
Toward Few Pixel Annotations for 3D Segmentation of Material from
Electron Tomography
Cyril Li
1
, Christophe Ducottet
1
, Sylvain Desroziers
2
and Maxime Moreaud
3
1
Universit
´
e de Lyon, UJM-Saint-Etienne, CNRS, IOGS, Laboratoire Hubert Curien UMR5516,
F-42023, Saint-Etienne, France
2
Manufacture Franc¸aise des Pneumatiques Michelin, 23 Place des Carmes D
´
echaux, 63000 Clermont-Ferrand, France
3
IFP Energies Nouvelles, Rond-point de L’
´
echangeur de Solaize BP 3, 69360 Solaize, France
Keywords:
Neural Network, Electron Tomography, Weakly Annotated Data, U-NET, Contrastive Learning,
Semi-Supervised Training.
Abstract:
Segmentation is a notorious tedious task, especially for 3D volume of material obtained via electron tomogra-
phy. In this paper, we propose a new method for the segmentation of such data with only few partially labeled
slices extracted from the volume. This method handles very restricted training data, and particularly less than
a slice of the volume. Moreover, unlabeled data also contributes to the segmentation. To achieve this, a combi-
nation of self-supervised and contrastive learning methods are used on top of any 2D segmentation backbone.
This method has been evaluated on three real electron tomography volumes.
1 INTRODUCTION
Electron tomography (ET) (Ersen et al., 2007) is a
powerful characterization technique for the recon-
struction of 3D nanoscale microstructure of material.
Volumes are reconstructed from sets of projections
from different angles acquired by a Transmission
Electron Microscope (TEM) providing a real three-
dimensional information at the nanometric scale. The
limited number of projections and the difficulty to
align them correctly (Frank, 2008) produce noisy data
with strong reconstruction artifacts (Figure 1). Stan-
dard segmentation approaches generally fail to pro-
duce accurate semantic segmentation of this kind of
data (Evin et al., 2021), or need intensive expertise of
the user (Fernandez, 2012; He et al., 2008; Volkmann,
2010).
Recently, deep learning (DL) based methods have
been successfully used in this field (Akers et al., 2021;
Khadangi et al., 2021; Genc et al., 2022), inheriting
from progresses in 2D or 3D image semantic segmen-
tation (Ronneberger et al., 2015; C¸ ic¸ek et al., 2016;
Milletari et al., 2016; Chen et al., 2018a; Sun et al.,
2019). The standard setup is first to train a DL model
on a fully labeled dataset, and then use this model on
the data at hand. However, this approach requires the
availability of a fully segmented set of 3D volumes,
which is a tedious preliminary task. In this paper,
we consider a more realistic semi supervised setup.
Figure 1: Slice of a volume of zeolite (resolution : 1
nm/voxel). Slices from the volume are noisy and contain
artifacts, that make segmentation tasks harder.
Given a 3D volume, we ask the user to manually seg-
ment only a few regions in a few slices (typically a
single one or two) of this 3D volume. This small
amount of annotated data is then used to train a DL
model, subsequently used to segment the whole 3D
volume.
124
Li, C., Ducottet, C., Desroziers, S. and Moreaud, M.
Toward Few Pixel Annotations for 3D Segmentation of Material from Electron Tomography.
DOI: 10.5220/0011658500003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 5: VISAPP, pages
124-131
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

The problem of few available training data is not
specific to ET and has been addressed in many com-
puter vision tasks, mainly using transfer learning (Pan
and Yang, 2009; Wurm et al., 2019). A promising ap-
proach is to rely on contrastive representation learn-
ing, whose goal is to learn an embedding space using
pairs of positive or negative samples (Hadsell et al.,
2006; Dosovitskiy et al., 2014; Zhao et al., 2021). In a
supervised setting, mainly used in Siamese networks
(Koch et al., 2015), positive pairs are taken from sam-
ples of the same class and negative ones are taken
from samples of different classes. The unsupervised
setting relies on positive pairs obtained from a single
sample subject to two independent random perturba-
tions. It was shown to be a powerful self-supervised
learning method (Chen et al., 2020; Chen and He,
2021).
In this article, we propose a new semi-supervised
approach to segment a full volume in ET with only
few annotated pixels. Our approach fully exploits the
labeled as well as the unlabeled pixels of the partially
annotated slices to learn a specific pixel-level embed-
ding space relevant for segmentation. More precisely,
given a DL model for semantic segmentation, we first
reshape the output to define a pixel-level embedding
space of dimension D. Then, for training, we rely
on contrastive learning and both a weakly-supervised
stream for labeled pixels and a self-supervised stream
for unlabeled ones (Figure 2). Supervised and self-
supervised contrastive learning are used together to
fully exploit partially-labeled data (Figure 3). The
final segmentation is obtained through a pixel-wise
classification layer operating in the embedding space
of dimension D.
Our principal contributions are:
• A new semi-supervised learning method for prac-
tical 3D image segmentation in ET, which takes
advantage of contrastive learning and self learning
principles to provide accurate volume segmenta-
tion using only few labeled regions of one or two
specific 2D slices.
