
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep
Learning Structures Worth It?
Mostafa Moussa
a
, Yahya Alzaabi and Ahsan Khandoker
Department of Biomedical Engineering, Khalifa University of Science and Technology, Abu Dhabi, 127788, U.A.E.
Keywords:
Depression, Electroencephalography, Electrocardiography, Breathing Signals, Gated Recurrent Unit Long
Short-Term Memory Networks.
Abstract:
The prevalence and severity of depression make it imperative to develop a means to automatically detect it, so
as to alleviate the associated mental effort and cost of seeing a dedicated professional. Depression can also
co-exist with other conditions, such as Obstructive Sleep Apnea Syndrome (OSAS). In this paper, we build
upon our previous work involving sleep staging, detection of OSAS, and detection of depression in OSAS
patients, but focus solely on the latter of the three. We use features extracted from EEG, ECG, and breathing
signals of 80 subjects suffering from OSAS and half of which also with depression, using 75 % of this 80-
subject dataset for training and 10-fold cross-validation and the remainder for testing. We train three models
to classify depression: a random forest (RF), a three-layer artificial neural network (3-ANN), and a gated-
recurrent unit long short-term memory (GRU-LSTM) recurrent neural network. Our analysis shows that, like
our previous work, the 3-ANN is still the best performing model, with the GRU-LSTM following closely
behind at an accuracy of 79.0 % and 78.6 %, respectively, but with a smaller F1-score at 80.0 % and 81.6 %.
However, we believe that the large increase in computation time and number of learnable parameters does not
justify the use of GRU-LSTM over a simple ANN.
1 INTRODUCTION
Major Depressive Disorder (MDD) is a common men-
tal disorder characterized by reduced production of
certain neurotransmitters in the brain that affects 10
% of the population (Gao et al., 2018). Patterns de-
scribed by Murray et al. in (Murray et al., 2012) show
that depression is consistently on the rise as a preva-
lent cause of morbidity or disability and its effects in-
clude but are not limited to, memory loss, irritabil-
ity, loss of interest, disordered sleep (insomnia or hy-
persomnia) and eating (weight loss or gain), tiredness
and lethargy, anxiety, reduced cognitive and/or mo-
tor performance, feelings of inadequacy, inability to
concentrate, self-harm or suicidal ideation or attempt,
and unexplained physical pain (Strock, 2002).
Obstructive Sleep Apnea Syndrome (OSAS) is a
condition characterized by cessation of breathing dur-
ing sleep specifically due to airway blockages pri-
marily caused by muscles, mainly the genioglossus.
Though OSAS is not as prevalent as depression and
has vastly differing causes, it can still occur in pa-
a
https://orcid.org/0000-0003-4977-355X
tients with depression, or vice versa. It is thus not un-
likely that a selected dataset of OSAS patients would
include those with depression as well, as is the case
in our previous works and this current one (Moussa
et al., 2022). Though depression was an important
part of these previous works, OSAS was the main fo-
cus and depression was classified as a comorbidity.
From the literature, we know sleep apnea and hy-
popnea are correlated with lower quality of life in
general including in large part psychological health.
That is to say depression is relatively prevalent in
people who suffer from OSAS (Yue et al., 2003;
Bj
¨
ornsd
´
ottir et al., 2016; Ejaz et al., 2011). In one
of the aforementioned works, Yue et al. found that
the 30 patients suffering from sleep apnea and hypop-
nea have higher scores for depression with a t-value
of 2.62 (P ¡ 0.05) (Yue et al., 2003).
In the literature, we have seen plenty of works
wherein the authors use electrophysiological signals,
such as ECG (Zang et al., 2022) or EEG (Mumtaz
et al., 2018; Hosseinifard et al., 2013) in classify-
ing major depressive disorder or depression. The
use of varying machine learning algorithms played
a critical role in classification in these works, which
Moussa, M., Alzaabi, Y. and Khandoker, A.
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep Learning Structures Worth It?.
DOI: 10.5220/0011655600003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 203-211
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
203

Figure 1: Summary of data extraction and feature selection, and representations of the A) random forest, B) 3-ANN, and C)
GRU-LSTM. The red crosses and subset symbol over the signal labels represent feature selection - no features selected from
ECG and breathing signals, and only 6 features selected from the EEG.
further supports our solution. Zang et al. use raw
ECG recordings of 74 subjects as input to their CNN
and obtain an accuracy of 93.96 %, a sensitivity of
89.43 %, a specificity of 98.49 %, and an F1-score
of 93.67 % (Zang et al., 2022). Mumtaz et al. use
support vector machines (SVM), logistic regression
(LR), and Naive Bayes (NB) with EEG synchroniza-
tion likelihood (SL) features from 64 subjects. They
obtained the best results at an accuracy of 98.00 %,
a sensitivity of 99.9. %, a specificity of 95.00 %
and an F1-score of 97.00 % with SVM with 10-fold
cross-validation (Mumtaz et al., 2018). Hosseinifard
et al. use K
th
Nearest Neighbor (KNN), linear dis-
criminant analysis (LDA), and LR with features like
average band powers, detrended fluctuation analysis
(DFA), Higuchi fractal dimension, correlation dimen-
sion and Lyapunov exponent extracted from the EEG
data of 90 subjects to diagnose MDD. Logistic Re-
gression yielded the best performance at an accuracy
of 90.00 %. They had used 2/3 of their set for train-
ing and leave-one-out cross-validation(Hosseinifard
et al., 2013).
