
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between
Valvular Heart Diseases
Ecem Erin
1
and Beren Semiz
2 a
1
Department of Physics, Bogazici University, Istanbul, Turkey
2
Department of Electrical and Electronics Engineering, Koc University, Istanbul, Turkey
Keywords:
Seismocardiogram, Cardiovascular Health Monitoring, Valvular Heart Disease, Biomedical Signal Processing.
Abstract:
Cardiovascular diseases are one of the top causes of mortality, accounting for a sizeable portion of all fatalities
globally. Among cardiovascular diseases, valvular heart diseases (VHDs) affect greater number of people
and have higher mortality rates. Current VHD assessment methods are cost-inefficient and limited to clinical
settings, therefore there is a compelling need for non-invasive and continuous VHD monitoring systems. In this
work, a novel framework was proposed to distinguish between aortic stenosis (AS), aortic valve regurgitation
(AR), mitral valve stenosis (MS), and mitral valve regurgitation (MR) using tri-axial seismocardiogram (SCG)
signals acquired from the mid-sternum. First, seismology domain knowledge was leveraged and applied to
SCG signals through ObsPy toolbox for pre-processing. From pre-processed signal segments, spectrogram,
wavelet, chromagram, tempogram and zero-crossing-rate features were extracted. Following p-value analysis
and variance thresholding, a multi-label/multi-class classification framework based on gradient boosting trees
was developed to distinguish between AS, AR, MS and MR cases. For all four VHDs, the accuracy, precision,
recall and f1-score values were above 95%, best performing axis being the dorso-ventral direction. Overall,
the results showed that spectral analysis of SCG signals can provide valuable information regarding VHDs
and potentially be used in the design of continuous monitoring systems.
1 INTRODUCTION
Today, diagnosis and treatment titration for any dis-
ease or injury are generally achieved with conven-
tional biomarkers, of which derivation requires fre-
quent hospital visits and expensive laboratory tests
(Meister et al., 2016). This causes an information
gap regarding the physiological changes occurring
between subsequent hospital visits, and it becomes
mandatory to resort to reactive treatment policies as
early diagnosis and intervention are often not possible
(Golubnitschaja et al., 2014). Thus, there is a com-
pelling need for wearable sensor systems and analysis
frameworks to digitize these indicators and to achieve
continuous health monitoring.
The World Health Organization’s 2020 report lists
cardiovascular diseases as one of the top causes of
mortality, accounting for a sizeable portion of all fa-
talities globally (WHO, 2020). Among cardiovascu-
lar diseases, valvular heart diseases (VHDs) affect
greater number of people and have higher mortal-
a
https://orcid.org/0000-0002-7544-5974
ity rates (Go et al., 2013). VHDs emerge from the
impairments of the heart valves, i.e. the pulmonary
valve, the tricuspid valve, the aortic valve, and the mi-
tral valve (Klabunde, 2011; Svensson, 2008). These
impairments can be listed under two main groups:
stenosis and regurgitation, which can be observed in
any of these four valves. In stenosis case, the valvu-
lar orifice narrows down, resulting in insufficient out-
flow of blood; whereas regurgitation corresponds to
the incompetence of the valve in preventing backflow
of blood (Svensson, 2008). Although the VHDs can
be monitored through magnetic resonance imaging,
echocardiography, computed tomography and cardiac
catheterization, these methods are cost inefficient and
limited to clinical settings (Svensson, 2008).
As physiological signals recorded from the hu-
man body directly emerge from the biological mech-
anisms, they can provide clinically useful informa-
tion about the underlying processes and pathological
conditions. Among these signals, seismocardiogram
(SCG) signal is one of the most popular ones lever-
aged in wearable system design. SCG represents the
mechanical activity of the heart and reflects the lo-
212
Erin, E. and Semiz, B.
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between Valvular Heart Diseases.
DOI: 10.5220/0011663900003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 212-219
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

Figure 1: The X, Y, and Z axes were corresponding to lat-
eral, head-to-foot, and dorso-ventral axes, respectively.
cal chest vibrations originating from the contraction
of the heart (Inan et al., 2014). Previous studies have
shown that the SCG signal can be leveraged in the
detection of aortic stenosis (Yang et al., 2019), heart
failure (Inan et al., 2018), and atrial fibrillation (Hur-
nanen et al., 2016), as well as estimating systolic time
intervals (Shandhi et al., 2019), predicting stroke vol-
ume values (Semiz et al., 2020) and assessing respi-
ration phases (Imirzalioglu and Semiz, 2022; Pandia
et al., 2012). To the best of our knowledge, there is no
study in the literature which focuses on distinguishing
between different VHDs using SCG signals.
The SCG signal has indeed have close similari-
ties with the seismograph signal, which represents the
recording of vibrations in the Earth to assess earth-
quake incidences (Lay and Wallace, 1995). As they
both measure vibrations, it can be proposed that the
pre-processing methods used in the analysis of seis-
mograph signal can facilitate the analysis of SCG sig-
nals. Accordingly, first the seismology domain anal-
ysis has been inherited through the ObsPy framework
for pre-processing (Beyreuther et al., 2010). In ad-
dition, recent studies have primarily focused on ana-
lyzing the SCG signals in time domain, i.e. analysis
of the peak- and valley-related features. However, the
frequency domain analysis of SCG signals has been
less studied. Thus, in the presented work, follow-
ing ObsPy analysis, the signals were additionally in-
vestigated in frequency domain through spectrogram,
wavelet, chromagram and tempogram analysis.
The contributions of this study can be listed as fol-
lows: (i) For the first time to the best of our knowl-
edge, a pipeline was presented to distinguish between
aortic stenosis (AS), aortic valve regurgitation (AR),
mitral valve stenosis (MS), and mitral valve regurgi-
tation (MR) using the tri-axial SCG signals collected
from the mid-sternum. In this novel analysis pipeline,
(ii) seismology domain knowledge was inherited and
applied to the SCG signals for pre-processing, and
(iii) spectral-domain analysis was leveraged to in-
vestigate the relationship between frequency domain
characteristics of the SCG signals and VHDs.
Table 1: Dataset Description.
