y.-T., Alraja, M. N., Chen, L.-S., Chang, Y.-H., Hu,
Y.-L., Huang, Y., Yu, C.-M., and Tsai, P.-Y. (2021).
(k, ε, δ)-anonymization: Privacy-preserving data re-
lease based on k-anonymity and differential privacy.
Serv. Oriented Comput. Appl., 15(3):175–185.
Wang, Y., Wu, X., and Hu, D. (2016). Using random-
ized response for differential privacy preserving data
collection. In Palpanas, T. and Stefanidis, K., edi-
tors, Proceedings of the Workshops of the EDBT/ICDT
2016 Joint Conference, EDBT/ICDT Workshops 2016,
Bordeaux, France, March 15, 2016, volume 1558 of
CEUR Workshop Proceedings.
Warner, S. L. (1965). Randomized response: A survey tech-
nique for eliminating evasive answer bias. Journal of
the American Statistical Association, 60(309):63–69.
Wu, W.-T., Li, Y.-J., Feng, A.-Z., Li, L., Huang, T., Xu,
A.-D., and Lyu, J. (2021). Data mining in clinical
big data: the frequently used databases, steps, and
methodological models. Military Med. Res., 8:44.
Ye, Y., Zhang, M., Feng, D., Li, H., and Chi, J. (2019). Mul-
tiple privacy regimes mechanism for local differential
HEALTHINF 2023 - 16th International Conference on Health Informatics
296

privacy. In Li, G., Yang, J., Gama, J., Natwichai, J.,
and Tong, Y., editors, Database Systems for Advanced
Applications, pages 247–263, Cham. Springer Inter-
national Publishing.
(, k)-Randomized Anonymization: -Differentially Private Data Sharing with k-Anonymity
297