A common thread among these discussed works
aside from the use of electrophysiological signals is
their goal; the authors aim to diagnose depression.
Our goal, and thus contribution, differs slightly, since
we focus only on depression in subjects we know
suffer from OSAS forming a novel dataset (Moussa
et al., 2022).
Figure 1 gives an abstract idea of our methodol-
ogy, as well as the architectures/algorithms we used
in our work. The contribution in our work lies
mainly in classification of depression in OSAS with
the novel dataset via machine learning and a simple
deep learning architecture, and gauging what would
make switching to deep learning worth the increase in
computational cost and subsequently, physical cost.
2 METHODOLOGY
2.1 Dataset and Processing
Seeing the extensive use of electrophysiological sig-
nals for classification of depression, we elected to use
electroencephalography (EEG), electrocardiography
(ECG), and breathing signals for that purpose. We fo-
cus particularly on depression in subjects that are suf-
fering from OSAS, so while our results may not nec-
essarily be applicable to the general population, they
can provide a suitable baseline for OSAS patients. For
the purpose of detecting depression alone in OSAS
patients, we use a subset of the dataset described in
our previous work (Moussa et al., 2022); instead of
using the electrophysiological signals of 118 subjects,
we use that of 80 subjects. These 80 subjects consist
of 40 with depression and OSAS and 40 with OSAS
alone, collected from the American Center of Psy-
chiatry and Neurology (ACPN) in Abu Dhabi, UAE,
meaning we omit the 6 healthy subjects from this set
and the 32 supplementary healthy subjects from the
STAGES dataset (Zhang et al., 2018). The 80 sub-
jects selected only from the original study, excluding
the STAGES healthy subjects, consist of 48 male sub-
jects and 32 female, all UAE Nationals between the
ages of 20 and 66 with a mean age of 44.2 ± 10.9
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
204

years-old at the time of the study. This study was ap-
proved by the Institutional Review Board (IRB) of the
ACPN on the 2
nd
of October, 2017 with IRB reference
number 0019.
Among the 80 subjects, 2 had an Apnea-
Hypopnea Index (AHI) less than 5, 27 had an AHI
between 5 and 15, 27 had an AHI between 15 and
30, and 24 had an AHI above 30. Since we know
the status of both depression and apnea, we can train
supervised machine learning models to classify our
subjects into one of two classes: depressed or not de-
pressed, both with OSAS. We can also better partition
them according to AHI, sleep stage, and depression
status to further investigate the effects of certain con-
ditions on classification performance in other works,
as we did for sleep stages in (Moussa et al., 2022).
As previously stated, we primarily use EEG, ECG,
and breathing signals, namely airflow, oxygen satura-
tion, and thoracic effort, in addition to other informa-
tion ”signals”, such as the hypnogram detailing sleep
stages. These are not the only recorded signals, how-
ever. The subjects undergo overnight polysomnog-
raphy, which conventionally include the aforemen-
tioned signals in addition to chin and leg electromyo-
graphy (EMG), electrooculography (EOG) for both
eyes, and abdominal effort. Chin EMG (Al-Angari,
2008; Moradhasel et al., 2021) could pave the way for
better detection of OSAS due to the more direct causal
effect between the condition and dilator muscles, and
could even facilitate the use of sensors directly with
the genioglossus muscle instead of chin placement.
The main five signals are recorded by means of
an 8-channel EEG cap for brain signals, an ECG for
heart signals, a spirometer for airflow, a pulse oxime-
ter for oxygen saturation, and a piezoelectric belt for
thoracic movement. The EEG channels used are O1,
O2, C3, C4, F3, and F4 with A1 and A2 according to
the 10-20 convention, as shown in Figure 2, and the
other signals are recorded via standard leads/sensors
and standard lead/sensor placement.
After obtaining the signals, some processing
would be required to ensure the data is clean and
ready for feature extraction, selection, and eventually,
classification. Since the EEG, ECG, and breathing
signals are sampled at 200 Hz, 100 Hz, and 10 Hz,
respectively. The EEG and ECG are also put through
a 50 Hz Notch filter to remove the power-line inter-
ference and all three signals are put through band-
pass filters in previous work to be published by Yahya
Alzaabi; the breathing signals and ECG at 0.1-0.4 Hz
and the EEG at 0.5-30 Hz to keep beta, theta, alpha,
and delta waves. Following filtering, the signals are
split into 5-minute intervals selected manually by in-
spection mainly based on whether or not an apnea has
Figure 2: EEG electrode configuration in which the green
electrodes are those used.
occurred, so as to avoid artifacts. This results in a
total of 1,424 intervals or observations from the 118
subjects, of which 1,005 observations are from our 80
subjects. For each of these observations, we compute
a set of 34 features, 24 from EEG signals, 6 from the
ECG signals/heart rate variability (HRV), 1 directly
from airflow, and 3 from the interaction between air-
flow and ECG/HRV, or more specifically R-R interval
(RRI) signals. The EEG features are simply average
powers extracted for each brain wave from each elec-
trode, with the exception of the reference electrodes,
the ECG features include the average very low fre-
quency, low frequency, and high frequency powers, a
normalized set of the latter two, and the ratio/division
between the latter two. The singular breathing sig-
nal/airflow feature is the respiratory frequency, and
the remaining three features are the respiratory sinus
arrhythmia (RSA), the normalized RSA, and the time-
dependent phase coherence between RSA and airflow
(phases extracted via Hilbert transform), also known
as lambda (λ).