Number of Subjects
Only MS 3
Only MR 13
Only AR 5
Only AS 16
MS and AS 3
AR and AS 1
MR and AR 6
MR and AS 3
MS and MR 1
MS, MR and AS 1
MS, MR and AR 1
MR, AR and AS 1
MS, MR, AR, and AS 2
2 METHODS
2.1 Dataset Description
In this paper, An Open-Access Database for the
Evaluation of Cardio-Mechanical Signals From Pa-
tients With Valvular Heart Disease was used (Yang
et al., 2021). Dataset includes SCG, gyrocardiogram
(GCG) and electrocardiogram (ECG) signals from
100 patients collected at Columbia University Med-
ical Center (USA), Stevens Institute of Technology
(USA), Southeast University (China), Nanjing Med-
ical University (China) and Nanjing Medical Univer-
sity (China). Subjects were diagnosed with various
conditions of valvular heart diseases (VHD), such as
aortic stenosis (AS), aortic valve regurgitation (AR),
mitral valve stenosis (MS), mitral valve regurgita-
tion (MR), and tricuspid valve regurgitation (TR). The
presence and non-presence of each VHD was labeled
with 1 and 0, respectively. It should be noted that
most of the subjects were labeled with more than 1
VHD. Subject demographics were as follows: age =
68 ± 14 years, height = 165 ± 9 cm, weight = 69 ±
13 kg.
To collect ECG, SCG and GCG signals, an off-
the-shelf sensor (Shimmer 3 ECG module, Shimmer
Sensing, United Kingdom) was used. Subjects were
asked to stay motionless in supine position while the
data was being recorded. For the analysis, only the
tri-axial SCG signal was used. As shown in Fig. 1,
the X, Y, and Z directions of the SCG signal were
corresponding to the lateral, head-to-foot, and dorso-
ventral directions, respectively. As there was no sub-
ject having only TR, subjects having TR in addition
to other VHD were excluded. Number of VHD cases
included in the analysis is summarized in Table 1.
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between Valvular Heart Diseases
213
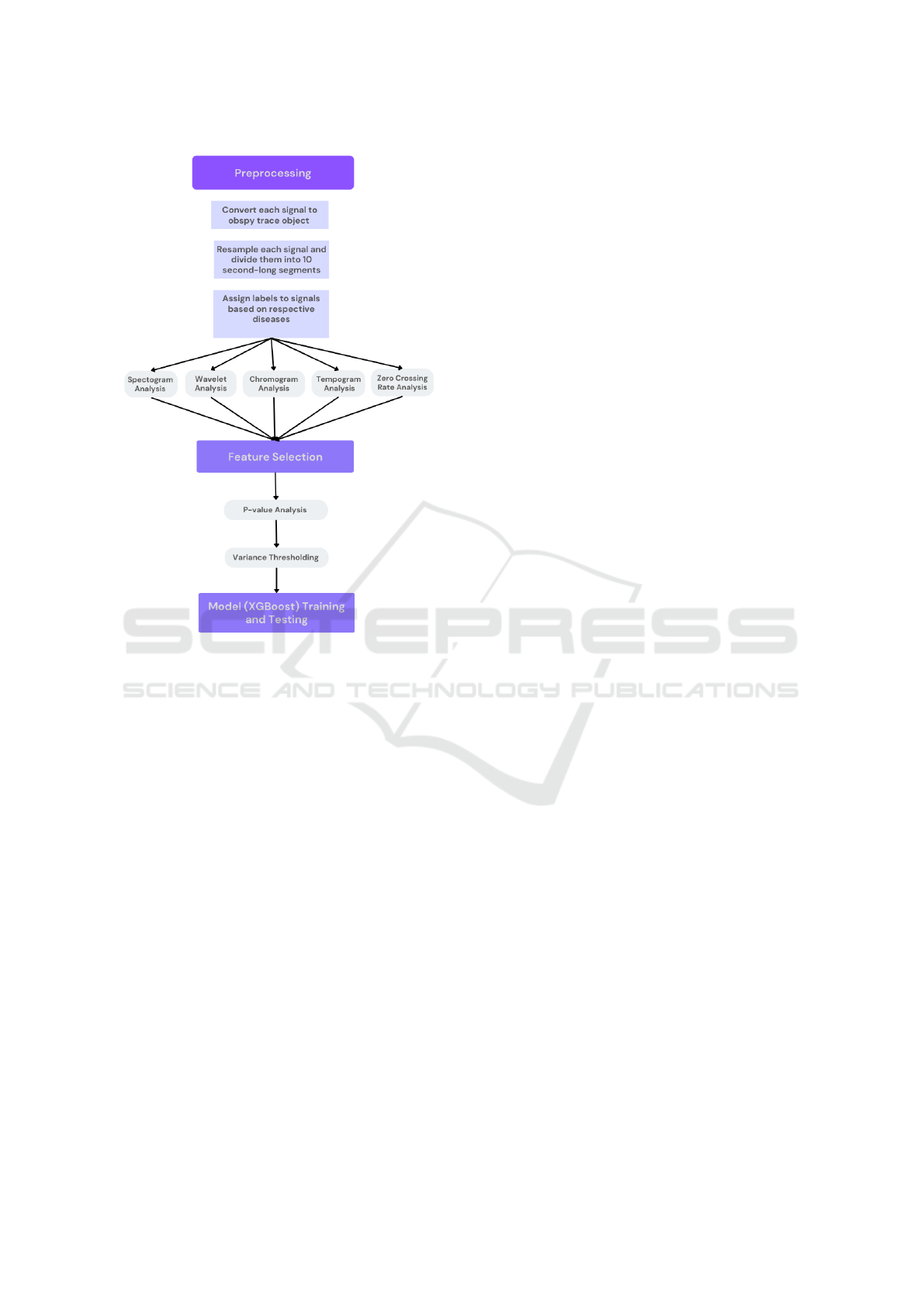
Figure 2: Block diagram for the proposed framework. First,
detrending, tapering, resampling and segmentation were ap-
plied on the signals. Spectrogram, wavelet, chromagram,
tempogram and zero crossing rate features were then ex-
tracted from each segment. Following p-value and vari-
ance thresholding, a multi-label/multi-output classification
model was trained and tested using the remaining features.
