
Novel Distributed Informatics Platform to Support Machine
Learning Discovery of Metabolic Biomarkers in Hypoxia
Predisposition
Anthony Stell
1
, Vedant Chauhan
1
, Sandra Amador
9
, Felix Beuschlein
2
, Judith Favier
3
, David Gil
4
,
Philip Greenwood
1
, Ronald de Krijger
5
, Matthias Kroiss
6
, Samanta Ortuno
10
,
Attila Patocs
7
and Axel Walch
8
1
School of Computing and Information Systems, University of Melbourne, Melbourne, Australia
2
Endokrinologie, Diabetologie und Klinische Ernährung, UniversitätsSpital Zürich (USZ) und Universität Zürich (UZH),
Zurich, Switzerland
3
Université Paris Cité, PARCC, INSERM, Equipe Labellisée par la Ligue contre le Cancer, Paris, France
4
Department of Computer Science Technology and Computation, University of Alicante, Alicante, Spain
5
Dept. of Pathology, University Medical Center Utrecht, and Princess Maxima Center for Pediatric Oncology,
Utrecht, The Netherlands
6
University Hospital Munich, Ludwig-Maximilians-Universität, München, Germany
7
ELKH Hereditary Cancer Research Group, Department of Laboratory Medicine, Semmelweis University and
Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary
8
Research Unit Analytical Pathology, Helmholtz Zentrum München, Oberschleissheim, Germany
9
Department of Computer Science and Systems, University of Murcia, Murcia, Spain
10
Health and Biomedical Research Institute of Alicante, Alicante, Spain
felix.beuschlein@usz.ch, judith.favier@inserm.fr, david.gil@ua.es, r.r.dekrijger@umcutrecht.nl,
matthias.kroiss@med.uni-muenchen.de, samanta.ortuno@gmail.com, patocs.attila@med.semmelweis-univ.hu,
axel.walch@helmholtz-muenchen.de
Keywords: Distributed Informatics Platform, Docker Orchestration, Content Delivery Networks.
Abstract: To realise the scientific and clinical benefits of machine learning (ML) in a multi-centre research collaboration,
a common issue is the need to bring high-volume data, complex analytical algorithms, and large-scale
processing power, all together into one place. This paper describes the detailed architecture of a novel platform
that combines these features, in the context of a proposed new clinical/bioinformatics project, Hypox-PD.
Hypox-PD uses ML methods to identify new metabolic biomarkers, through the analysis of high-volume data
including mass spectrometry and imaging morphology of biobank tissue. The platform features three
components: a content delivery network (CDN); a standardised orchestration application; and high-
specification processing power and storage. The central innovation of this platform is a distributed application
that simultaneously manages the workflow between these components, provides a virtual mapping of the
domain data dictionary, and presents the project data/metadata in a FAIR-compliant external interface. This
paper presents the detailed design specifications of this platform, as well as initial test results in establishing
the benchmark challenge of current direct transfer times without any specialised support. An initial costing of
CDN usage is also presented, which indicates that significant performance improvement may be achievable
at a reasonable cost to research budgets.
1 INTRODUCTION
A common feature of modern clinical and
bioinformatics research is the need to co-locate large
volumes of data, and the high processing power
necessary to analyse it over a feasible timescale. With
the increased interest in machine learning (ML) since
the early 2010’s, and a commensurate surge in “Big
Data” applications, what was once considered a
diminishing issue – Moore’s Law dictating the ever-
increasing availability of memory – has now taken on
new relevance. The scale of ML requirements has, in
Stell, A., Chauhan, V., Amador, S., Beuschlein, F., Favier, J., Gil, D., Greenwood, P., de Krijger, R., Kroiss, M., Ortuno, S., Patocs, A. and Walch, A.
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition.
DOI: 10.5220/0011665100003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 105-116
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
105
turn, also surged, often beyond the capabilities of the
current internet infrastructure, and innovations are
once again required to achieve results over a feasible
timescale.
In this paper, a clinical scientific endeavour is
presented that features exactly such requirements, and
presents an opportunity to consider a novel method to
support the overall issue of “science-at-a-distance”.
The Hypox-PD project is a recently proposed multi-
centre research collaboration that aims to identify
metabolites and morphological features in tissue that
indicate a predisposition to hypoxia. This is primarily
achieved through the analysis of two linked datasets:
mass spectrometry (matrix-assisted laser desorption
ionization, abbreviated as MALDI-MSI) and
morphological image pathology slides, both derived
from biobank tissues. The MSI and morphological
analysis, along with the original tissue samples will
be used as inputs into various machine learning
algorithms, to identify metabolic biomarkers that help
predict a predisposition of hypoxia. This
identification will then be validated against the
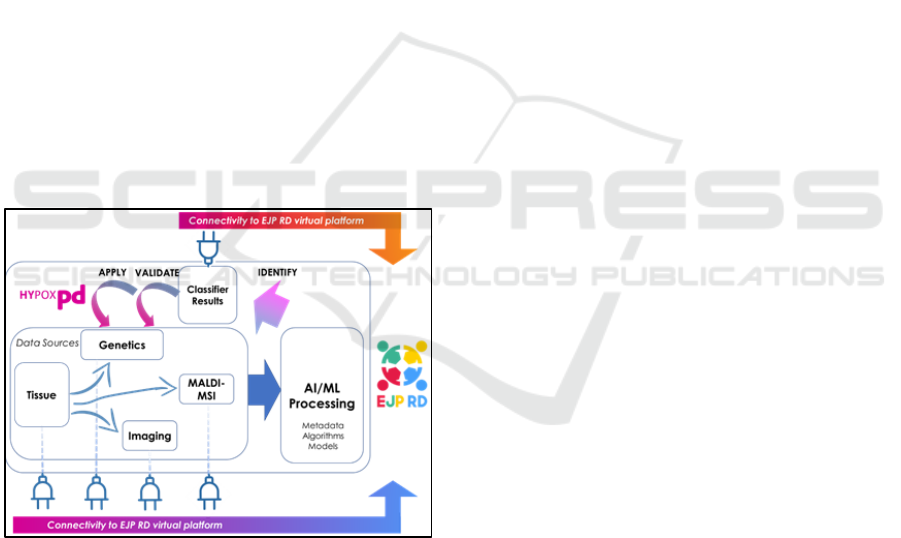
associated genetic information. Figure 1 shows a
high-level abstract overview of the workflow of the
project, along with it’s connection to the (separate)
EJP RD (European Joint Programme – Rare Disease)
“virtual platform” (Kaliyaperumal, 2022).
Figure 1: Data work-flow of Hypox-PD project.
Though the novel platform presented in this paper
is intended to demonstrate a generalised
technological solution to unifying the three tech
components across large distances, it is worth
highlighting the geographically disparate nature of
this particular consortium, to emphasise it’s practical
utility. In this case, Hypox-PD has wide geographical
reach, bringing together research expertise from
Germany, France, Switzerland, Spain, the
Netherlands, Hungary and Australia. Therefore, the
main global regions to consider transport data to and
from are Europe and Asia-Pacific.