After taking care of noise with filtering and man-
ual selection of intervals and extracting our feature
set, we fill in missing values using shape-preserving
piecewise cubic spline interpolation (Fritsch and Carl-
son, 1980; Kahaner et al., 1989), also known as
Pchip, then follow that by Softmax normalization,
Box-Cox transform (Box and Cox, 1964) to ensure
normal probability distribution, and z-score normal-
ization (Moussa et al., 2022). These processing steps
are described in Equations 1-3, where Data1 is the
Softmax normalized data, Data2 is Data1 with prob-
ability distribution made approximately normal, and
DataFinal is centered and standardized Data2. Box-
Cox transform is a non-linear power transform that
makes the data probability distribution approximately
normal by finding an optimal value of an exponent (λ)
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep Learning Structures Worth It?
205

that results in the best normal distribution approxima-
tion. Looking at Equation 2, we can conclude that
Box-Cox transformation would require the input data
to be positive, which we achieve via Softmax normal-
ization before applying the power transform.
Data1 =
1
1 + exp
mean(Data)−Data
std(Data)
(1)
Data2(λ) =
(
Data1
λ
−1
λ
, i f λ ̸= 0
log(Data1), i f λ = 0
(2)
DataFinal =
Data2 − mean(Data2)
std(Data2)
(3)
Despite extracting 34 features for each observa-
tion, we do not use the full feature set in this work.
As we have seen in (Moussa et al., 2022), using
Chi
2
to select features whose importance score is
greater than or equal to the average feature impor-
tance score, along with the bi-layer artificial neu-
ral network (ANN) yielded the best classification re-
sult for depression compared to other feature selec-
tion algorithms including sequential feature selec-
tion, neighborhood and principal component analysis,
maximum relevance minimum redundancy and Reli-
efF algorithms, so we opt to directly apply Chi
2
in
feature selection, ending up with six features out of
the thirty four. The six selected features by this tech-
nique are all extracted from EEG signals, also surpris-
ingly from only two channels. These features include
the average powers of beta, theta, and alpha waves
from channels F3 and F4. In the context of feature se-
lection on MATLAB, the function examines whether
each of our 34 features is independent of the depres-
sion status using individual Chi
2
tests. The score out-
put from this function is the negative of the common
logarithm of the p-value, and we know a small p-value
indicates that the corresponding feature is dependent
on the label is an important feature. This score would
approach infinity as the p-value approaches zero. Our
analysis concluded that the aforementioned six fea-
tures have an infinite score, hence were selected as
our features.
Now that signal processing has concluded, we
have a clean dataset of 1,005 observations each with
six features with an approximately normal probability
distribution and no missing values. The 80 subjects
are then split into two sets, one for training and 10-
fold cross-validation and comprises the observations
of 75 % of the subjects, and the other set for testing
and comprises the observations of the remaining 25 %
of the subjects. The labels are likewise partitioned in
the same manner, culminating in a partitioned dataset
ready to be input to machine learning algorithms.
2.2 Classifiers and Performance
Evaluation
As we saw in Section 1, machine learning is com-
monly used in detecting depression in the literature,
due to its automated nature, the simplicity of its met-
rics, and the insights it could help us derive regard-
ing the nature of the condition, the widely established
methods of diagnosing depression, or the features
used in classification. In addition, it has social ben-
efits as it reduces the need for human interaction in
diagnosis.
Artificial neural networks (ANNs) and deep learn-
ing techniques use the back-propagation algorithm to
minimize a loss function, and to automatically extract
features with the major difference being an added
function or layer. In convolutional neural networks,
the added function would be convolutional layers,
which, as their name suggests, convolve the input to
reduce its size, producing a smaller feature map. In
gated recurrent unit long short-term memory (GRU-
LSTM or GRU) networks, the added function(s) are
an update and reset gates that control the flow of in-
formation (Erdenebayar et al., 2019).