2.2 Pre-processing of the SCG Signals
As the seismological data and vibrations on the chest
wall have a similar mechanism, SCG signals show
close resemblance to the seismograph signal. Hence,
biomedical signal processing knowledge was com-
bined with the tools available for seismograph anal-
ysis. To that end, the use of ObsPy toolbox, which
allows manipulation and processing of seismograph
signals (Beyreuther et al., 2010), was leveraged. In-
deed, using ObsPy let the SCG signal characteristics
(such as sampling rate, number of points and dura-
tions) be more easily accessible. More importantly,
employment of manual and heuristic pre-processing
and feature extraction methods could be avoided.
Previous studies have shown that SCG analysis
highly benefits from the features extracted from all
three axes (dorso-ventral, lateral and head-to-foot);
hence, in this study, all three axes were included in
the analysis (Semiz et al., 2020). While transform-
ing these signals into traces, each signal was first de-
trended linearly to remove unwanted baseline oscilla-
tions. Detrended waveforms were then tapered with
“Hann” window having a decimal percentage of 0.05
to prepare them for frequency analysis. In the dataset,
some signals had a sampling rate of 512 Hz while the
sampling rate for others were 256 Hz. As a solution,
all sampling rates were set to 256 Hz using Obspy’s
resampling method. As the final step, each signal was
divided into 10-second long segments to increase the
number of instances. This resulted in a total of 2393
segments (instances).
2.3 Feature Extraction
Following aforementioned pre-processing steps, fea-
ture extraction was performed on the SCG signals. To
that aim, spectrogram, wavelet, chromagram and tem-
pogram features were extracted from each 10-second-
long X, Y, Z segment. Additionally, zero crossing rate
analysis was leveraged as zero crossings were found
correlated with the spectral centroid and dominant
frequencies (Koutroumbas and Theodoridis, 2008).
2.3.1 Spectrogram Analysis
A spectrogram is a two-dimensional function of fre-
quency and time that depicts how a non-stationary
signal’s frequency content changes over time (Mc-
Clellan et al., 2003). Hence, spectrogram analysis
can provide useful information regarding the time-
dependent variances observed in the spectrum.
In this work, spectrogram analysis was employed
using ObsPy’s spectrogram function, with a slight
modification in the output. The original function was
outputting the spectrogram images, i.e., the combi-
nation of coefficients, frequency values and time in-
stances. However, for this study, only the spectro-
gram coefficients were required to be used as features.
That is why, time average of the coefficients for each
frequency band was calculated and stored as a vector.
This was repeated for each of the 10-second-long seg-
ment, i. In the end, a matrix S
i
= (S
x,i
, S
y,i
, S
z,i
) was
generated, which includes time-averaged spectrogram
coefficients for each of the lateral, head-to-foot and
dorso-ventral axes of segment i.
2.3.2 Wavelet Analysis
Physiological signals show time-varying statistics due
to their dynamic nature. As wavelet transform al-
lows for the analysis of non-stationary signals (e.g.
biomedical signals) at multiple scales, it was also
found to be an effective approach for this study. In
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
214
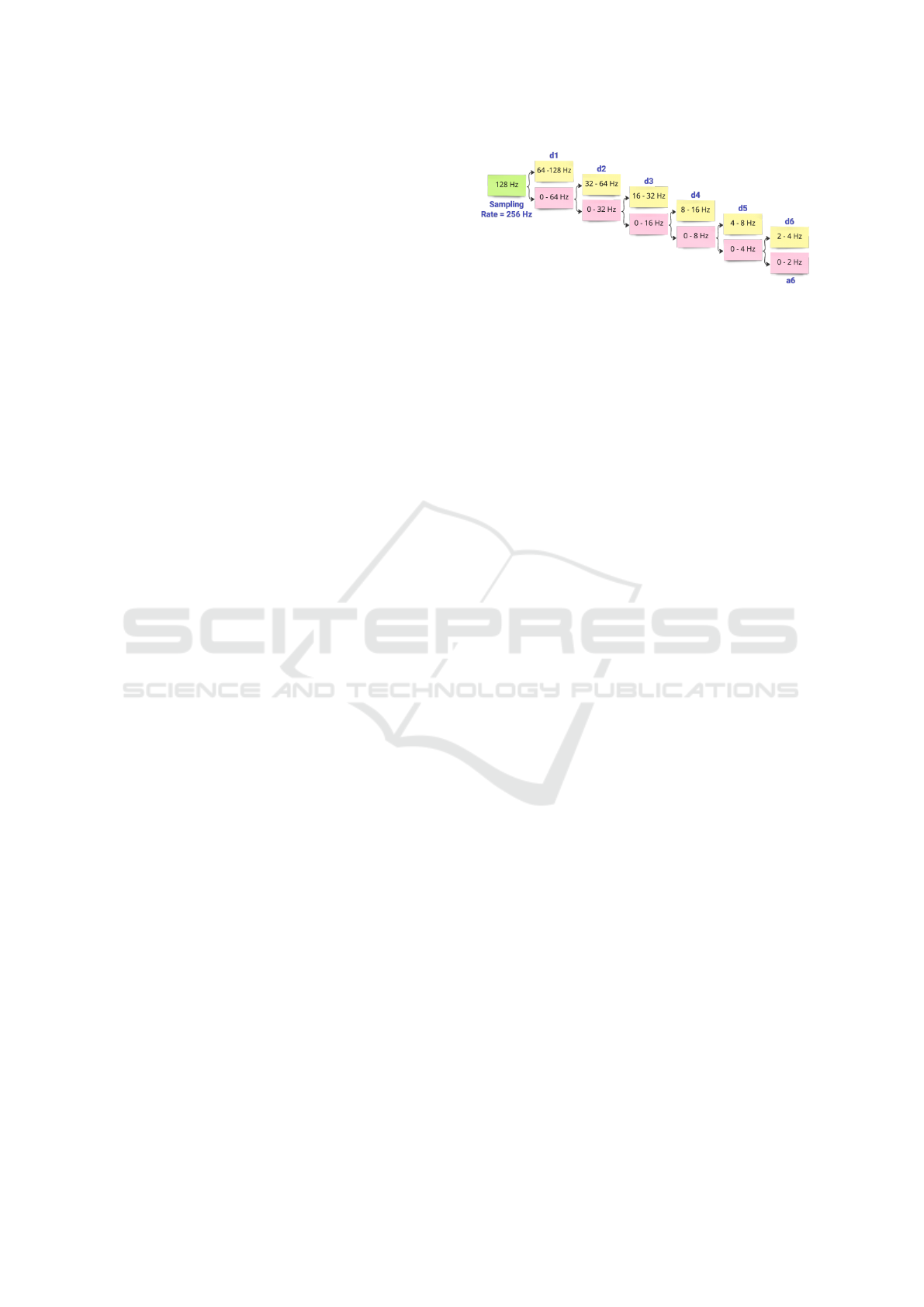
wavelet analysis, the signal is decomposed into a col-
lection of basis functions using a finite-length func-
tion called mother wavelet. Indeed, there are many
different mother wavelets available depending on the
application. The width and central frequency of the
mother wavelet can be adjusted by moving it through
the signal-of-interest. These shifted and scaled ver-
sions of the mother wavelet are also known as daugh-
ter wavelets (Polikar, 1996).