Given the scientific requirements of the project,
the key practical issue in terms of technology is the
need to bring together three components: data,
algorithms and processing power. In the specific case
of the Hypox-PD project, the data are largely located
in Utrecht (Netherlands – pathology slides) and
Munich (Germany – MALDI-MSI), the machine
learning algorithms are located in Alicante (Spain),
and the processing power is located in Melbourne
(Australia).
As this is a relatively typical “science-at-a-
distance” problem, the actual locations of the
components are not necessarily important. The point
to note is that they are geographically disparate
enough to make the transport of data, analysis and
results to each of the partner nodes – whilst
maintaining security, accessibility and availability –
a non-trivial challenge.
In the commercial domain (as opposed to
research), a well-established technique in performing
large-scale data transportation is to use a content
delivery network (CDN). Using a network like this,
data are uploaded to a local gateway, then is mirrored
to various sites around the world, connected to the
same network. The aim of this network is to provide
copies of the data, close to the users of that data,
wherever they may be located in the world. This
mirroring occurs as a background task, across large-
bandwidth channels (indexed to cost) and is separate
from individual transfer requests, making it an
effective asynchronous method of transport. In other
words, a CDN is – in effect – a network of cache
proxies that aim to speed up the delivery of web assets
to its intended audience (Content Delivery Network,
2022).
The second component to the novel mechanism
proposed in this paper, is the automated transport of
algorithms and libraries to other points within the
consortium, with as little configuration requirement
as possible. Building on the work done by the Global
Alliance for Genomics Health (GA4GH) (Rehm,
2021), the main idea is to wrap the necessary
analytical tools within an orchestration mechanism –
in this case Docker – so if a scientist with a local store
of high-volume data, also has access to the necessary
infrastructure power, then they can co-locate the data
and processing locally (described in summary by
GA4GH as “taking code to the data”).
Finally, the last component is the access to high-
performance hardware upon which to run the data and
algorithms. Again, the provision of this hardware in
general could be located anywhere. However as the
HEALTHINF 2023 - 16th International Conference on Health Informatics
106

Hypox-PD partner with this provision is located in
Melbourne, Australia, this allows an edge-case of
inter-region data transfer to be explored (specifically
between Europe and Asia-Pacific). To that end, a
cost-benefit analysis of different CDN providers will
allow an evaluation of whether the use of CDNs in
general is now within the reach of research networks.
As will be outlined in the architecture section of
this paper, it is this general combination of a CDN,
orchestration software, and hardware processing
power that allows research partners to shorten the
time required to share data (input and results),
algorithms and processing, to a feasible timescale for
repeatable validation of scientific outcomes.
The rest of this paper is structured as follows: a
discussion of the background literature that has led to
this proposition; detail of the architecture and
implementation design of the platform (including
security, robustness, and FAIR-compliance); some
initial prototype tests and results (remote data transfer
times and CDN costs); and finally a discussion about
the viability of the platform in general.
2 BACKGROUND LITERATURE
A literature review was performed to explore the
background landscape of combining high-volume
data, analysis tools and processing power in
bioinformatics research. The aim was to highlight the
core issues in the area, to ascertain the latest state-of-
the-art solutions, and to evaluate whether these would
support the requirements of this project.
The overall strategy of this review was to ask the
following research question:
“What is the state of the art in technology solutions
to support bioinformatics and clinical research,
where high-volume data and complex analysis
require geographical co-location?”
The eligibility criteria were:
• Articles published in English between 2010 and
2022 (2010 being identified as the more recent
advent of ML research)
• Articles which had a clinical or bioinformatics
application
The exclusion criteria were:
• Poster abstracts
• Articles that exclusively used commercial
closed-source implementations
The following terms and key-words were searched:
• "ga4gh workflow execution service"
• "content delivery network bioinformatics" +
“cdn” + “cost” + “2022”
• "machine learning data network platform
bioinformatics" + “2022” + “volume” +
“transporting”
This set of keywords were searched once on the 10
th
September 2022, then repeated again on the 17
th
October 2022. All papers referenced appeared in the
first search, and were confirmed in the second. No
new papers were found in the second search.
The main sources of academic output were:
• Google Scholar
• IEEE Xplore Digital Library
• Science Direct
• Pubmed
Throughout the search, various themes repeatedly
appeared, which concerned the accuracy, consistency
and provenance of platforms that could be re-used to
perform high-volume, high-processing requirements.
Spanning the decade from 2010 to 2020, it appeared
that the “Big Data” phase of internet evolution had
begun by identifying the possibilities of micro web-
services to provide re-usable, pooled knowledge and
information, to support research into bioinformatics
(Weizhong, 2015) (Hubbard, 2002). However, as the
maintenance requirements of such services became
unsustainable (Kern, 2020), in common with other
features of the internet, such as search engines and
social media, by the close of the decade, the provision
of these services had moved towards a pattern of
centralised uniform platforms (Sheffield, 2022). This
has some relatively large and negative consequences
for open, shared science, where reproducibility and
transparency becomes increasingly limited, due to the
“walled garden” nature of many service providers.
The idea of a data-sharing network to support
research clinical and bioinformatic science, appears
to have been posited in different forms through this
period (Parker, 2010), but has often been hampered
by the raw cost of data transport required to achieve
the ends of the bioinformatics initiatives. (Parker,
2010) is representative of the cost-benefit
considerations of high-volume bioinformatics
projects circa 2010. However, as technology
efficiencies increased with time, a paper written in
2016 (Williams, 2017), expressed a vision of a
“Research CDN” as a viable concept, using public
university resources (in the United States in this
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition
107
example)
1
. In 2016, this was still expressed as a vision
statement only (i.e. it didn’t exist) and as far as can be
found in 2022, has still yet to be realised as a general
implementation.
A large initiative that appeared to be most active
until around 2017, was the Global Alliance for
Genomics Health (GA4GH). It deserves special
attention for the focus on repeatability in large-scale
scientific work-flows, using a robust design (Rehm,
2021). Two features are particularly salient to the
area discussed in this paper: the application usage of
their workflow execution service (WES) (Suetake,
2022), and the development of the “Dockstore”
(O’Conner, 2017) (Yuen, 2021).
The WES allows the re-use of a work-flow engine
to provide a repeatable pipeline of analysis, thus
allowing the output of large-scale analytics to be
compared with others that have followed a known,
and proveably repeatable, method. Some flexibility in
parameter specification is allowed by the method of
invocation: a schema description written in YAML
(“Yet Another Markup Language”), allowing the tool
to provide an avenue for generalised specification.
The “Dockstore” itself, connects to these engines
by allowing the components (implemented in
Docker) to be constructed, again providing a balance
between flexibility and constraint, in a process that
previously would be almost completely
unconstrained.