As we have previously tested out numerous classi-
fiers in (Moussa et al., 2022), we opt to directly com-
pare the best-performing model in that work (ANN),
with a deep learning technique- a GRU-LSTM net-
work, and getting the results with random forest as
some form of baseline. This is because random
forests are known for their generally robust perfor-
mance and relative simplicity compared to deep learn-
ing techniques. The random forest (RF) used was the
same as the previous work; bagged trees with sur-
rogate decision split and 200 learning cycles. How-
ever, some changes were made to the ANN model
to better optimize it for the problem. The model,
named 3-ANN, now consists of three hidden layers
instead of two with 100 units each and a regulariza-
tion term (lambda) of 0.01 instead of 0 in between
the input and output layers. The GRU-LSTM model
is new, as it would have been difficult to employ
prior to hardware upgrade from a machine with the
Nvidia GTX 1050Ti GPU to one with RTX 3080,
and consists of a total of 15 layers, as shown in Fig-
ures 1 and 3. These layers begin with a feature in-
put layer of size 6 × Number of training samples,
followed by a gated recurrent unit of size 10 and a
40 % dropout layer. Afterwards, we have three fully
connected layers with 50 units with batch normaliza-
tion and reLu following each one. Then finally, we
have our ”output” layer, which consists of a fully con-
nected layer with 2 units, a SoftMax layer, and the
actual output layer, since our entire analysis and net-
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
206
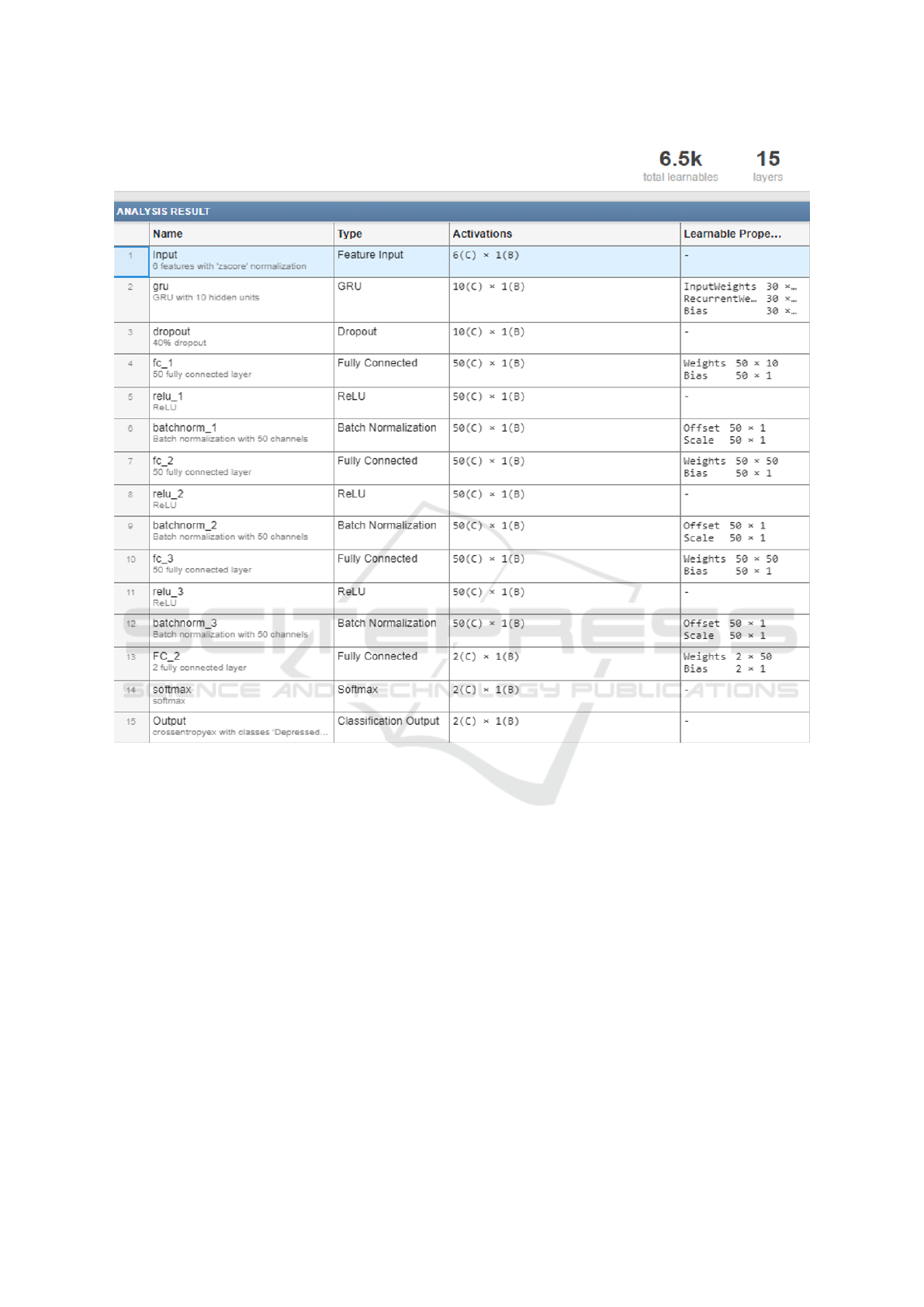
Figure 3: Layer descriptions and number of learnable parameters of the GRU-LSTM model. Each layer has the number of
units under its name and/or any additional options (i.e. normalization, dropout, number of channels).
work design are done on MATLAB. The weights are
initialized via the Glorot initializer (Glorot and Ben-
gio, 2010). The 3-ANN uses the Limited-memory
Broyden–Fletcher–Goldfarb–Shanno (LBFGS) algo-
rithm in training while the GRU uses stochastic gra-
dient descent with a momentum of 0.8. The addi-
tional training options for the GRU include a mini-
batch size of 32, a fixed learn rate of 0.01, an L2-
regularization term of 0.005, a validation frequency
of 1 (each epoch), and a maximum number of epochs
of 1000. Figure 3 shows a brief description of each
layer and gives an idea about the number of associ-
ated computations.
We use 10-fold cross-validation, as stated earlier,
to ensure our models do not overfit. With the deep
learning model on MATLAB, this is implemented by
training a model using 9 folds and the last fold for val-
idation, and repeating while changing the validation
fold until all folds have been used for validation, and
then the model with the best validation performance
is taken. With the other two models, the implemen-
tation on MATLAB is more automatic than having to
select a model based on validation performance algo-
rithmically.