• The model can be easily built on top of any 2D
segmentation DL model.
• We provide detailed experimentation on several
real ET data, and we show that an accurate seg-
mentation is possible with only one slice and 6%
of annotated pixels in this slice.
2 RELATED WORKS
Electron Tomography Segmentation. Due to low
SNR and reconstruction artifacts, segmentation of to-
mograms is a difficult task and manual segmentation
still remains the prevalent method (Fernandez, 2012)
often used in interaction with the user through visu-
alization tools (He et al., 2008), sometimes combined
with various image processing methods as watershed
transform (Volkmann, 2010). Following the promis-
ing development of DL in general semantic segmen-
tation tasks (Ronneberger et al., 2015; C¸ ic¸ek et al.,
2016; Milletari et al., 2016; Chen et al., 2018a; Sun
et al., 2019) some recent works have investigated DL
based techniques in 2D electron microscopy (Akers
et al., 2021; Khadangi et al., 2021). In these works,
the bottleneck of the availability of labeled training
data is addressed either by a semi-supervised few-
shot approach (Akers et al., 2021) or by a scalable
DL model, which requires only small- and medium-
sized ground-truth datasets (Khadangi et al., 2021).
In 3D, we only found a first investigation of U-Net
model to multi-class semantic segmentation of a γ-
alumina/Pt catalytic material in a class imbalance sit-
uation (Genc et al., 2022). In this work, 30 labeled
slices and data augmentation are needed to train the
model. To the best of our knowledge, our method is
the first one providing accurate 3D segmentation with
only few annotated pixels in a single slice.
Contrastive Learning. Contrastive learning aims to
exploit labels better, usually with Siamese networks
(Zhao et al., 2021). The goal is to construct a la-
tent space in which objects with the same label are
close to each other, and objects with different labels
are far from each other. Positive and negative pairs are
formed. Positive pairs are composed of two objects of
the same class, whereas negative ones are composed
of two objects of different classes. A contrastive loss
function is used during training to bring positive pairs
together and negative pairs far from each other. Con-
trastive learning has shown excellent results in image
classification (Chen et al., 2020; Grill et al., 2020;
Khosla et al., 2020; Chen and He, 2021). For se-
mantic segmentation, pairs of pixels can be consid-
ered, and these methods can also train a model with-
out labeled data (Chaitanya et al., 2020). When pairs
are created without the knowledge of the class, only
positive ones can be created. Input images are trans-
formed, and the transformed pixels are compared to
their original version. The images, even transformed,
represent the same object and a similarity function
can be minimized during training.
Semi-Supervised Methods in Semantic Segmenta-
tion. The goal of semi-supervised methods in seman-
tic segmentation is to optimize how labeled and unla-
beled data are exploited together to learn a segmen-
tation task. Transformations can be used in Siamese
network to mix a self-supervised loss with a standard
Toward Few Pixel Annotations for 3D Segmentation of Material from Electron Tomography
125
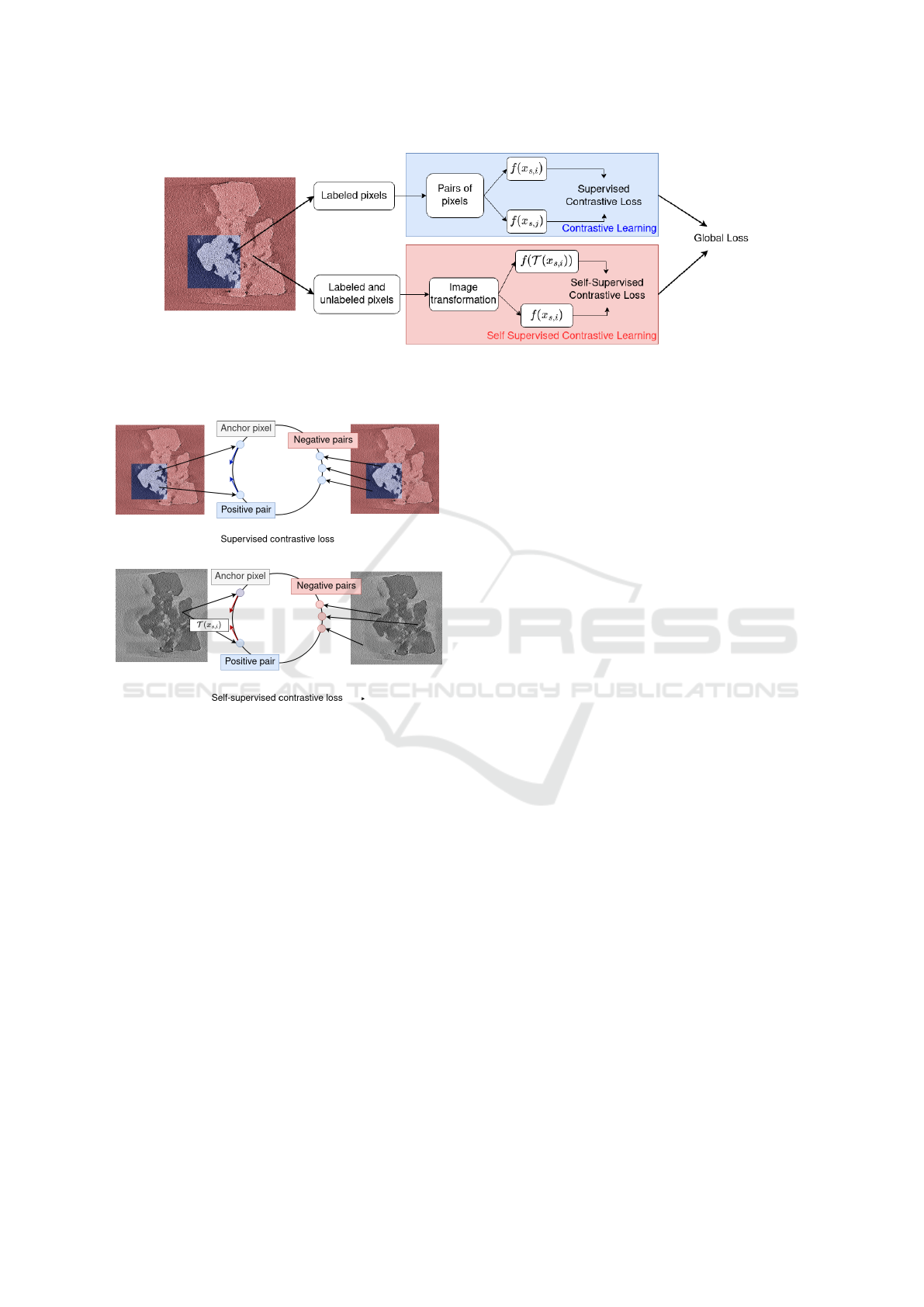
Figure 2: The loss calculation is different depending on pixel labeling. For labeled pixels, pairs of pixels are constructed and
are used in a contrastive loss. For labeled and unlabeled pixels, the training image and the transformed version of itself are