Whilst no direct reporting of the practical utility
and adoption of these tools could be found, the overall
principle of this part of the GA4GH initiative was that
of “taking code to the data”. The underpinning idea
is that the often complex libraries and analytical tools
are independent entities from both the data and the
processing power, which goes a long way to
providing truly generalised tools that allow complex
machine learning work-flows to be executed
remotely, repeatedly.
However, without specifying hardware
processing requirements in detail, it does still leave
the underlying logistical issues of hardware power
and large-scale data transfer to be solved. Similarly,
though a general idea of using CDNs to solve the data
transfer problem have been mooted in literature, the
barrier – either perceived or actual – has almost
always been one of cost. Given the progress of
technology efficiency over the period studied, it is
entirely possible that the cost-efficiency is now within
reach of a “reasonable” research budget, but this idea
1
This author (A.S.) speculates that this is possibly an echo of the
development of the ARPAnet in the late 1960s which became the
precursor to the modern-day Internet.
has yet to be challenged or implemented directly “in-
situ”.
This cost consideration and the separation of
concerns that the GA4GH initiative puts forward,
does lead somewhat naturally to the question of how
to combine those three features – data, analysis,
hardware – in a generalised platform (as presented in
this paper). And it appears to be a solution that does
not readily exist in any current or accessible form.
Therefore this is what informs the approach that is
being taken for the Hypox-PD project. The solution
proposed in this paper attempts to combine three
aspects in a form of transient virtual network: a CDN,
orchestration software to transmit ML libraries,
supported by hardware acceleration.
3 ARCHITECTURE
The overall goal of this construction is to bring
together data, algorithms and processing power, as
seamlessly and as usefully as possible for the goals of
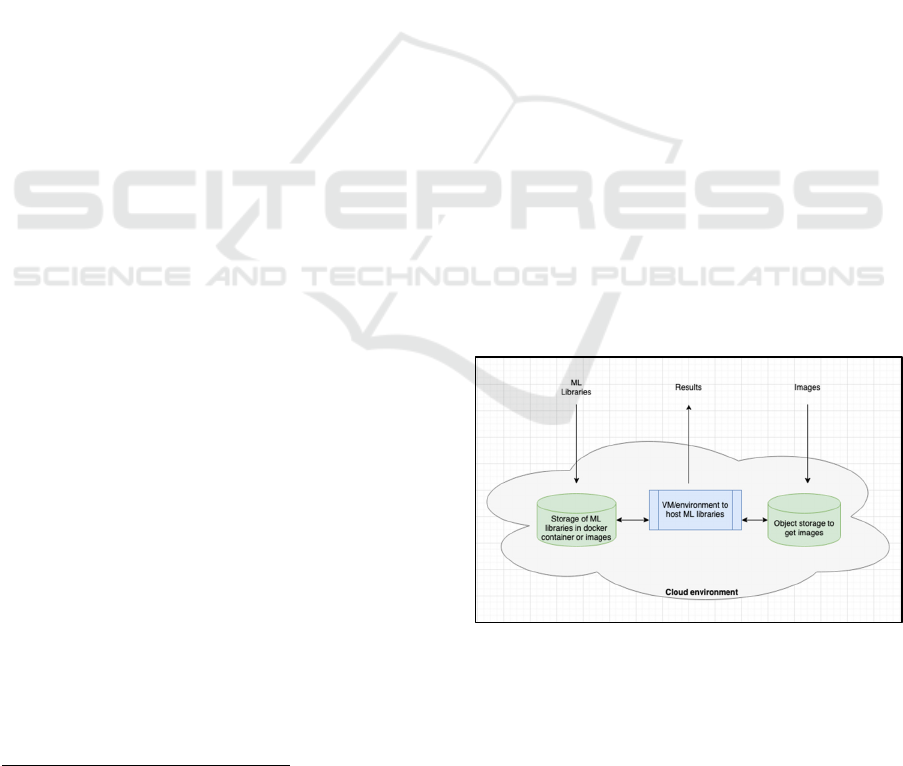
a clinical and bio-informatics research project. Figure
1 (above) shows the specific work-flow
considerations for the Hypox-PD project. Figure 2
(below) shows a high-level abstract entity – a
platform – that we wish to build so that the workflow
can operate. This could be considered a private cloud
where high-volume data (e.g. images) and complex
libraries are input, classifier results are output, and the
complexity of distributed management and high
processing power is largely hidden from the key
stakeholders, at the entry and exit points of this cloud.
Figure 2: High-level abstract diagram of the private cloud
to be constructed.
However, beneath this abstraction is a more closely
specified virtual network, shown in figure 3. Here, the
HEALTHINF 2023 - 16th International Conference on Health Informatics
108
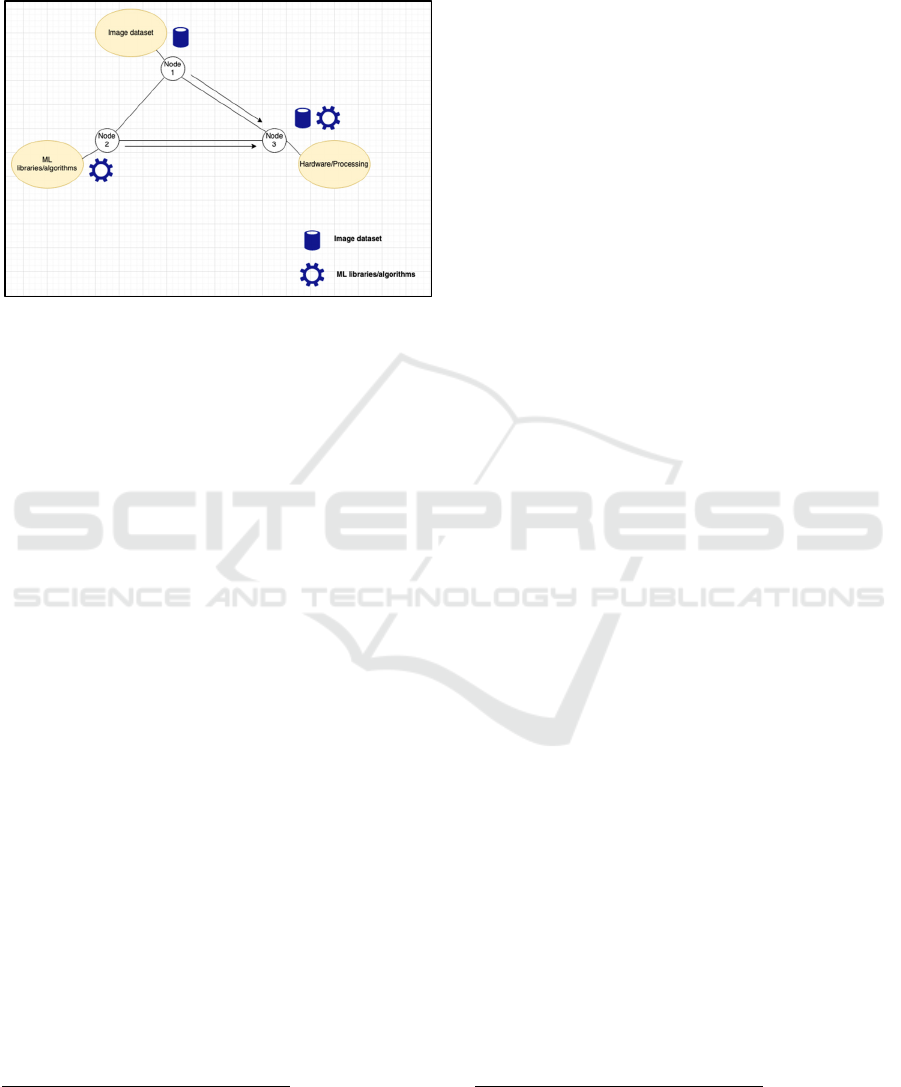
diagram shows three nodes, one relating to each
feature. In practice, the number of nodes is unbound
– and indeed, that is the case with eight partner nodes
contributing to the Hypox-PD project.