After the models are trained and validated, we
measure their performance with the testing set. Clas-
sification performance is measured by accuracy, sen-
sitivity, specificity, precision, F1-score, the area un-
der receiver operating characteristics (ROC) curve
(AUC), Cohen’s κ coefficient (Cohen, 1960), and
Matthews correlation coefficient (Matthews, 1975).
Accuracy measures how many instances were cor-
rectly classified, sensitivity measures the number of
instances correctly classified positive out of the actual
positive instances, specificity measures the number of
instances correctly classified negative out of the ac-
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep Learning Structures Worth It?
207
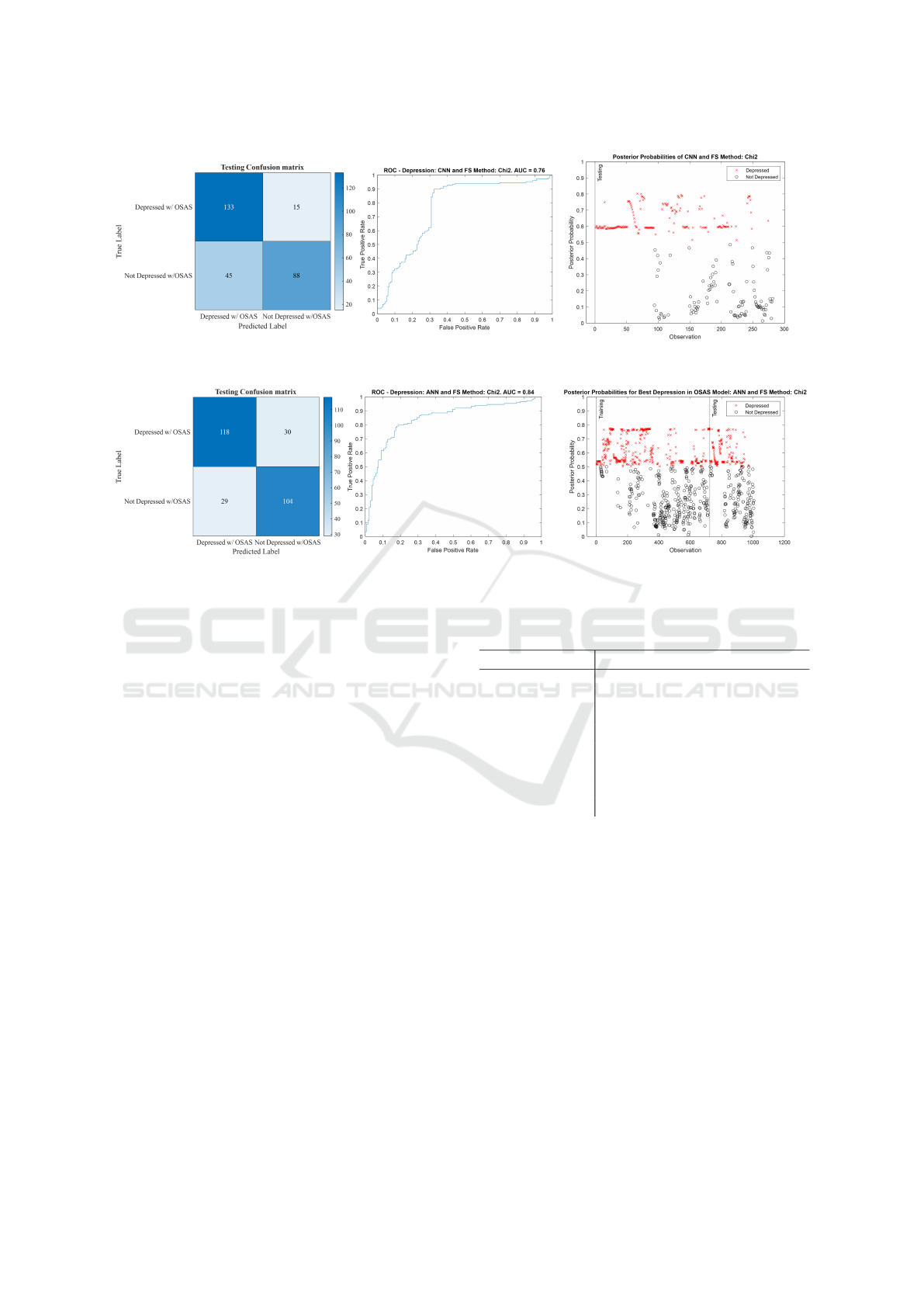
Figure 4: Testing confusion matrix, receiver operating characteristics (ROC) curve, and posterior probability plot of the 3-
ANN model.
Figure 5: Testing confusion matrix, receiver operating characteristics (ROC) curve, and posterior probability plot of the GRU
model.
tual negative instances, precision measures the num-
ber of correctly classified positive instances out of the
total number of classified positive instances, and F1-
score is a harmonic mean defined as twice the prod-
uct of sensitivity and precision divided by their sum.
The AUC is a measure of class separability or essen-
tially how useful the model is at distinguishing the
classes, whereas Cohen’s κ coefficient is a measure of
how much the model’s accuracy is better than chance
based on class distribution, and Matthews correlation
coefficient is a correlation coefficient similar to the
F1-score, and is generally known as the most infor-
mative measure of the quality of a binary classifier.