used as a positive pair in a self-learning contrastive loss. Both contrastive loss and similarity loss are added for the global loss.
Figure 3: The supervised contrastive loss is computed for
each labeled pixels. Given a labeled pixel considered as
an anchor pixel, pixels of the same class as this anchor
pixel (positive pairs) are pushed together, while pixels of
a different class (negative pairs) are pushed aside. The self-
supervised loss is computed for both labeled and unlabeled
classes. Given an anchor pixel, a single positive pair is ob-
tained by applying a transform to it, while negative pairs
are obtained from all other pixels. The anchor pixel and its
transformed version are pushed together, while other pixels
are pushed aside from the anchor pixel.
supervised methods (Li et al., 2018). Another ap-
proach is to apply an adversarial method, where two
networks are trained on labeled images. Unlabeled
images are fed into both networks and a similarity
function is minimized (Peng et al., 2020). In (Ouali
et al., 2020), a standard encoder-decoder is used on la-
beled images. Unlabeled images are fed into several
decoders, a slightly modified version of the decoder
used for the labeled data. The output of all the de-
coders is then compared to the output of the main de-
coder. In this case, the network is transformed. How-
ever, these methods have all their images either fully
labeled or fully unlabeled. In our case, images them-
selves can have unlabeled pixels.
3 PROPOSED METHOD
Our method relies on a 2D representation model
trained with contrastive-learning and self-learning
principles. This model is used to segment each slice
of the volume to provide the final 3D segmentation.
Indeed, although several fully 3D convolutional neu-
ral networks (CNN) have been proposed (Milletari
et al., 2016), they were shown to be more resource in-
tensive than 2D models without providing convincing
gains (Kern et al., 2021). The overview of the method
is summarized in Figure 2. The whole architecture
is composed of an encoder-decoder f used to project
pixels of the slice in a pixel level embedding space
and a classification layer (a linear model classifier) h
providing the semantic segmentation. The represen-
tation model f is trained using positive and negative
pairs of labeled pixels in the contrastive path, and pos-
itive pairs of labeled and unlabeled pixels in the self-
supervised path. Note that there is a single embedding
space for both labeled and unlabeled pixels. The clas-
sification layer is subsequently trained using labeled
pixels after freezing the representation model. A stan-
dard weighted cross-entropy loss is used to train this
classification layer.
3.1 Formalization
The input volume V is a set of S slices {X
s
}
s=1,...S
such that each slice X
s
∈ R
W ×H×1
where W is its
width, H its height and 1 is because the slice is a
grayscale image. We denote x
s,i
the graylevel at pixel
i ∈ I of slice X
s
, I being the image spatial support.
We then have X
s
= (x
s,i
)
i∈I
. Each slice is processed
independently. The output segmentation of slice s is
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
126

ˆ
Y
s
= h( f (X
s
)). The encoder-decoder f transforms X
s
onto Z
s
= (z
s,i
)
i∈I
= f (X
s
) ∈ R
W ×H×D
where D is the
dimension of the latent space. The classification layer
h performs the pixel-wise class prediction
ˆ
Y
s
= h(Z
s
)
such that
ˆ
Y
s
∈ R
W ×H×C
, where C is the number of
classes.