Figure 3: Virtual network detail supporting nodes within the
private cloud.
The perimeter of this virtual network could be
defined by the different components available, such
as participation in the CDN, or the transmission of the
algorithms through the Docker “containerisation”
service (building on the GA4GH workflow execution
service). However, the preferred perimeter definition
is the to use the interfaces that provide the data
sharing capability (an application service that
provides data and meta-data according to the FAIR
criteria – Findable Accessible Interoperable Reusable
– www.go-fair.org). This service would take the form
of an API that serves the contributory project data and
meta-data using a shared and accessible standard,
JSON-LD (Linked Data) – this is described in more
detail in section 4.2.1.
Though in a generalised solution the nodes would
be interchangeable, for the purposes of illustration,
the three nodes in figure 3 are designated as follows:
node 1 provides data, node 2 provides algorithms,
node 3 provides the processing.
The two arrows in figure 3 indicate what can be
considered the inputs in data workflow of data (node
1) and complex ML libraries (node 2). This would
then be processed by node 3 and returned to nodes 1
and 2 in the form of results of the processing.
For more accurate definitions, the inputs and
outputs need to be defined as "external" or "internal".
For instance, an input external to the CDN would be
data (e.g. image files) or ML libraries uploaded into
the CDN at nodes 1 and 2. An internal input would be
2
In the jargon of, for instance, the Google Cloud CDN, the inputs
and outputs internal to the CDN are known as "intra/inter-region
cache transfer", while the external inputs are known as "ingress"
the transfer of these - once on the CDN - to node 3 for
processing. Similarly, an internal output would be the
transmission of results from node 3 to nodes 2 and 1.
An external output would be the downloading of
these results from nodes 1 and 2 to their local users
2
The application software that brings all this
together would have a variety of features: it would
interact primarily with the CDN for transmission
around the virtual network; it would be designed as a
general-purpose service so that nodes can
interchangeably offer to receive data or algorithms as
inputs into the CDN and receive results from the
processing at other nodes. In this way the virtual
network itself would have many of the features
common to a typical cloud service.
The network perimeter providing “FAIRified”
data and meta-data would also be provided as a
service, implementing an on-demand translation
wrapper of the contained project data and meta-data
in the JSON-LD standardised format.
Finally, the application will perform the necessary
functions of mapping structured data (e.g. biobank
tissue information), linking this with other aspects,
such as clinical records, and providing the necessary
security in terms of data encryption and signatures.
The details of these aspects will now be described in
the implementation section.
4 IMPLEMENTATION
The implementation detail of this architecture is
broadly divided into primary and secondary
components. The primary ones are those that form the
main features considered in this paper: the CDN,
orchestration and hardware. The secondary ones
include those that indirectly support the primary
function of the platform including data, security, and
the external data-sharing ability (“FAIRification”).
Figure 4 shows another network diagram, at a still
lower level than figures 1-3, with an outline of the
logical network components, their various internal
and external interfaces, and data formats specific to
the Hypox-PD project.
A full explanation of the network diagram is as
follows:
• Tissue, mass spectrometry and imaging data
enters the application from their external sources
(left of diagram)
and external outputs are known as "egress". Note that the costs
associated with CDNs mainly revolve around the actions of
cache transfer and egress.
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition
109
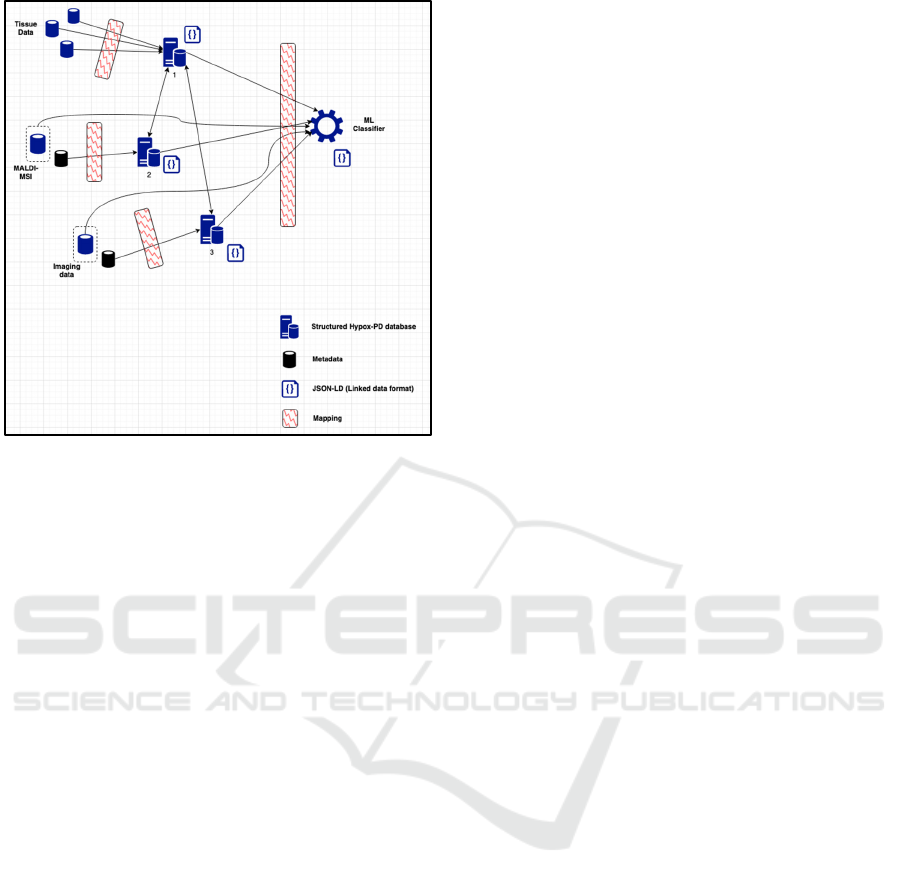
Figure 4: Lower-level logical component network diagram.