In this work, 1D multilevel discrete wavelet trans-
formation with a level-6 sym5 wavelet was employed.
Having 6 levels ensured that the signal was decom-
posed up until the respiration band (Pandia et al.,
2012). On the other hand, sym5 was chosen as it
shows morphological similarities with the SCG sig-
nals and has been one of the most popular mother
wavelets used in biomedical signal analysis (Jahromi
et al., 2017).
The output was including the approximation co-
efficients (a
6
) and 6-levels of detail coefficients (d
1
,
d
2
, d
3
, d
4
, d
5
and d
6
). Considering that the sam-
pling rate was set to 256 Hz, the maximum frequency
available in the signals was 128 Hz. Thus, the fre-
quency ranges of the detail coefficients were 64-128
Hz, 32-64 Hz, 16-32 Hz, 8-16 Hz, 4-8 Hz, and 2-4
Hz, which correspond to d
1
, d
2
, d
3
, d
4
, d
5
and d
6
co-
efficients, respectively (Fig. 3). On the other hand,
a
6
was representing the 0-2 Hz frequency band. The
detail and approximation coefficients were extracted
for all three axes and concatenated together. In the
end, a matrix W
i
= (W
x,i
, W
y,i
, W
z,i
), was generated,
which includes concatenated approximation and de-
tail coefficients for each of the lateral, head-to-foot
and dorso-ventral axes of segment i.
2.3.3 Chromagram Analysis
In recent years, there have been great advancements in
audio analysis thanks to the semantic analysis (Shah
et al., 2019). Chroma features have found to be pow-
erful representations of any audio as they can be used
to generate audio fingerprints. Chroma features mea-
sure the dominance of the characteristics of a certain
pitch (C, C#, D, D#, E, F, F#, G, G#, A, A# or B)
in signal-of-interest. To compute chroma features,
instantaneous frequency estimates from the spectro-
gram were taken to obtain high-resolution chroma
profiles (Giannakopoulos and Pikrakis, 2014).
Recently, it has been shown that biomedical sig-
nal processing can highly benefit from audio anal-
ysis methods, more specifically from chroma fea-
tures (Hersek et al., 2017). Similarly, in this work,
chroma features corresponding to aforementioned 12
pitch values were extracted from each signal segment.
After extracting pitch values, their time-average was
Figure 3: 6-Level wavelet decomposition. 6-Levels of detail
coefficients: d
1
, d
2
, d
3
, d
4
, d
5
and d
6
. 6
th
Level approxima-
tion coefficient: a
6
.
computed to have one single value for each pitch.
These steps were repeated for all three axes. In the
end, a matrix C
i
= (C
x,i
, C
y,i
, C
z,i
), was generated
which includes time-averaged pitch values for each
of the lateral, head-to-foot and dorso-ventral axes of
segment i.
2.3.4 Tempogram Analysis
As mentioned in Section 2.3.3, it has been shown
that biomedical signal processing can highly benefit
from audio analysis methods (Hersek et al., 2017).
That is why in addition to chromagram features, tem-
pogram analysis was also leveraged in the analysis.
Tempo and beat, which are components of rhythm,
can be used to distinguish audio from each other.
Unlike beat, tempo can vary locally within an au-
dio. Hence using tempogram, information regarding
tempo at each time instance can be obtained.
From each signal segment, tempogram features
were computed using a hop length and sampling rate
of 256 Hz each. Tempogram coefficients were then
averaged within each window. These steps were again
repeated for all three axes. In the end, a matrix T
i
=
(T
x,i
, T
y,i
, T
z,i
), was generated which includes aver-
aged tempogram values for each of the lateral, head-
to-foot and dorso-ventral axes of segment i.
2.3.5 Zero-Crossing Rate Analysis
Zero crossing rate (ZCR) represents the rate of sign-
changes of the signal-of-interest (Giannakopoulos
and Pikrakis, 2014). It is generally used to evaluate
the signal’s noise level, i.e., ZCR values tend to in-
crease as noise levels increases. On the other hand,
ZCR was found to be correlated with the spectral
centroid and dominant frequencies (Koutroumbas and
Theodoridis, 2008). Hence, ZCR value for each seg-
ment was included as a feature in the analysis. For
the 10-second-long segments, ZCR was calculated for
each of the X, Y, Z axes. In the end, a matrix Z
i
= (z
x,i
,
z
y,i
, z
z,i
) was generated where each row refers to one
10-second-long segment, i, and (z
x,i
, z
y,i
, z
z,i
) repre-
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between Valvular Heart Diseases
215

sent the ZCR values of X, Y, Z axes for segment i.
2.3.6 Data Frame Generation
After the aforementioned features were extracted
from each segment, one single data frame was formed
for ease of feature selection and model training. First,
the VHD labels were organized to make the data ready
for multi-label classification. To that aim, correspond-
ing VHD label vectors in the form [AR, AS, MR,
MS] were generated and assigned to each 10-second
long segment. The presence of each VHD was la-
beled with 1 and non-presence was indicated with 0.
For example, let us assume that the subject had AR
and MR, however did not have AS or MS. Then the
corresponding label vector was formed as [1,0,1,0].