Each voxel of the volume is either labeled or un-
labeled. Let Y
s
be the pixel-wise class label map of
the slice s, then Y
s
= (y
s,i
)
i∈I
with y
s,i
∈ {
/
0, 1, 2, ..., C}
where 1, 2, .., C are the labeled classes and
/
0 repre-
sents unlabeled pixels. We note L
s
the set of labeled
pixels and U
s
the set of unlabeled pixels:
L
s
= {i ∈ I , y
s,i
̸=
/
0}, U
s
= {i ∈ I , y
s,i
=
/
0} (1)
The loss function is computed differently if the
pixel is labeled or unlabeled. Pixels in L
s
follow the
contrastive path, whereas pixels in L
s
and U
s
follow
the self-learning path. For shake of simplicity, in the
following subsections, we will consider the case of a
single labeled slice X
s
for training, but it can be easily
generalized to any number of training slices by sum-
ming the corresponding losses.
3.2 Contrastive Loss
A contrastive loss is computed for labeled pixels
(Khosla et al., 2020). This loss aims to learn a repre-
sentation space where pixels from the same label are
close to each other in that space, while pixels from
different labels are far from each other in that space.
The contrastive loss is relevant in our case because the
model can be fed with only a few number of labeled
pixels and the contrastive loss can fully exploit each
pixel by forming positive and negative pairs. For each
labeled pixel x
s,i
, i ∈ L
s
, positive pairs P
+
s,i
and nega-
tive pairs P
−
s,i
are constructed. As the label is known
for this kind of pixels, given an anchor pixel, posi-
tives pairs are built by choosing pixels of the same
class as this anchor pixel, whereas negatives pairs are
composed of pixels of a different class (Figure 3). We
have:
P
+
s,i
= { j ∈ L
s
, i ̸= j, y
s,i
= y
s, j
} (2)
P
−
s,i
= { j ∈ L
s
, i ̸= j, y
s,i
̸= y
s, j
} (3)
Let P
s,i
= P
+
s,i
∪ P
−
s,i
be the set of pairs that are
formed for each pixel x
s,i
. To balance positive and
negative pairs and limit the computational complex-
ity, we randomly choose an equal number of positive
and negative pairs such that the total number of pairs
per pixel is a given constant value N
P
.
The supervised contrastive loss (Khosla et al.,
2020) associated to one labeled slice is defined as:
L
1
(Z
s
) =
−1
N
L
s
∑
i∈L
s
1
N
P
∑
j∈P
+
s,i
log
exp(sim(z
s,i
, z
s, j
))
∑
k∈P
s,i
exp(sim(z
s,i
, z
s,k
))
(4)
with N
L
s
the number of pixels in L
s
and sim is the
cosine similarity, defined as:
sim(u, v) =
u.v
||u||.||v||
(5)
3.3 Self-Supervised Contrastive Loss
A self-supervised method is used for labeled and un-
labeled pixels. It is especially useful for unlabeled
data, as the information on labels is unknown. To
train without labels, the training slice is transformed,
and a self-supervised contrastive loss is computed be-
tween each transformed image (Chen et al., 2020).
The corresponding pixels, even from slightly trans-
formed images, should have their feature vector close
to each other in the representation space. These two
pixels are considered as a positive pair. All other com-
binations are considered as negative pairs (Figure 3).
Let T be a random transformation of the pixels of the
original slice x
s,i
, i ∈ I. The output Z
s
of the orig-
inal slice and the output of its transformed version
˜
Z
s
= f (T (X
s
)) = (˜z
s,i
)
i∈I
are compared.
The self-supervised contrastive loss associated to
slice X
s
is defined as:
L
2
(Z
s
) =
−1
N
I
∑
i∈I
log
exp(sim(z
s,i
, ˜z
s,i
))
∑
j∈I,i̸= j
exp(sim(z
s,i
, z
s, j
))
(6)
where N
I
is the number of pixels in I.
The final loss is a combination of both losses :
L(Z
s
) = L
1
(Z
s
) + L
2
(Z
s
) (7)
4 EXPERIMENTS AND RESULTS
4.1 Implementation Details
For the encoder-decoder f , a U-Net like model is
used to project pixels in the embedding space (Fig-
ure 4). The encoder is composed of three downsam-
pling steps. A downsampling step is composed of two
3 × 3 convolution layers followed by a ReLU layer
and a 2 × 2 Max Pooling layer with a stride 2 for
downsampling. The input image resolution is halved
and the number of channels is doubled for each down-
sampling step. The decoder is also composed of three
steps, with an upsampling layer followed by a 2 ×
Toward Few Pixel Annotations for 3D Segmentation of Material from Electron Tomography
127

Figure 4: Our model is based on U-Net, with three stages. The output is the projection for each pixel in the embedding space.
A classification layer is trained to transform the feature vectors into a segmentation map.
Table 1: Random transformations used to compute the self-
supervised loss.