3 RESULTS AND DISCUSSION
As stated earlier, we compute the testing accuracy,
sensitivity, specificity, precision, F1-score, AUC, Co-
hen’s κ coefficient, and Matthews correlation coeffi-
cient (MCC) for the three classifiers. Although we
compute all metrics, we mainly look at the accuracy,
F1-score, κ, and MCC in comparison in order to make
a conclusion regarding the best classifier for detecting
depression in OSAS patients with our dataset and pro-
cessing steps.
Figure 4 and Figure 5 show the testing confusion
matrix, ROC, and posterior probability plots of both
Table 1: Testing performance of the three classifiers in clas-
sification of depression in OSAS patients.
Model RF 3-ANN GRU-LSTM
AUC 0.71 0.84 0.76
Accuracy (%) 67.6 79.0 78.6
Sensitivity (%) 56.8 79.7 89.9
Specificity (%) 79.7 78.2 66.2
Precision (%) 75.7 80.3 74.7
F1-Score (%) 64.9 80.0 81.6
κ 0.36 0.58 0.57
MCC 0.37 0.58 0.58
of these models to better visualize the difference in
performance. Table 1 shows comparable performance
between the 3-ANN and the GRU-LSTM and shows
both beating the random forest model in all metrics
but specificity. The 3-ANN has a higher AUC, accu-
racy, specificity, precision, and κ than the GRU, but
the GRU has a higher sensitivity, F1-score and they
both have almost the same value of the Matthews cor-
relation coefficient.
The reasons the performance of the two neural
network models is similar could include the relatively
small size of the available dataset, the use of only
6 out of the 34 features, the simplicity of the se-
lected features, or the simplicity of the supposedly
more complex model (GRU). The first reason is sim-
ple enough; artificial neural networks generally re-
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
208
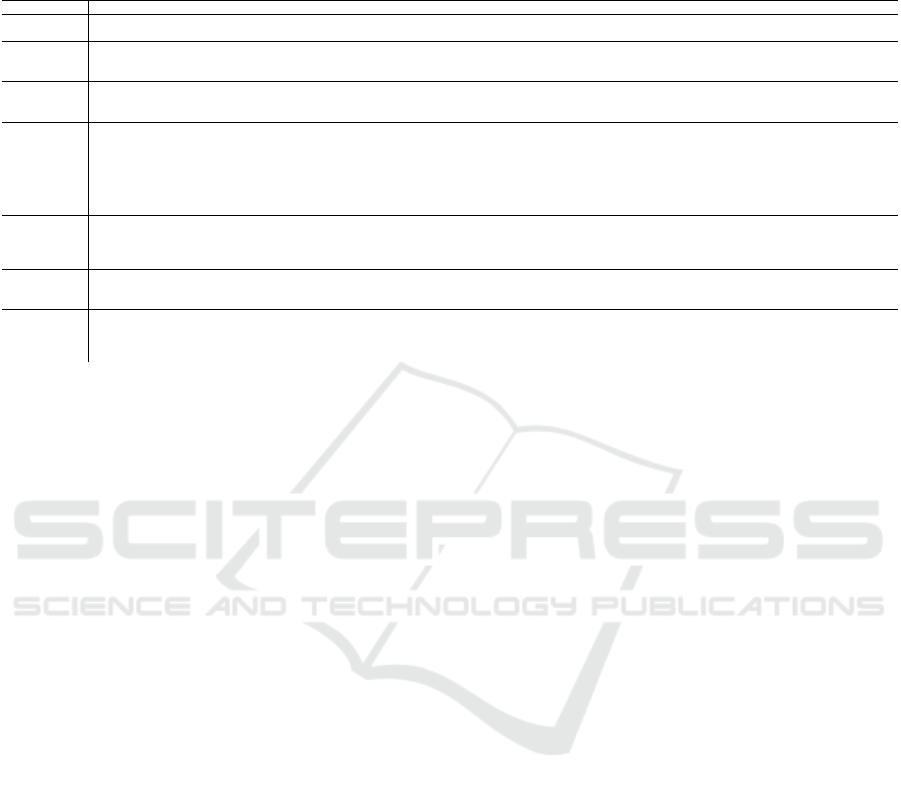
Table 2: Comparison between our model and works focused on detecting depression. OSAS: Obstructive Sleep Apnea Syn-
drome, EEG: Electroencephalography, ECG: Electrocardiography, ANN: Artificial Neural Networks, NB: Naive Bayes, LR:
Logistic Regression, KNN: K-th Nearest Neighbor, SVM: Support Vector Machine, RF: Random Forest, CNN: Convolutional
Neural Network, GRU-LSTM: Gated Recurrent Unit Long Short-Term Memory Network, LDA: Linear Discriminant Analysis.