• Mass spectrometry and imaging also have
associated meta-data components, which index
the detail of the high-volume data (meta-data
indicated on diagram as smaller databases
outside the dashed line for each source)
• With the exception of the two high-volume
“raw” databases, data from all three of these
sources come through the application mapping
code, and are input into the structured internal
databases (numbered 1, 2 and 3)
• These internal databases are (internally) linked
from database 1 to database 2 and from database
1 to database 3, to specifically connect the data
between MSI/imaging and tissue
• The data from databases 1, 2, 3, and the “raw”
mass-spectrometry/imaging data all goes
towards the ML classifier section (right of
diagram) through another application mapping
layer
• The three internal databases and the ML
classifier section have adjacent APIs that provide
the data to a public external interface in JSON-
LD (Linked Data) format. This is used as the
logical defining boundary of the network.
A key implementation point will be the need to
agree a data dictionary for the internal databases.
Also, specifically for Hypox-PD, clinical and genetic
information would be part of this dataflow (see figure
1), but the inputs detailed in figure 4 directly impact
the primary components of this paper.
4.1 Primary Components
4.1.1 Content Delivery Network (CDN)
The specific function of most CDNs operates as
follows: the interactions with the input I/O layer
provide an upload URI that sits in front of the gateway
to the CDN. Wherever this URI is accessed in the
world, the CDN intercepts the input and redirects to
the nearest local CDN server, thus reducing the
travel-time to upload. These files are then replicated
through the rest of the CDN network. Depending on
the speed available in that CDN network, the initial
throughput time may still be high, but repeat
executions after that initial phase will be much faster.
An initial cost analysis of CDN usage has
performed with the results shown in section 5.3. A
group of CDNs were chosen to cover representation
of coverage, price, commercial vs research audience
targeting, and accessibility. A common feature of the
smaller offerings (e.g. CacheFly, Fastly) was that
costing was done on a bespoke basis. Unfortunately,
through this process, it was found that the small scale
of a research project was not cost-effective for
companies of that small scale.
Therefore, the most immediately available
options were from the larger companies, in this case
AWS CloudFront (Low-latency content delivery
network, 2022) and Google Cloud CDN (Cloud CDN:
content delivery network, 2022). The cost
considerations are three-fold:
• Cache egress – where the data leaves the
network
• Inter-region cache fill – the transfer of data
between different global regions of the network
• HTTP/HTTPS hits – the number of user
downloads of the data (event number rather than
data size)
With approximate size estimates of these three
components, a monthly running cost can be
established as a benchmark. This cost can then be
justified against the overall operational cost of the
research project, and the scientific value gained (often
being fed back into the translation pipeline of
scientific discovery through to clinical application).
4.1.2 Orchestration
The following ML libraries are required for the
Hypox-PD project, interrogating the datasets to best
fit models predicting metabolites that indicate
hypoxia predisposition. They are primarily based in
the Python programming language and include
HEALTHINF 2023 - 16th International Conference on Health Informatics
110

tensorflow, keras, data-augmentation, transfer-
learning for deep-learning, and scikit-learn.
In reference to the way that the Workflow
Execution Service (WES) in GA4GH (Suetake, 2022)
operates, these packages would be incorporated into
a separate programmable workflow including pre-
processing, test and training of models, and iterative
feedback changes. This workflow would be invoked
remotely using a YAML schema descriptor and will
be transferred along with a Dockerfile invocation
(mimicking the “taking code to the data” method of
GA4GH).
4.1.3 Hardware - Processing
The hardware processing provided in this
implementation is currently a virtual machine,
provisioned from the OpenStack implementation at
the Melbourne Research Cloud (MRC) based at the
University of Melbourne, Australia. The specific
virtual machine runs the Ubuntu 22.04 (Jammy)
operating system, with 64Gb of RAM, 16 vCPUs
with NVIDIA vGPU acceleration, and 50Gb of disk
storage. However, the concept behind the application
design is that this hardware could be easily swapped
for another VM, that would have either higher
specifications for greater performance, or lower for
greater resource usage efficiency.
4.1.4 Hardware - Storage
The raw disk storage within this implementation is
50Gb. The interactive I/O layer is managed by MinIO
(Multi-cloud object storage, 2022), which provides
access to the use of S3 object storage. The use of S3
objects focuses the management of data on purely
efficiency concerns (c.f. relational data management,
such as with SQL). In effect, MinIO manages the
application layer I/O of the stored data to and from
the CDN.
4.2 Secondary Components
4.2.1 Data
Broadly, the data considerations can be split into two
varieties: structured and unstructured. Typically –
though not always – structured will have low-volume
(e.g. clinical data or biobank meta-data), and
unstructured will be high-volume (e.g. imaging).
In the case of Hypox-PD, the structured data
needs to be mapped from the different sources and
stored in internal databases (identified as databases 1,
2 and 3 in figure 4). This mapping will follow a data
dictionary as agreed by the project partners.
This unstructured data is one of the primary
motivations for the technical solution proposed in this
paper. The main types that require transfer are:
• Pathology slides: an estimated size of these files
is approximately 1.5Gb per file. With a full
sample set, this would be approximately x20 per
sample. As the number of estimated samples in
the project is ~400, there is an anticipated upper
limit of 12Tb total data size.
• MALDI-MSI: these files include .imzML
metadata (several Kb in size) and .ibd raw binary
files (approximately 1-2 Gb each). Similar to the
image files, the same multipliers can be applied
(1 per sample, with 400 samples) and an upper
limit of the order of 10Tb total data size can be
anticipated.
Typically, for the pathology slides, the formats
available (e.g. .ndpi, or .svs) can be deconstructed to
.tiff formats, with the different zoom layers extracted.
This is a development that would be part of the
analytical processing performed in concert with the
ML libraries (e.g. pre-process data cleaning).
Therefore, though removing different zoom layers
could help in reducing the file volume size, it is likely
that the ML processing would require all layers, so
the capacity to transmit and store all of the associated
data would still need to be available.
Finally, as already noted, there are also clinical,
biobank tissue and genetic data to be considered and
combined into the overall project architecture.
4.2.2 Security
A key point to the above considerations is that even
though the CDN is a secure network, with a dedicated
channel for the Hypox-PD project, it still uses
infrastructure based in the open Internet, and relies on
commercial providers (e.g. Google or AWS).
Therefore, given the potentially sensitive nature of
the data involved, it would be prudent to assume at
least two layers of encryption to establish the security
and integrity of the data between the nodes.
First is the obvious pipeline encryption of the
CDN, which would use the common TLS/SSL
standards to ensure that the point-to-point
communication between nodes is secure. This comes
as standard with any CDN.