Transformation Probability Parameters
Gaussian noise 1 N (0, [0.01;0.06])
Gray level shift 0.5 [−0.01;0.01]
Gaussian blur 0.5 σ = [0.5 − 1.5]
2 convolution layer that halves the number of chan-
nels, a concatenation with the corresponding feature
map from the encoder and two 3 x3 convolution lay-
ers followed by a ReLU. As a result, the number of
filters for each layer of the encoder is 16, 32 and 64.
For each layer of the decoder, the image resolution is
doubled and the number of channels is halved. The
resulting number of layers are 64, 32 and 16. At the
end of the network, a feature map at the size of the
input is obtained. A 1 × 1 convolutional layer is ap-
plied with 16 filters to project the result in the em-
bedding space. The loss function is computed, de-
pending upon the fact that the pixel is labeled or un-
labeled. To compute the final segmentation result, a
pixel-wise classification layer h is trained on the out-
put of the training data. This layer is composed of a
single convolutional layer with a kernel of size 1 ×
1. The classification layer is trained with a weighted
standard cross-entropy loss, where unlabeled pixels’
weight is set to 0 (C¸ ic¸ek et al., 2016).
For each labeled pixel, 10 positive pairs and 10
negative pairs are made, and thus N
P
= 20.
Table 1 details the random transformation used
on the training slice in order to compute the self-
supervised loss, as well as their probability of occur-
ring and their respective parameters. As each pixel is
associated to a corresponding transformed pixel, only
transformations that do not change the structure of the
slice are chosen. The parameters are chosen so that
the resulting image corresponds visually to a realistic
image.
The Intersection over Union (IOU) is computed
on the whole volume V to assess our results:
IOU(V ) =
∑
S
s=1
ˆ
Y
s
∩Y
s
∑
S
s=1
ˆ
Y
s
∪Y
s
(8)
Figure 5: Example of a partially labeled slice (r = 0.5) used
to train the model. Blue pixels represent the object, purple
pixels, the background and yellow pixel are unlabeled data.
The closer the IOU to 1, the better the 3D semantic
segmentation result.
4.2 Data
Zeolites are used in several chemical processes in the
energy field (Flores et al., 2019) and are composed
of nanoscale cavities that are challenging to segment
properly (Figure 1). Our methodology is illustrated
on ET volume of hierarchical zeolite, NaX Siliporite
G5 from Ceca-Arkema (Medeiros-Costa et al., 2019).
The size of the volume is 592×600×623. 10 slices
are selected as the pool of possible training slices. A
5-fold cross validation is used, where 1 slice for train-
ing and 1 for validation are chosen for each fold. All
other slices of the volume are used for test. The an-
notation for the training slice is artificially hidden to
provide partially labeled data to the network. Each
slice is divided into 100 equally sized patches. A ran-
dom number of patches are set to be hidden. At least
one patch with object pixels and one patch with back-
ground pixels are selected to ensure that positive pairs
and negative pairs can be created. An example of a
partially annotated slice can be seen in Figure 5. The
number of hidden patches depends on a labeling rate
denoted r.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
128
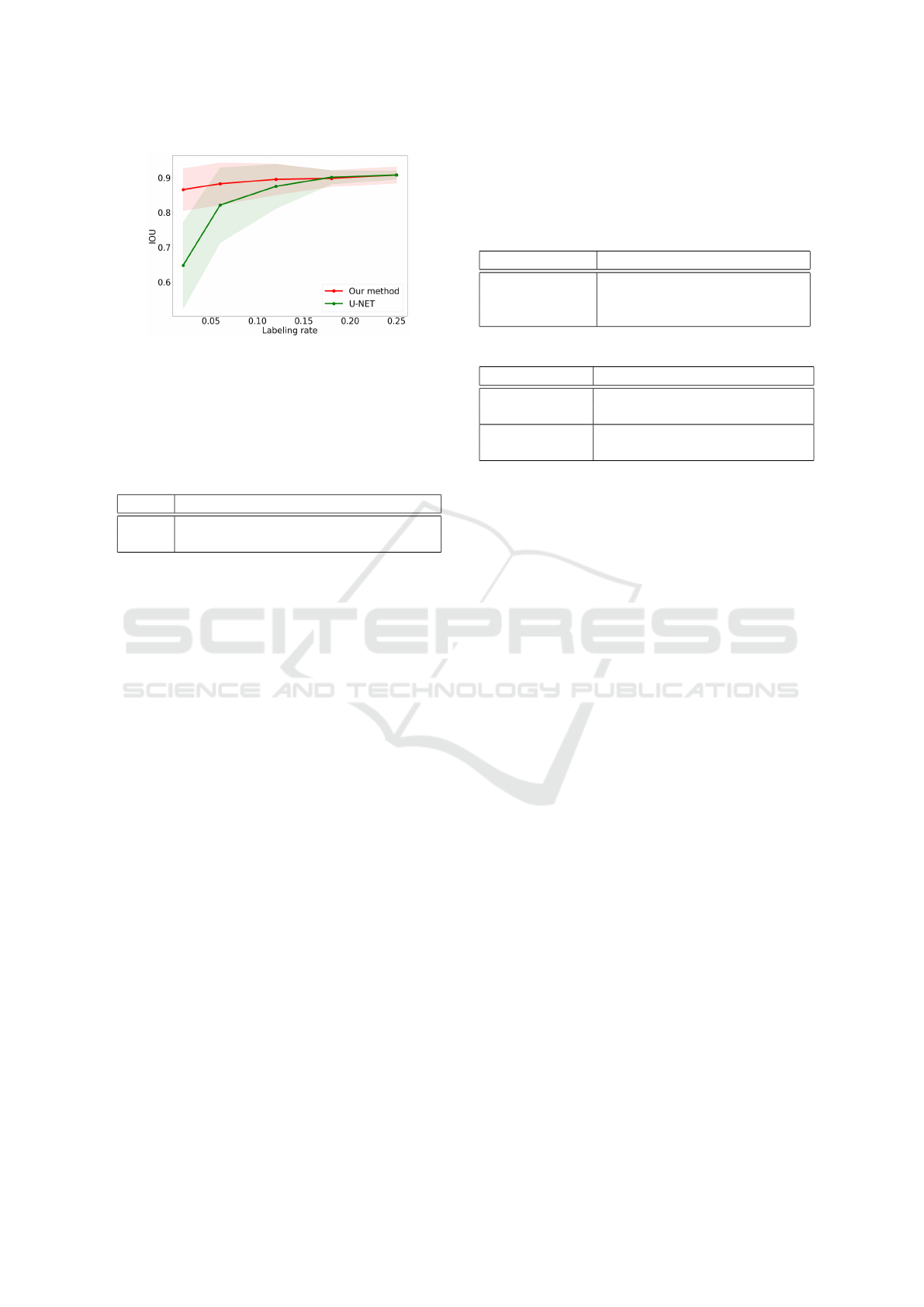
Figure 6: Mean IOU of our method (red) and U-Net (green)
when reconstructing the segmentation mask for the volume
with one partially labeled training slice for different labeling
rate. The green and red areas represent the standard devi-