Similarly if a subject had MS, bu no other VHDs, the
vector was formed as [0,0,0,1]. These label vectors
were generated for each of the 10-second-long seg-
ments, eventually leading to a matrix L, where each
row represents the label vector for one segment.
Following label generation, previously generated
feature matrices and the label matrix were concate-
nated altogether to have one single frame where each
row corresponds to one 10-second long segment. In
the end, the dataframe had the following structure:
[S
i
, W
i
, C
i
, T
i
, Z
i
, L
i
], where S
i
, W
i
, C
i
, T
i
, Z
i
, are the
spectrogram, wavelet, chromagram, tempogram and
zero crossing rate features, respectively; and L
i
is the
corresponding VHD vector for segment i.
2.4 Feature Selection
Following feature extraction, the data frame was in-
cluding 7497 features in total. However this would
cause curse of dimensionality, i.e., the vector space
would be sparser due to an increase in dimension-
ality (Chen, 2009). This would potentially hurt the
training/learning phase and lead to overfitting; hence,
multi-step feature selection was employed. First,
Mann-Whitney U test with an alpha level of 0.05 was
applied to the generated dataframe. In other words,
any feature having a p-value smaller than 0.05 was
accepted statistically significant. After removing the
features having p > 0.05, variance thresholding was
employed with a threshold of 0.0001. Both methods
are detailed in the following subsections.
2.4.1 p-value Analysis
For p-value-based feature selection, Mann-Whitney
U test with an alpha value of 0.05 was implemented.
Mann-Whitney U test falls under non-parametric hy-
pothesis tests, i.e., it does not make any assumption
regarding the distribution of the data unlike paramet-
ric tests, which assume normal distribution in the
data. Hence, Mann-Whitney U test was determined
as the feature selection method (Sedgwick, 2015).
While performing the test, instead of comparing
the feature variances of the diseases altogether, fea-
tures corresponding to different diseases were com-
pared pair-wise. For example, the spectrogram coeffi-
cients of AR were compared with the spectrogram co-
efficients of AS, MR and MS one by one (e.g. ([S
AR
i
]
vs. [S
AS
i
]), ([S
AR
i
] vs. [S
MR
i
]), ([S
AR
i
] vs. [S
MS
i
]),
etc.). This comparison was repeated for all combina-
tions. During each comparison, any feature column
with p value > 0.05 was eliminated and the remain-
ing columns were used in the analysis. As a result,
total number of columns decreased from 7497 to 763.
2.4.2 Variance Thresholding
Following p-value analysis, variance thresholding
was applied on the remaining 763 columns to elim-
inate the redundant features further. Variance thresh-
olding is one of the filter-based methods and aims to
drop the features having variance values below a pre-
determined threshold (Bommert et al., 2020). Fea-
tures with lower variance indeed carry less informa-
tion, since their variance is proportional to the level
of predictive power they have. Accordingly, the vari-
ance of each feature was calculated and a threshold of
0.0001 was applied. As a result, the total number of
features was decreased from 763 to 308.
2.5 Model Training and Validation
2.5.1 Classification Framework and
Performance Metrics
To establish a multi-label/multi-output classification
framework, label vectors were generated as previ-
ously explained in Section 2.3.6. The framework was
built using One vs. Rest approach. As the classifi-
cation model, Extreme Gradient Boosting Trees (XG-
Boost) was leveraged. XGBoost falls under ensemble
methods, i.e., instead of a single estimator, multiple
estimators are simultaneously used to predict a vari-
able. During training, multiple decision trees are iter-
atively trained, so that residual errors from the previ-
ous iteration can be predicted and improved over time
(Dietterich et al., 2002).
In this work, 80% of the data was used for training
process and 20% was used for testing. During train-
ing, the objective function was determined to be bi-
nary:logistic, whereas other parameters were kept in
their default values. The performance of the model
was assessed using accuracy, precision, recall, and
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
216
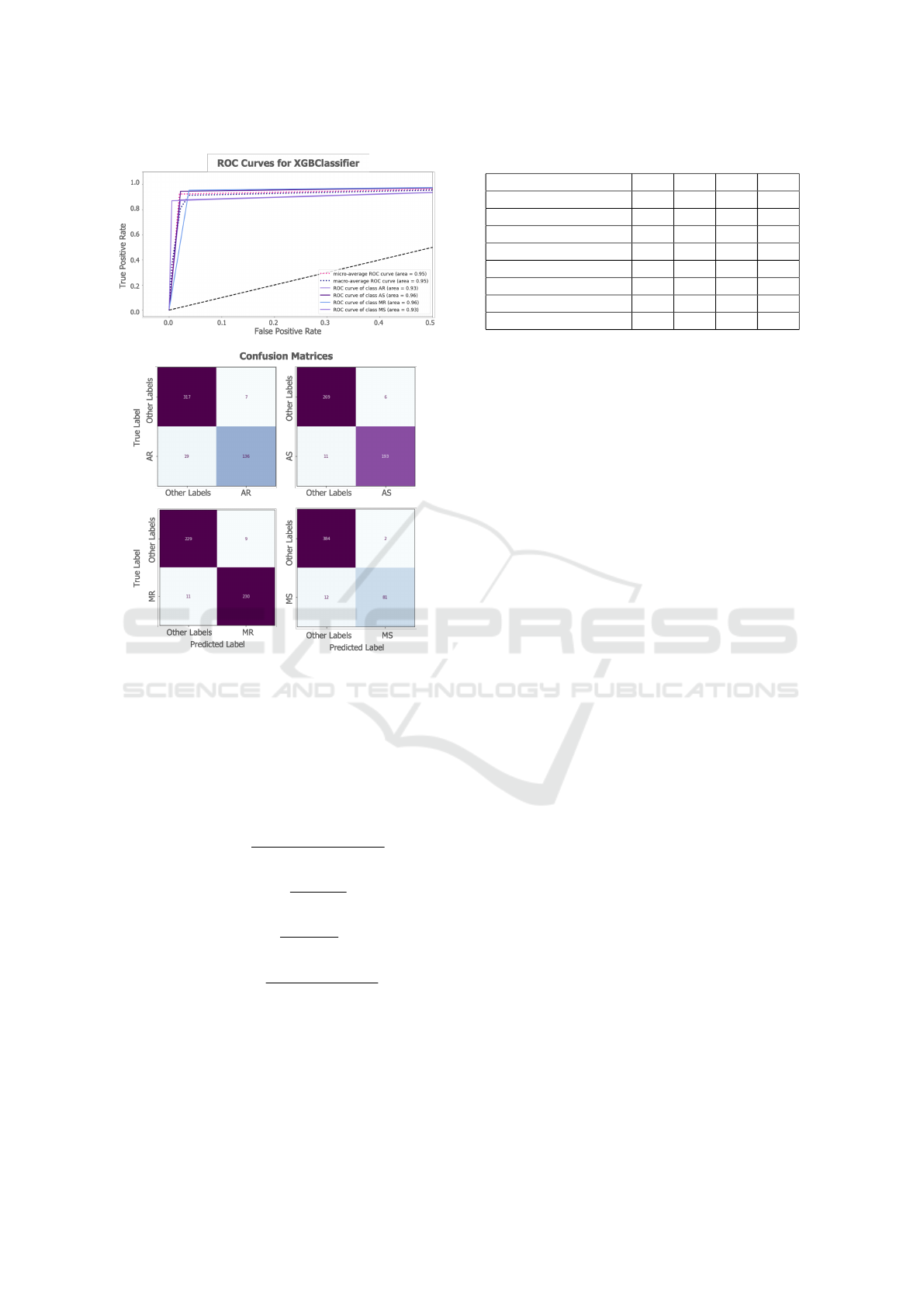
Figure 4: ROC curves and confusion matrices.