Second, would be a more involved aspect that
would need to be written as part of the distributed
application: actual encryption of the individual data
files before ingress and decryption after egress to the
network, using a AES256 cipher (at minimum). This
would likely increase the size of each data file and
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition
111

would possibly then necessitate some form of
compression technology to be invoked as well. Some
benchmarking tests have been run to get an initial
sense of this consideration, presented in section 5.4.
4.2.3 FAIR Criteria
An increasingly popular feature of research initiatives
is the desire to publish data and meta-data for long-
running research projects, so that open science can
support the entire research community, and work can
be made genuinely and openly repeatable. One of the
primary initiatives to encourage this, is the FAIR
project (www.go-fair.org), that looks to make all
research data Findable, Accessible, Interoperable and
Reusable. The Hypox-PD project aims to adhere to
this initiative, and will implement the following
features to achieve FAIR compliance:
• Render as much data and meta-data as possible
in JSON-LD (presented through a REST API)
• Provide provenance, licensing and context
metadata (e.g. how the data was generated,
authorship listing, and associated type of license)
• Provide universal identifiers where appropriate
(e.g. DOI numbers)
In terms of the mechanics of this interface, the
channels of data sourcing, mapping and onward
transmission to the ML analysis, will be “intercepted”
by presenting an open query interface to the internal
database. The endpoint results will be rendered as
JSON-LD (www.json-ld.org) based on parameterised
(but comprehensive) queries of that internal database.
It is also proposed that this endpoint will provide
rudimentary quality metrics (e.g. the overall coverage
of a key data point, such as a universal identifier),
which will also help external parties understand the
quality of the returned data more deeply. Any
sensitive data – either from an intellectual property
protection or clinical subject point of view – will be
restricted.
Figure 5: Simple example of JSON-LD format (parameters
indicated with “@” are linkable across other domains
similar to the operation of Semantic Web constructs).
5 FEASIBILITY TEST RESULTS
To explore and understand both scale of the challenge
and the feasibility of the architecture of this proposal,
the following four initial tests have been run:
1) Large-volume file transfer between local
(intra-region) systems
2) Large-volume file transfer between remote
(inter-region) systems
3) A costing estimate of the CDN usage
4) Analysis of time and size of encryption of
large-volume files
5.1 Large-Volume Local File Transfer
Using representative pathology image files, four
operations were run:
1) Remote to local system download
2) Local to remote system upload
3) Introduction of application-level
management software (OpenKM)
4) Introduction of application-level
management software, focused on S3
storage (MinIO)
The two representative files were one .svs file (an
Aperio TIFF file) and one .ndpi file (Hamamatsu
TIFF file). The .svs file was 537.2 Mb in size, and the
.ndpi file was 1.7 Gb in size. The protocol used for
transfer was SFTP (Secure File Transfer Protocol)
and the transfer was run at 11.30pm in the evening
(AEDT time-zone) between a 70 Mb/s wifi
connection download and 15 Mb/s upload (Australian
National Broadband Network) and a resource at the
University of Melbourne (210 Mb/s Ethernet
download, 110 Mb/s upload). Tables 1 to 6 show the
results of these tests (with tables 3 and 4 transferred
in-machine).
Table 1: Remote to local (download).
File
format
Time
taken
Avg transfer speed
.svs 1m 50s
15ms
4.8 MB/s
.ndpi 5m 18s
45ms
5.4 MB/s
Table 2: Local to remote (upload).
File
format
Time
taken
Avg transfer speed
.svs 4m 20s 2.1 MB/s
.ndpi 15m 4s 1.9 MB/s
HEALTHINF 2023 - 16th International Conference on Health Informatics
112
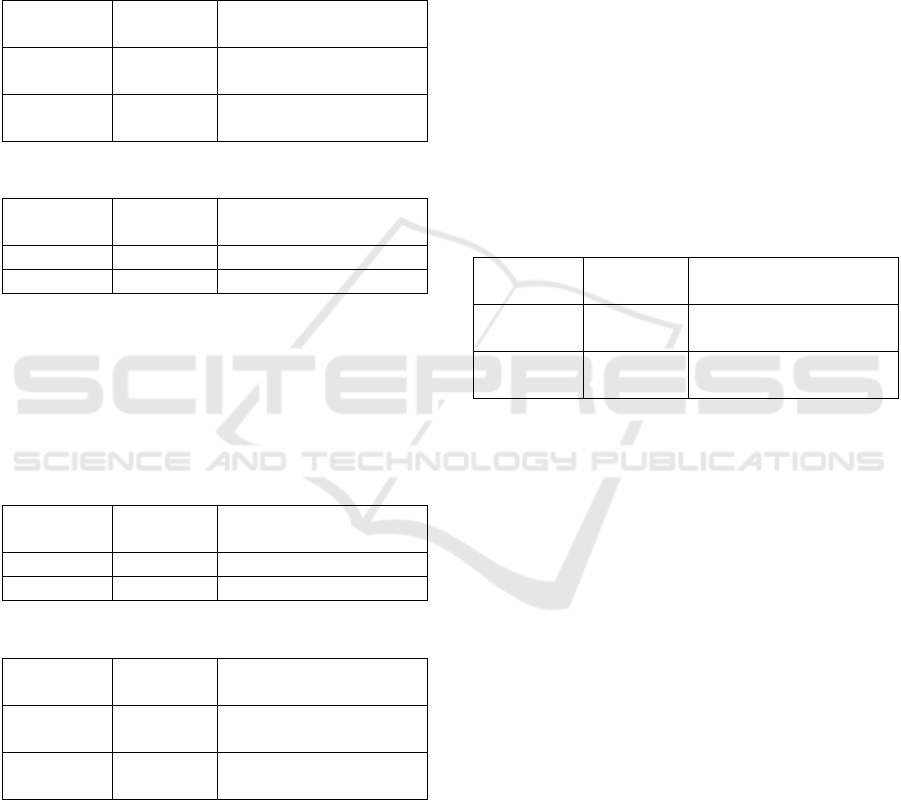
Tables 1 and 2 were inverse operations. At first, the
expectation was that the times would be
approximately equal. However, the skewed times and
speeds appear to be a result of the large differential in
upload speed between the commercial NBN endpoint
and dedicated fibre-optic channel at the University.
These results serve as a benchmark for the intra-
region testing.
Table 3: Local to local transfer (using OpenKM).
File
format
Time
taken
Avg transfer speed
.svs 0m 21s
82ms
25.5 MB/s
.ndpi 1m 21s
01ms
21.0 MB/s
Table 4: Local to local transfer (using MinIO).
File
format
Time
taken
Avg transfer speed
.svs 4s 86ms 131.4 MB/s
.ndpi 14s 48ms 120.7 MB/s
The tests outlined in tables 3 and 4 indicate local-to-
local testing i.e. conducted in a single machine
between segregated applications (native file explorer
to the download management application). At first
exploratory, it was observed that the introduction of
MinIO greatly increased the transfer speed.
Table 5: Local to remote transfer (upload – using MinIO).