ation of the results with U-Net and with our method (each
experiment is a 5-fold cross validation, repeated 5 times).
Table 2: Comparison of IOU between our method with both
loss combined and a U-Net architecture.
r 0.02 0.06 0.12 0.18 0.25
U-Net 0.648 0.821 0.875 0.902 0.908
Ours 0.866 0.883 0.895 0.898 0.908
4.3 Results
Study of Parameter r. We compare our method with
U-Net as a reference for different labeling rates to il-
lustrate the benefit of learning an embedding space.
In addition to the 5-fold cross validation, each exper-
iment is repeated 5 times, resulting in a total of 25
realizations by experiment. The mean and standard
deviation are computed across these realizations. The
U-Net network is modified in order to make it com-
patible with partially labeled data like in C¸ ic¸ek et al.’s
work (C¸ ic¸ek et al., 2016) where a weighted cross-
entropy is used and unlabeled pixels’ weights are set
to 0. The results are shown in Figure 6 and in Table 2.
Our method performs significantly better than U-Net
for small labeling rates, and has similar performance
for higher learning rate. The results are already perti-
nent, only using 2% of one labeled slice for training.
Comparison Study Between Supervised and Self-
Supervised Losses An ablation study was made to
study, for each labeling rate r the effect of activat-
ing either the supervised or the self-supervised loss
alone or the combination of both. As shown in Ta-
ble 3, when only the self-supervised loss is used,
the results are good even for a small labelling rate:
the latent space is well learned with only few anno-
tated pixels. As the labelling rate grows, only the
classification layer benefits from the supplementary
data. This results in little improvement as the la-
belling rate increases. The contrastive loss does not
perform well when only few data are provided, but
Table 3: Comparison of obtained mean IOU for indepen-
dent trainings by activating either the self-supervised, the
supervised or both losses (each experiment is a 5-fold cross
validation, repeated 5 times and the average is reported in
the table). Note that the classification layer h is always
trained with supervision.
r 0.06 0.12 0.18 1.00
Self-Supervised 0.907 0.869 0.891 0.916
Supervised 0.773 0.875 0.861 0.926
Combined 0.883 0.895 0.898 0.927
Table 4: Results of our method for several volumes.
Zeolite 1 Zeolite 2 Alumina
U-Net r = 0.06 0.821 0.219 0.128
Ours r = 0.06 0.883 0.443 0.533
U-Net r = 0.25 0.908 0.392 0.196
Ours r = 0.25 0.908 0.595 0.729
as there are more labelled data, the supervised loss
performs better due to more diverse pairs of pixels.
When combining both supervised and self-supervised
loss, good results are obtained with a small labelling
rate, while performing better as the amount of labelled
data raises. Both supervised and self-supervised con-
trastive loss are required to obtain the best segmen-
tation. The whole reconstructed volume with both
losses combined can be seen in Figure 7.
Generalization to Other Volumes. Another vol-
ume of the same kind of zeolite (size 512×512×100)
and a volume of γ-alumina (Gay et al., 2016) (size
592×840×296) have also been segmented with our
approach. The results can be seen in Table 4. We
compared our method with both losses combined
against a U-Net like network. Our model performs
better than U-Net in most cases. Moreover, our
method reaches maximum values at a smaller label-
ing rate than U-Net, and performs better than U-Net
with very small values of r . The reconstruction of the
segmentation of the volume can be seen in Figure 7.