f1-score metrics for each of the AR, AS, MR, and
MS classes. The corresponding equations were listed
in Equations 1, 2, 3, 4, respectively (TP: true pos-
itives, FP: false positives, TN: true negatives and
FN: false negatives). In addition, the areas under the
receiver operating characteristics curve (ROC AUC)
were computed for each disease.
Accuracy =
T P + T N
T P + T N + FP + FN
(1)
Precision =
T P
T P + FP
(2)
Recall =
T P
T P + FN
(3)
f
1
score = 2 ∗
precision ∗ recall
precision + recall
(4)
2.5.2 Axis Interpretation
As an additional analysis, the X, Y and Z axes fea-
tures were investigated individually to assess the con-
tribution of each axis. To that aim, similar feature
extraction, feature selection and model training steps
Table 2: Classification Results.
AR AS MR MS
Accuracy 0.95 0.96 0.96 0.97
Precision (macro) 0.95 0.97 0.96 0.97
Precision (weighted) 0.95 0.96 0.96 0.97
Recall (macro) 0.93 0.96 0.96 0.93
Recall (weighted) 0.95 0.96 0.96 0.97
f1-score (macro) 0.94 0.96 0.96 0.95
f1-score (weighted) 0.95 0.96 0.96 0.97
ROC AUC 0.93 0.96 0.96 0.93
were leveraged, i.e., (i) The spectrogram, wavelet,
tempogram, chromagram and zero crossing rate fea-
tures were first assessed using p-value analysis and
variance thresholding. (ii) A multi-label/multi-output
model was then trained and tested using the remain-
ing features. These steps were employed on each of
the X, Y and Z axes. The performance of the models
were again assessed using accuracy, precision, recall,
f1-score and ROC AUC metrics.
3 RESULTS AND DISCUSSION
3.1 Feature Selection
In the first part of the analysis, the combination of the
features extracted from all three SCG axes was used.
First, the number of features was decreased from 7497
to 308 using a cascaded feature selection pipeline in-
cluding p-value analysis and variance thresholding.
One important observation was that out of the wavelet
features, only the approximation coefficients, which
correspond to the 0 - 2 Hz, could reject the null hy-
pothesis and be included in the final feature set. This
band primarily represents the respiration information
as the frequencies below 1 Hz are associated with the
chest movements originating from exhalation and in-
halation phases (Pandia et al., 2012). In the literature,
it has been shown that respiration has an indisputable
effect on cardiac murmurs as the change in murmur
intensity during breathing can provide valuable infor-
mation regarding the origin and pathological signif-
icance of the murmur (Levin et al., 1962). Hence,
these findings were indeed consistent with the under-
lying physiological processes. Another important ob-
servation was that all zero-crossing-rate features from
all three axes could reject the null hypothesis, thus
it can be deduced that different VHDs induce differ-
ent amounts of sign change on the SCG signals. This
difference can be attributed to either the differences
in induced noise or effects on spectral components as
previously discussed (Giannakopoulos and Pikrakis,
2014; Koutroumbas and Theodoridis, 2008).
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between Valvular Heart Diseases
217
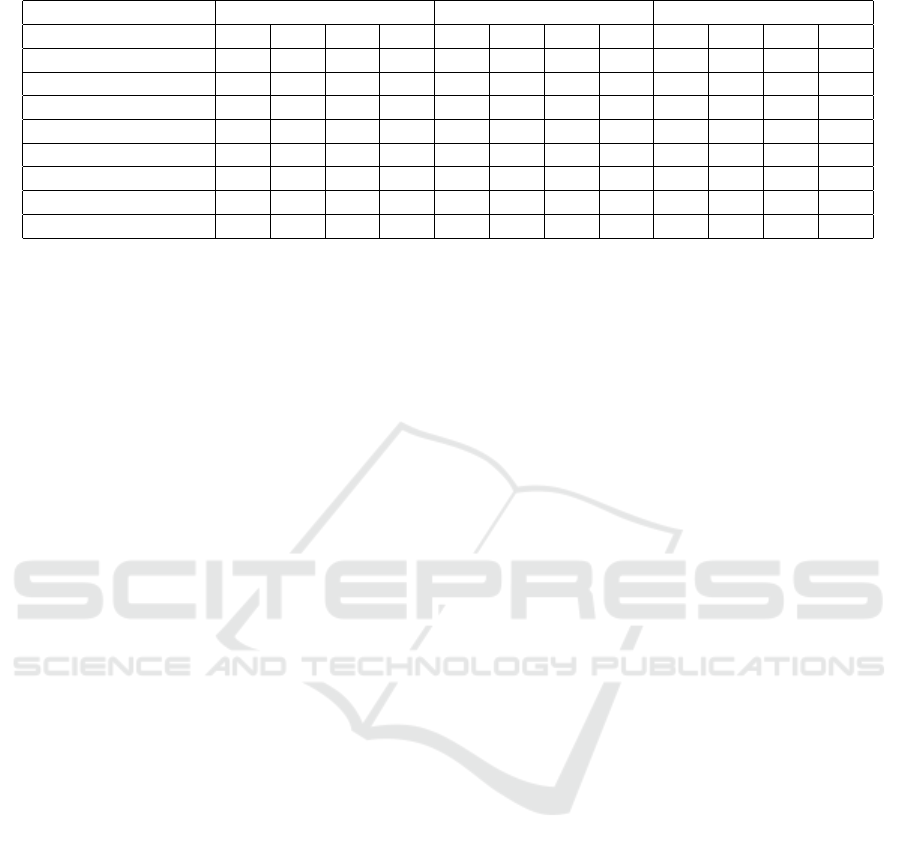
Table 3: Performance Metrics for X, Y, Z axes individually.