File
format
Time
taken
Avg transfer speed
.svs 4m 20s 2.1 MB/s
.ndpi 14m 14s 2.0 MB/s
Table 6: Remote to local transfer (download – using MinIO).
File
format
Time
taken
Avg transfer speed
.svs 1m 11s
40ms
7.6 MB/s
.ndpi 4m 9s
75ms
6.8 MB/s
Tables 5 and 6 repeated the upload/download
between local and remote servers, but with MinIO
managing the interaction at both sides (both client and
server). Local to remote upload appeared unaffected,
but an increase in transfer speed on remote to local
was observed (approx. 25-50%).
Though tests run in such situations are subject to
the varying environment of network conditions, and a
clear topology of the route would need to be
established (e.g. using tools such as WireShark or
traceroute) an approximate benchmark was
established and the use of MinIO to manage the
application transfer was clearly indicated.
5.2 Large-Volume Remote File
Transfer
A test for both files between the Europe and Asia-
Pacific regions (specifically between the UK and
Australia), was conducted to understand current
travel time estimates, without CDN usage.
The test was conducted on the same 210 Mb/s
ethernet connection from the University of
Melbourne, Australia at 3pm AEDT (5am British
summertime), but with unknown connection speed at
the UK endpoint (limited access to remote VM). The
results are shown in table 7.
Table 7: Local to remote transfer (Asia-Pacific to Europe).
File
format
Time
taken
Avg transfer speed
.svs 2 hrs
40mins
0.193 MB/s
.ndpi 4 hrs 47
mins
0.199 MB/s
Though the speed of the Internet connection at the UK
endpoint was unknown, the resulting travel times
were not unexpected. The average transfer speed
reduced by several orders of magnitude and the time
taken correspondingly increased beyond the realms of
feasibility for short-scale feedback (~of the order of
hours). Further testing to establish exact parameters
would be required, but this initial test does confirm
the expected result that high-volume transfer between
global regions, without support, is a significant
barrier to collaborative work.
5.3 Cost Estimate of CDN Usage
An approximate cost calculation was performed on
the usage of two large-scale commercial CDNs: AWS
CloudFront (Low-latency content delivery network,
2022) and Google CDN (Cloud CDN: content
delivery network, 2022).
The calculation used the following figures: a
typical image file was considered to be 1.7 Gb in raw
size. If there were approximately 300 samples for the
Hypox-PD project, this would be 1020 Gb in total
(300 * 1.7 * 2), which – according to the proposed
architecture on one run - would egress from the
network twice and would transmit from one region to
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition
113
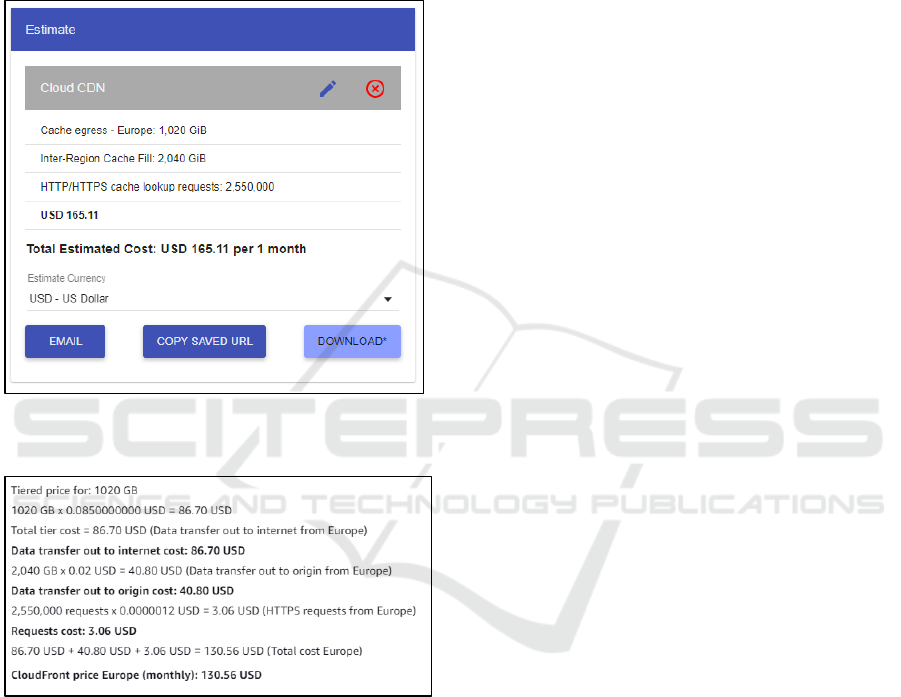
the other four times (Europe to Asia-Pacific and back,
twice round). An approximate rule of thumb for the
count of HTTPS hits is to multiply the overall egress
size by 5000, resulting in 2.55 million hits. All of this
as a monthly running cost is US$165.11 with Google
and US$130.56 with AWS. Figures 6 and 7 show the
detail of this calculation using each provider’s
calculator.
Figure 6: CDN usage estimate calculation from Google
Cloud CDN.
Figure 7: CDN usage estimate calculation from Amazon
CloudFront CDN.
Whilst these calculations likely need further
refinement, it appears that since the cost
considerations outlined in the background literature
were expressed, the increase of the efficiency of
technology, has possibly brought the use of CDNs to
support large-scale research bioinformatics within
reach.
5.4 File Encryption Time/Size
A local test of the effects of encryption on the large-
volume files was also conducted. The main aims were
to look at how quickly the files were encrypted and
what the resulting file size was. For the .svs file
(537.2 Mb), time to encryption was 4s 30ms; for the
.ndpi file (1.7 Gb), time to encryption was 9s 87ms.
The size of the files were unaffected (a minor change
in single byte counts). The cipher used was AES, as
provided in the Python pycryptodomex library, and
the local hardware supporting this was an Apple M10
Pro processor (8 core) with 16Gb RAM.
From this test, it is a reasonable assumption that
the encryption part of the proposed application will
be required before entry and after the exit points of
the network but will not appreciably affect either file
size or time to encrypt.
6 DISCUSSION
The results from section 5 establish an initial
benchmark for the various barriers that currently exist
to the feasibility of a remote multi-center research
collaboration. They show typical transfer times of the
files involved locally (intra-region – Asia-Pacific)
and remotely (inter-region – Asia-Pacific to Europe).
These produce results that are not unexpected, such
as inter-region transfer speeds dropping to ~10
-1
Mb/s
even when supported by large endpoint connections.
However, an important next step would be a full
forensic examination of the entire network paths
involved using tools such as WireShark (WireShark,
2022) or the network diagnostic command traceroute.
The results also indicate the effects of secure AES
encryption on the files in terms of resulting size and
the time taken to encrypt on a typical workstation
setup, as well as a costing of the usage of a CDN in
the context of the proposal architecture.