Work (Zang et al., 2022) (Mumtaz et al., 2018) (Hosseinifard et al., 2013) Proposed Method
Main Objective Classify Depression Classify Depression
Classify MDD
Classify Depression in
OSAS patients
Dataset
74 subjects’
raw ECGs
64 subjects’
EEGs
90 subjects’
EEGs + 4 non-linear
features
1,005 observations extracted
from EEG, ECG, and
breathing signals of 80 subjects
Machine
Learning
Algorithms
CNN
LR, SVM,
and NB
KNN,
LDA,
and LR
Random Forest, 3- ANN,
GRU-LSTM
Significance
Simplicity of
methodology: The authors
use raw ECG signals
with CNNs in their analysis
Thorough analysis for
some classic machine
learning algorithms and
features used are promising
The authors present a thorough
description of a robust methodology
to classify depression in general,
describing in detail their features,
machine learning models and cross-
validation schemes, as well as their
novel dataset
Compares best
depression in OSAS classification
method in (Moussa et al., 2022) with deep learning
Limitations
Using CNNs with raw signals
is inconvenient in
resource-restricted environments
No significant limitations found,
though we would be interested to see
how this setup performs with other datasets
Only the accuracy
is reported
Deep learning not thoroughly
explored, and no automatic
hyperparameter optimization via grid-
search or Bayesian optimization
Best Model
Convolutional
Neural
Network
Support Vector
Machines
Logistic Regression 3-ANN
Accuracy (%) 93.96 SVM: 98.00 LR: 90.00 ANN:79.00
Sensitivity (%) 89.43 99.90 N/A 79.70
Specificity (%) 98.49 95.00 N/A 78.20
F1-Score (%) 93.67 97.00 N/A 80.00
quire large amounts of data to train most optimally.
This also feeds into the second reason, the features
selected by the Chi
2
algorithm may be too few, and
as we have seen in (Moussa et al., 2022), the ANN
has performed worse with all 34 features, and when
other feature selection algorithms were used, but that
is not an indicator as to how the GRU would perform
with them. That introduces the need to test the GRU
model with the other feature selection configurations
for future work. The third reason does not refer to
the number of the features, but rather the extracted
features to begin with; is the average power of each
EEG channel and brain wave the best singular fea-
ture we could extract? Wavelet decomposition and
entropy, for example, are features extracted from elec-
trophysiological signals seen in literature (Khandoker
et al., 2008; Srinivasulu et al., 2021). The final pos-
sible cause simply refers to the use of few units in
the GRU layer and subsequent fully connected layers.
While the performance of a deep neural network does
not necessarily improve as it gets more complicated,
only one architecture of GRU was explored, even if
the aforementioned parameters are optimized. Using
different architectures, like more GRU layers, adding
LSTM layers, adding pooling layers, changing reLU
into some other activation function, or cascading with
a convolutional network or transformer could all pos-
sibly improve performance. Having the dropout lay-
ers does help with keeping the number of parameters
under control, but increasing still
Due to the similarity of the MCC in particular, and
the closeness of the accuracy, F1-score, and κ values,
we cannot accurately say that one model outclasses
the other for classification of depression in OSAS pa-
tients with this dataset. Instead, we can compare the
resources required for each model and select the opti-
mal one based on the less computationally expensive
and less time-consuming one.
Despite similar performance, we see from Fig-
ure 3 that the number of learnable parameters is
6,500 for the GRU, comparatively smaller than that
of 3-ANN at 21,102 (Weights + Biases: [(6×100) +
(100×100) + (100×100) + (100×2)] + [(100×1) +
(100×1) + (100×1) + (2×1)]). Despite that, it takes
only 77.8 seconds to compute with a NVIDIA 1050
Ti GPU and significantly less with the NVIDIA 3080
GPU, whereas the GRU takes upwards of an hour to
train with the latter. This could be attributed to the
small size of mini-batches coupled with the large it-
eration/epoch limit, and the GRU layer itself. This
makes the 3-ANN more suited for this problem, as it
takes less time to train and is less demanding in terms
of resources. Table 2 compares our work with similar
works in the literature.
4 CONCLUSION
To sum up, the main goal of this work was to classify
depression in OSA patients and investigate whether
using deep learning over classic machine learning
techniques is a worthy endeavor. The dataset included
overnight EEG, ECG, and breathing signal record-
ings from 80 subjects, 40 of which were depressed
with OSAS and 40 were not depressed but had OSAS.
Afterwards, we extract 1,005 intervals from the sig-
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep Learning Structures Worth It?
209

nals depending on the status of obstructive apnea oc-
currence, in addition to depression status and sleep
stage. We then process the data to ensure it is clean,
has an approximately normal distribution, and is z-
score normalized before we partition and input it into
our three classifiers. We train three classifiers using
the intervals or observations of 75 % of the subjects
and perform 10-fold cross-validation on the same set,
then test classifier performance with the data of the
remaining 25 % of subjects. Using the Chi
2
algo-
rithm to select the six most important features and
ANN for classification yielded the best performance
with an accuracy of 79.00 %, F1-score of 80.00 %, a
κ of 0.58, a Matthews correlation coefficient of 0.58
and an AUC of 0.84, while also considering the low
computational cost compared to the GRU-LSTM. The
performance is promising, and we believe further pre-
processing of the data, as well as further optimizing
network architectures and hyperparameters and us-
ing more novel approaches like transformers could
improve classification performance. In addition, im-
plementing explainability metrics, like SHAP and de-
scriptions would certainly make our work more ac-
cessible to clinical personnel, or even laypersons.
ACKNOWLEDGEMENTS
The authors would like to thank the American Cen-
ter for Psychiatry and Neurology (ACPN) in Abu
Dhabi for their invaluable contribution in sharing the
polysomnography data and acknowledge the support
of the biomedical engineering department and the
Healthcare Engineering Innovation Center (HEIC) at
Khalifa University of Science and Technology. The
authors would also like to highlight the importance
of the KAU-KU Joint Research Program, in partic-
ular, project DENTAPNEA between Khalifa Univer-
sity and King Abdulaziz University, in particular the
advice of Dr. Angari, Dr. Balamesh, Dr. Khraibi, and
Dr. Marghalani.