X (lateral) Y (head-to-foot) Z (dorso-ventral)
AR AS MR MS AR AS MR MS AR AS MR MS
Accuracy 0.92 0.91 0.90 0.93 0.91 0.90 0.90 0.93 0.92 0.93 0.92 0.95
Precision (macro) 0.92 0.91 0.90 0.91 0.92 0.90 0.90 0.90 0.93 0.93 0.92 0.95
Precision (weighted) 0.92 0.91 0.90 0.93 0.91 0.90 0.90 0.92 0.92 0.93 0.92 0.95
Recall (macro) 0.89 0.90 0.90 0.86 0.87 0.90 0.90 0.86 0.90 0.92 0.92 0.88
Recall (weighted) 0.92 0.91 0.90 0.93 0.91 0.90 0.90 0.93 0.92 0.93 0.92 0.95
f1-score (macro) 0.91 0.90 0.90 0.89 0.89 0.90 0.90 0.88 0.91 0.93 0.92 0.91
f1-score (weighted) 0.92 0.91 0.90 0.93 0.90 0.90 0.90 0.93 0.92 0.93 0.92 0.95
ROC AUC 0.89 0.90 0.90 0.86 0.87 0.90 0.90 0.86 0.89 0.92 0.92 0.88
3.2 Classification Results
Following feature elimination, an XGBoost-based
multi-label/multi-class classification framework
leveraging One vs. Rest strategy was built. To assess
the model performance, accuracy, precision, recall,
f1-score and ROC AUC metrics were calculated.
The multi-label/multi-class classification results,
corresponding ROC curves and confusion matrices
for each VHD are presented in Table 2 and Fig. 4.
As seen, for all four VHDs, the accuracy, weighted
precision, weighted recall and weighted f1-score
values were above 95%. Additionally, the average
ROC AUC value for all four VHDs was calculated
as 0.95 (Fig. 4). Obtaining superior performance
values shows that the spectral features and the
XGBoost-based multi-label/multi-class classification
framework work at sufficiently high performance.
3.3 Axis Interpretation
As the last analysis, the aforementioned analysis
pipeline was repeated on the X, Y and Z axes indi-
vidually. Initially, there were 2499 features extracted
from each of the lateral (X), head-to-foot (Y), and
dorso-ventral (Z) axes. For the X axis, this num-
ber decreased to 252 following p-value analysis and
to 143 after variance thresholding. Similarly for Y
and Z axes, these values were 256 to 131 and 253 to
159, respectively. For each axis, a separate model was
trained and tested. The multi-label/multi-class classi-
fication results are presented in Table 3. As expected,
the classifier performances were lower compared to
the case where features from all three axes were used,
however still most of the performance values were
above 90%. The best performing axis was found to
be the dorso-ventral direction (Z), which has indeed
been the most commonly used axis in cardiovascular
health assessment (Taebi et al., 2019).
4 CONCLUSIONS
In this paper, an XGBoost-based multi-label/multi-
class classification framework was proposed to dis-
tinguish between aortic stenosis (AS), aortic valve
regurgitation (AR), mitral valve stenosis (MS), and
mitral valve regurgitation (MR) using the tri-axial
SCG signals collected from the mid-sternum. First,
seismology domain knowledge was leveraged and
applied to SCG signals through ObsPy toolbox for
pre-processing. From pre-processed signal segments,
spectrogram, wavelet, chromagram, tempogram and
zero-crossing-rate features were extracted. Following
p-value analysis and variance thresholding, a multi-
label/multi-class classification framework based on
gradient boosting trees was developed to distinguish
between AS, AR, MS and MR cases.
For all four VHDs, the accuracy, weighted pre-
cision, weighted recall and weighted f1-score values
were above 95%. In addition, when the SCG axes
were investigated individually, it was found that the
features extracted from the dorso-ventral direction (Z
axis) shows superior performance compared to other
two axes. Overall, the results showed that spectral
analysis of SCG signals can provide valuable infor-
mation regarding VHDs and potentially be used in
the design of continuous monitoring systems. Future
work will focus on validating these findings in larger
datasets and improving the current pipelines further
to achieve real-time VHD detection and assessment.
REFERENCES
Beyreuther, M., Barsch, R., Krischer, L., Megies, T., Behr,
Y., and Wassermann, J. (2010). Obspy: A python tool-
box for seismology. Seismological Research Letters,
81(3):530–533.
Bommert, A., Sun, X., Bischl, B., Rahnenf
¨
uhrer, J., and
Lang, M. (2020). Benchmark for filter methods
for feature selection in high-dimensional classifica-
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
218

tion data. Computational Statistics & Data Analysis,
143:106839.
Chen, L. (2009). Curse of dimensionality. encyclopedia of
database systems.
Dietterich, T. G. et al. (2002). Ensemble learning.
The handbook of brain theory and neural networks,
2(1):110–125.
Giannakopoulos, T. and Pikrakis, A. (2014). Introduction
to audio analysis: a MATLAB® approach. Academic
Press.