5 CONCLUSION
In this paper, we introduce a new semi-supervised
learning method for 3D image segmentation in elec-
tron tomography. Our model can achieve accurate
segmentation of electron tomography volumes with
only less than a slice for training data by using both
labeled and unlabeled data. Specifically, we com-
bine a contrastive path for labeled voxels and a self-
supervised path for unlabeled ones. This strategy
tends to maximize the possible use of all the informa-
tion available from the data. As the model can be built
on any 2D segmentation DL model, more modern ar-
Toward Few Pixel Annotations for 3D Segmentation of Material from Electron Tomography
129

(1) (2) (3)
Figure 7: 3D reconstructions of the segmentation map of zeolites (1) (2) and γ-alumina (3). The volume (1) is cut to render
the inner structure of the volume. 6% of one slice has been taken to train the model used for each volume.
chitecture such as DeepLabV3+ (Chen et al., 2018b)
or UNet++ (Zhou et al., 2018) can be investigated for
future work. We have illustrated our strategy on elec-
tron tomography volume of material, but it is not lim-
ited to this type of data and acquisition, and could be
used in other fields as well, such as medical applica-
tions.
ACKNOWLEDGEMENTS
This work was supported by the LABEX MILYON
(ANR-10-LABX-0070) of Universit
´
e de Lyon, within
the program ”Investissements d’Avenir” (ANR-11-
IDEX- 0007) operated by the French National Re-
search Agency (ANR).
REFERENCES
Akers, S., Kautz, E., Trevino-Gavito, A., Olszta, M.,
Matthews, B. E., Wang, L., Du, Y., and Spurgeon,
S. R. (2021). Rapid and flexible segmentation of elec-
tron microscopy data using few-shot machine learn-
ing. npj Computational Materials, 7(1):1–9.
Chaitanya, K., Erdil, E., Karani, N., and Konukoglu, E.
(2020). Contrastive learning of global and local fea-
tures for medical image segmentation with limited an-
notations. Advances in Neural Information Processing
Systems, 33:12546–12558.
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K.,
and Yuille, A. L. (2018a). Deeplab: Semantic im-
age segmentation with deep convolutional nets, atrous
convolution, and fully connected crfs. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence,
40(4):834–848.
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., and
Adam, H. (2018b). Encoder-decoder with atrous sep-
arable convolution for semantic image segmentation.
Computer Vision – ECCV 2018, pages 833–851.
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020).
A simple framework for contrastive learning of visual
representations. International conference on machine
learning, pages 1597–1607.
Chen, X. and He, K. (2021). Exploring simple siamese rep-
resentation learning. Proceedings of the IEEE/CVF
Conference on Computer Vision and Pattern Recogni-
tion (CVPR), pages 15750–15758.
C¸ ic¸ek,
¨
O., Abdulkadir, A., Lienkamp, S. S., Brox, T.,
and Ronneberger, O. (2016). 3d u-net: Learning
dense volumetric segmentation from sparse annota-
tion. Medical Image Computing and Computer-
Assisted Intervention – MICCAI 2016, pages 424–
432.
Dosovitskiy, A., Springenberg, J. T., Riedmiller, M., and
Brox, T. (2014). Discriminative unsupervised fea-
ture learning with convolutional neural networks. Ad-
vances in neural information processing systems, 27.
Ersen, O., Hirlimann, C., Drillon, M., Werckmann, J., Ti-
hay, F., Pham-Huu, C., Crucifix, C., and Schultz, P.
(2007). 3D-TEM characterization of nanometric ob-
jects. Solid State Sciences, 9(12):1088–1098.
Evin, B., Leroy,
´
E., Baaziz, W., Segard, M., Paul-Boncour,
V., Challet, S., Fabre, A., Thi
´
ebaut, S., Latroche,
M., and Ersen, O. (2021). 3D Analysis of Helium-
3 Nanobubbles in Palladium Aged under Tritium by
Electron Tomography. Journal of Physical Chemistry
C, 125(46):25404–25409.
Fernandez, J.-J. (2012). Computational methods for elec-
tron tomography. Micron, 43(10):1010–1030.
Flores, C., Zholobenko, V. L., Gu, B., Batalha, N., Valtchev,
V., Baaziz, W., Ersen, O., Marcilio, N. R., Ordomsky,
V., and Khodakov, A. Y. (2019). Versatile Roles of
Metal Species in Carbon Nanotube Templates for the
Synthesis of Metal–Zeolite Nanocomposite Catalysts.
ACS Applied Nano Materials, 2(7):4507–4517.
Frank, J. (2008). Electron tomography: methods for three-
dimensional visualization of structures in the cell.
Springer Science & Business Media.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
130

Gay, A.-S., Humbert, S., Taleb, A.-L., Lefebvre, V.,
Dalverny, C., Desjouis, G., and Bizien, T. (2016).
Combined characterization of cobalt aggregates by
haadf-stem electron tomography and anomalous x-
ray scattering. European Microscopy Congress 2016:
Proceedings, pages 39–40.