Overall, the indication that the proposed
architecture is a feasible one is positive: the fixed
processing time costs (e.g. encryption) are
“reasonable”. The file transfer time inter-region is
obviously the primary challenge to overcome, but the
financial cost of using of a single aspect of a
commercial CDN (c.f. using Google’s entire GPU
processing service) appears to be within reach for a
typical research project budget. Based on the
background literature review, this appears to be a
significant step forward in the progress of performing
remote bioinformatics research projects.
Though the proposal attempts to present a general
consideration, the fact that partners between Europe
and Australasia are co-ordinating in this project does
provide significant insight. This relationship means
that an extreme travel distance can be evaluated to
“stress test” the proposed platform. However, it also
HEALTHINF 2023 - 16th International Conference on Health Informatics
114

is likely to be un-representative of most research
collaborations. Though the funding bodies, such as
the EJP RD (European Joint Programme – Rare
Diseases) encourage collaborations at distances such
as these, it may still be the case that local relationships
still predominate precisely because of the logistical
challenges involved in transferring data, processing
libraries and hardware over such large distances
(though the same challenges no doubt exist intra-
region too, for instance between individual European
countries, with national infrastructures at varying
stages of development).
This is also a consideration when it comes to the
regional jurisdictions in terms of data sharing laws.
Whilst the security of the data in transit has been
addressed in this proposal, there must be agreement
at a legal level of the usage and privacy laws at each
endpoint of the network. Again, this is a technical
proposal that attempts to be general in scope – but this
is a specific consideration that would always be
relevant in a network like the one proposed here.
Equivalence with the European GDPR legislation is
generally considered the gold-standard in this regard,
and Australia is amongst the various developed
nations that is pursuing this equivalence nationally
(Review of the Privacy Act, 2020).
In terms of generalised re-usability, most – but not
all – aspects are covered in this proposal. It builds
upon the idea presented by GA4GH of a generalised
ML workflow, made accessible by specifying Docker
execution scripts using the YAML specification. The
other prominent feature of sharing and repeatability is
the presentation of all internal data and meta-data in
JSON-LD interfaces, to make the data accessible
according to the FAIR principles. These
standardisations are untested, and it may yet be the
case – even once fully implemented – that their
adoption may be limited. Only the test of time and re-
use will prove this.
It is also the case that providing concrete features
such as commercial CDN usage and hardware for
processing are not easy to generalise. The most that
can be provided is the “gateway” schema descriptions
that allow these to be integrated into a project as
easily as possible. However, the apparently
reasonable cost of CDN usage, does appear to be a
significant step change in the mode of operation of
high-volume data research projects, one that appears
to be generally un-reported. There may be unforeseen
barriers or consequences to the use of these
commercial offerings that will only become apparent
as wider scale usage increases. However, this is an
option that will appear to serve the specific needs of
the Hypox-PD project well in the short- to medium-
term and could perhaps be submitted for
consideration as a step towards a general “Research
CDN”.
7 CONCLUSIONS
A novel mechanism has been presented in this paper,
facilitating the exchange of data, algorithms and
processing when developing a multi-centre
clinical/bioinformatics research project. It has the
potential to significantly improve the feasibility of
transfer of data, analysis and results between
geographically disparate partner nodes, but has
potential limitations of budget (with the content
delivery network), complex orchestration and
synchronisation. As the Hypox-PD project
progresses, the development of this infrastructure will
continue to be reported as an outcome of the research,
additional to the clinical and bioinformatics outputs
of metabolite identification.
ACKNOWLEDGEMENTS
The members of the Hypox-PD consortium
acknowledge the support obtained during the
development of the proposal for the European Joint
Program Rare Diseases (EJP-RD) through the
European Advanced Translational Research
Infrastructure in Medicine (EATRIS).
REFERENCES
Suetake H, Tanjo T, Ishii M et al., (2022), Sapporo: A
workflow execution service that encourages the reuse
of workflows in various languages in bioinformatics.
F1000Research, 11:889
O'Connor BD, Yuen D, Chung V, Duncan AG, Liu XK,
Patricia J, Paten B, Stein L, Ferretti V., (2017), The
Dockstore: enabling modular, community-focused
sharing of Docker-based genomics tools and
workflows. F1000Res. 2017 Jan 18;6:52.
Yuen D., et al, (2021), The Dockstore: enhancing a
community platform for sharing reproducible and
accessible computational protocols, Nucleic Acids
Research, Volume 49, Issue W1, 2 July 2021, Pages
W624–W632
Rehm L. H., et al., (2021), GA4GH: International policies
and standards for data sharing across genomic
research and healthcare, Cell Genomics, Volume 1,
Issue 2, 2021, 100029, ISSN 2666-979X
Novel Distributed Informatics Platform to Support Machine Learning Discovery of Metabolic Biomarkers in Hypoxia Predisposition
115

Kern F., et al, (2020) On the lifetime of bioinformatics web
services, Nucleic Acids Research, Volume 48, Issue 22,
16 December 2020, Pages 12523–12533
Parker, A., Bragin, E., Brent, S. et al., (2010), Using
caching and optimization techniques to improve
performance of the Ensembl website. BMC
Bioinformatics 11, 239 (2010)
Williams, J.J. and Teal, T.K. (2017), A vision for
collaborative training infrastructure for
bioinformatics. Ann. N.Y. Acad. Sci., 1387: 54-60
Sheffield, N.C., Bonazzi, V.R., Bourne, P.E. et al., (2022),
From biomedical cloud platforms to microservices:
next steps in FAIR data and analysis. Sci Data 9, 553
(2022)
Weizhong Li, et al., (2015), The EMBL-EBI bioinformatics
web and programmatic tools framework, Nucleic Acids
Research, Volume 43, Issue W1, 1 July 2015, Pages
W580–W584
Hubbard T., et al., (2002), The Ensembl genome database
project, Nucleic Acids Research, Volume 30, Issue 1, 1
January 2002, Pages 38–41
Kaliyaperumal, R., Wilkinson, M.D., Moreno, P.A. et al.,
(2022), Semantic modelling of common data elements
for rare disease registries, and a prototype workflow
for their deployment over registry data. J Biomed
Semant 13, 9 (2022)
“Review of the Privacy Act 1988”, Australian government
white paper (2020) - https://www.ag.gov.au/integrity/
consultations/review-privacy-act-1988 [last accessed -
28th October 2022]
“Content Delivery Network” - https://en.wikipedia.org/
wiki/Content_delivery_network [last accessed - 30th
October 2022]
“Low-latency content delivery network” -
https://aws.amazon.com/cloudfront/ [last accessed –
30th October 2022]
“Cloud CDN: content delivery network” -
https://cloud.google.com/cdn/ [last accessed – 30th
October 2022]
“Multi-cloud object storage” - https://min.io/ [last accessed
– 30th October 2022]
“Wireshark – Go Deep” - https://www.wireshark.org [last
accessed – 30th October 2022]
HEALTHINF 2023 - 16th International Conference on Health Informatics
116