Go, A. S., Mozaffarian, D., Roger, V. L., Benjamin, E. J.,
Berry, J. D., Borden, W. B., Bravata, D. M., Dai, S.,
Ford, E. S., Fox, C. S., et al. (2013). Heart disease
and stroke statistics—2013 update: a report from the
american heart association. Circulation, 127(1):e6–
e245.
Golubnitschaja, O., Kinkorova, J., and Costigliola, V.
(2014). Predictive, preventive and personalised
medicine as the hardcore of ‘horizon 2020’: Epma po-
sition paper. EPMA Journal, 5(1):1–29.
Hersek, S., Pouyan, M. B., Teague, C. N., Sawka, M. N.,
Millard-Stafford, M. L., Kogler, G. F., Wolkoff, P.,
and Inan, O. T. (2017). Acoustical emission analysis
by unsupervised graph mining: A novel biomarker of
knee health status. IEEE Transactions on Biomedical
Engineering, 65(6):1291–1300.
Hurnanen, T., Lehtonen, E., Tadi, M. J., Kuusela, T.,
Kiviniemi, T., Saraste, A., Vasankari, T., Airaksinen,
J., Koivisto, T., and P
¨
ank
¨
a
¨
al
¨
a, M. (2016). Auto-
mated detection of atrial fibrillation based on time–
frequency analysis of seismocardiograms. IEEE jour-
nal of biomedical and health informatics, 21(5):1233–
1241.
Imirzalioglu, M. and Semiz, B. (2022). Quantifying respira-
tion effects on cardiac vibrations using teager energy
operator and gradient boosted trees. In 2022 44th An-
nual International Conference of the IEEE Engineer-
ing in Medicine & Biology Society (EMBC), pages
1935–1938. IEEE.
Inan, O. T., Baran Pouyan, M., Javaid, A. Q., Dowling, S.,
Etemadi, M., Dorier, A., Heller, J. A., Bicen, A. O.,
Roy, S., De Marco, T., et al. (2018). Novel wearable
seismocardiography and machine learning algorithms
can assess clinical status of heart failure patients. Cir-
culation: Heart Failure, 11(1):e004313.
Inan, O. T., Migeotte, P.-F., Park, K.-S., Etemadi, M.,
Tavakolian, K., Casanella, R., Zanetti, J., Tank, J.,
Funtova, I., Prisk, G. K., et al. (2014). Ballistocardio-
graphy and seismocardiography: A review of recent
advances. IEEE journal of biomedical and health in-
formatics, 19(4):1414–1427.
Jahromi, M. G., Parsaei, H., Zamani, A., and Dehbozorgi,
M. (2017). Comparative analysis of wavelet-based
feature extraction for intramuscular emg signal de-
composition. Journal of biomedical physics & engi-
neering, 7(4):365.
Klabunde, R. (2011). Cardiovascular physiology concepts.
Lippincott Williams & Wilkins.
Koutroumbas, K. and Theodoridis, S. (2008). Pattern
recognition. Academic Press.
Lay, T. and Wallace, T. C. (1995). Modern global seismol-
ogy. Elsevier.
Levin, H. S., Runco, V., Goodwin, R. S., Ryan, J. M., et al.
(1962). The effect of respiration on cardiac murmurs:
An auscultatory illusion. The American Journal of
Medicine, 33(2):236–242.
McClellan, J. H., Schafer, R. W., and Yoder, M. A. (2003).
Signal processing first. Pearson education Upper Sad-
dle River, NJ.
Meister, S., Deiters, W., and Becker, S. (2016). Digital
health and digital biomarkers–enabling value chains
on health data. Current Directions in Biomedical En-
gineering, 2(1):577–581.
Pandia, K., Inan, O. T., Kovacs, G. T., and Giovangrandi, L.
(2012). Extracting respiratory information from seis-
mocardiogram signals acquired on the chest using a
miniature accelerometer. Physiological measurement,
33(10):1643.
Polikar, R. (1996). The wavelet tutorial second edition
part i. Fundamental Concepts & An Overview of The
Wavelet Theory.
Sedgwick, P. (2015). A comparison of parametric and non-
parametric statistical tests. BMJ, 350.
Semiz, B., Carek, A. M., Johnson, J. C., Ahmad, S.,
Heller, J. A., Vicente, F. G., Caron, S., Hogue, C. W.,
Etemadi, M., and Inan, O. T. (2020). Non-invasive
wearable patch utilizing seismocardiography for peri-
operative use in surgical patients. IEEE Journal
of Biomedical and Health Informatics, 25(5):1572–
1582.
Shah, A., Kattel, M., Nepal, A., and Shrestha, D. (2019).
Chroma feature extraction. Chroma Feature Extrac-
tion using Fourier Transform.
Shandhi, M. M. H., Semiz, B., Hersek, S., Goller, N.,
Ayazi, F., and Inan, O. T. (2019). Performance
analysis of gyroscope and accelerometer sensors for
seismocardiography-based wearable pre-ejection pe-
riod estimation. IEEE journal of biomedical and
health informatics, 23(6):2365–2374.
Svensson, L. (2008). Aortic valve stenosis and regurgita-
tion: an overview of management. Journal of Cardio-
vascular Surgery, 49(2):297.
Taebi, A., Solar, B. E., Bomar, A. J., Sandler, R. H., and
Mansy, H. A. (2019). Recent advances in seismocar-
diography. Vibration, 2(1):64–86.
WHO (2020). The top 10 causes of death. Geneva: World
Health Organization. https://www.who.int/news-
room/fact-sheets/detail/the-top-10-causes-of-death
(visited: 2022-09).
Yang, C., Aranoff, N. D., Green, P., and Tavassolian, N.
(2019). Classification of aortic stenosis using time–
frequency features from chest cardio-mechanical sig-
nals. IEEE Transactions on Biomedical Engineering,
67(6):1672–1683.
Yang, C., Fan, F., Aranoff, N., Green, P., Li, Y., Liu, C., and
Tavassolian, N. (2021). An open-access database for
the evaluation of cardio-mechanical signals from pa-
tients with valvular heart diseases. Frontiers in Phys-
iology, 12.
Spectral Analysis of Cardiogenic Vibrations to Distinguish Between Valvular Heart Diseases
219