Genc, A., Kovarik, L., and Fraser, H. L. (2022). A deep
learning approach for semantic segmentation of un-
balanced data in electron tomography of catalytic ma-
terials. arXiv preprint arXiv:2201.07342.
Grill, J.-B., Strub, F., Altch
´
e, F., Tallec, C., Richemond,
P. H., Buchatskaya, E., Doersch, C., Pires, B. A., Guo,
Z. D., Azar, M. G., Piot, B., Kavukcuoglu, K., Munos,
R., and Valko, M. (2020). Bootstrap Your Own Latent:
A new approach to self-supervised learning. Neural
Information Processing Systems.
Hadsell, R., Chopra, S., and LeCun, Y. (2006). Dimen-
sionality reduction by learning an invariant mapping.
2006 IEEE Conference on Computer Vision and Pat-
tern Recognition (CVPR’06), 2:1735–1742.
He, W., Ladinsky, M. S., Huey-Tubman, K. E., Jensen,
G. J., McIntosh, J. R., and Bj
¨
orkman, P. J. (2008).
Fcrn-mediated antibody transport across epithelial
cells revealed by electron tomography. nature,
455(7212):542–546.
Kern, D., Klauck, U., Ropinski, T., and Mastmeyer, A.
(2021). 2D vs. 3D U-Net abdominal organ segmenta-
tion in CT data using organ bounds. Medical Imaging
2021: Imaging Informatics for Healthcare, Research,
and Applications, 11601:192 – 200.
Khadangi, A., Boudier, T., and Rajagopal, V. (2021). Em-
net: Deep learning for electron microscopy image seg-
mentation. 2020 25th International Conference on
Pattern Recognition (ICPR), pages 31–38.
Khosla, P., Teterwak, P., Wang, C., Sarna, A., Tian, Y.,
Isola, P., Maschinot, A., Liu, C., and Krishnan, D.
(2020). Supervised contrastive learning. Advances
in Neural Information Processing Systems, 33:18661–
18673.
Koch, G., Zemel, R., and Salakhutdinov, R. (2015).
Siamese neural networks for one-shot image recogni-
tion. ICML deep learning workshop, 2.
Li, X., Yu, L., Chen, H., Fu, C., and Heng, P. (2018). Semi-
supervised skin lesion segmentation via transforma-
tion consistent self-ensembling model. British Ma-
chine Vision Conference 2018, BMVC 2018, Newcas-
tle, UK, September 3-6, 2018, page 63.
Medeiros-Costa, I. C., Laroche, C., P
´
erez-Pellitero, J., and
Coasne, B. (2019). Characterization of hierarchi-
cal zeolites: Combining adsorption/intrusion, electron
microscopy, diffraction and spectroscopic techniques.
Microporous and Mesoporous Materials, 287:167–
176.
Milletari, F., Navab, N., and Ahmadi, S.-A. (2016). V-
net: Fully convolutional neural networks for volumet-
ric medical image segmentation. 2016 fourth interna-
tional conference on 3D vision (3DV), pages 565–571.
Ouali, Y., Hudelot, C., and Tami, M. (2020). Semi-
supervised semantic segmentation with cross-
consistency training. Proceedings of the IEEE/CVF
Conference on Computer Vision and Pattern Recogni-
tion, pages 12674–12684.
Pan, S. J. and Yang, Q. (2009). A survey on transfer learn-
ing. IEEE Transactions on knowledge and data engi-
neering, 22(10):1345–1359.
Peng, J., Estrada, G., Pedersoli, M., and Desrosiers, C.
(2020). Deep co-training for semi-supervised image
segmentation. Pattern Recognition, 107:107269.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. Medical Image Computing and Computer-
Assisted Intervention – MICCAI 2015, pages 234–
241.
Sun, K., Xiao, B., Liu, D., and Wang, J. (2019). Deep high-
resolution representation learning for human pose es-
timation. Proceedings of the IEEE/CVF Conference
on Computer Vision and Pattern Recognition, pages
5693–5703.
Volkmann, N. (2010). Methods for segmentation and in-
terpretation of electron tomographic reconstructions.
Methods in enzymology, 483:31–46.
Wurm, M., Stark, T., Zhu, X. X., Weigand, M., and
Taubenb
¨
ock, H. (2019). Semantic segmentation of
slums in satellite images using transfer learning on
fully convolutional neural networks. ISPRS Journal
of Photogrammetry and Remote Sensing, 150:59–69.
Zhao, X., Vemulapalli, R., Mansfield, P. A., Gong, B.,
Green, B., Shapira, L., and Wu, Y. (2021). Con-
trastive learning for label efficient semantic segmen-
tation. Proceedings of the IEEE/CVF International
Conference on Computer Vision, pages 10623–10633.
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N., and
Liang, J. (2018). Unet++: A nested u-net architecture
for medical image segmentation. In Deep learning in
medical image analysis and multimodal learning for
clinical decision support, pages 3–11. Springer.
Toward Few Pixel Annotations for 3D Segmentation of Material from Electron Tomography
131