REFERENCES
Al-Angari, H. (2008). Evaluation of chin emg activity at
sleep onset and termination in obstructive sleep apnea
syndrome. In 2008 Computers in Cardiology, pages
677–679. IEEE.
Bj
¨
ornsd
´
ottir, E., Benediktsd
´
ottir, B., Pack, A. I., Arnardot-
tir, E. S., Kuna, S. T., Gislason, T., Keenan, B. T.,
Maislin, G., and Sigurdsson, J. F. (2016). The preva-
lence of depression among untreated obstructive sleep
apnea patients using a standardized psychiatric inter-
view. Journal of Clinical Sleep Medicine, 12(1):105–
112.
Box, G. E. and Cox, D. R. (1964). An analysis of trans-
formations. Journal of the Royal Statistical Society:
Series B (Methodological), 26(2):211–243.
Cohen, J. (1960). A coefficient of agreement for nominal
scales. Educational and psychological measurement,
20(1):37–46.
Ejaz, S. M., Khawaja, I. S., Bhatia, S., and Hurwitz, T. D.
(2011). Obstructive sleep apnea and depression: a re-
view. Innovations in clinical neuroscience, 8(8):17.
Erdenebayar, U., Kim, Y. J., Park, J.-U., Joo, E. Y., and
Lee, K.-J. (2019). Deep learning approaches for au-
tomatic detection of sleep apnea events from an elec-
trocardiogram. Computer methods and programs in
biomedicine, 180:105001.
Fritsch, F. N. and Carlson, R. E. (1980). Monotone piece-
wise cubic interpolation. SIAM Journal on Numerical
Analysis, 17(2):238–246.
Gao, S., Calhoun, V. D., and Sui, J. (2018). Machine learn-
ing in major depression: From classification to treat-
ment outcome prediction. CNS neuroscience & thera-
peutics, 24(11):1037–1052.
Glorot, X. and Bengio, Y. (2010). Understanding the diffi-
culty of training deep feedforward neural networks. In
Proceedings of the thirteenth international conference
on artificial intelligence and statistics, pages 249–
256. JMLR Workshop and Conference Proceedings.
Hosseinifard, B., Moradi, M. H., and Rostami, R. (2013).
Classifying depression patients and normal subjects
using machine learning techniques and nonlinear fea-
tures from EEG signal. Computer methods and pro-
grams in biomedicine, 109(3):339–345.
Kahaner, D., Moler, C., and Nash, S. (1989). Numerical
methods and software. Prentice-Hall, Inc.
Khandoker, A. H., Palaniswami, M., and Karmakar, C. K.
(2008). Support vector machines for automated recog-
nition of obstructive sleep apnea syndrome from ECG
recordings. IEEE transactions on information tech-
nology in biomedicine, 13(1):37–48.
Matthews, B. W. (1975). Comparison of the predicted and
observed secondary structure of t4 phage lysozyme.
Biochimica et Biophysica Acta (BBA)-Protein Struc-
ture, 405(2):442–451.
Moradhasel, B., Sheikhani, A., Aloosh, O., and Dabanlou,
N. J. (2021). Chin electromyogram, an effectual and
useful biosignal for the diagnosis of obstructive sleep
apnea. Journal of Sleep Sciences, 6(1-2):32–40.
Moussa, M. M., Alzaabi, Y., and Khandoker, A. H. (2022).
Explainable computer-aided detection of obstructive
sleep apnea and depression. IEEE Access, 10:110916–
110933.
Mumtaz, W., Ali, S. S. A., Yasin, M. A. M., and Malik,
A. S. (2018). A machine learning framework involv-
ing EEG-based functional connectivity to diagnose
major depressive disorder (mdd). Medical & biologi-
cal engineering & computing, 56(2):233–246.
Murray, C. J., Vos, T., Lozano, R., Naghavi, M., Flax-
man, A. D., Michaud, C., Ezzati, M., Shibuya, K.,
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
210

Salomon, J. A., Abdalla, S., et al. (2012). Disability-
adjusted life years (dalys) for 291 diseases and injuries
in 21 regions, 1990–2010: a systematic analysis for
the global burden of disease study 2010. The lancet,
380(9859):2197–2223.
Srinivasulu, A., Mohan, S., Harika, T., Srujana, P., and Re-
vathi, Y. (2021). Apnea event detection using ma-
chine learning technique for the clinical diagnosis of
sleep apnea syndrome. In 2021 3rd International
Conference on Signal Processing and Communication
(ICPSC), pages 490–493. IEEE.
Strock, M. (2002). Depression. national institute of mental
health. Technical Report 02-3561, NIH Publication.
Yue, W., Hao, W., Liu, P., Liu, T., Ni, M., and Guo,
Q. (2003). A case—control study on psychological
symptoms in sleep apnea-hypopnea syndrome. The
Canadian Journal of Psychiatry, 48(5):318–323.
Zang, X., Li, B., Zhao, L., Yan, D., and Yang, L. (2022).
End-to-end depression recognition based on a one-
dimensional convolution neural network model using
two-lead ECG signal. Journal of Medical and Biolog-
ical Engineering, pages 1–9.
Zhang, G.-Q., Cui, L., Mueller, R., Tao, S., Kim, M.,
Rueschman, M., Mariani, S., Mobley, D., and Red-
line, S. (2018). The national sleep research resource:
towards a sleep data commons. Journal of the Amer-
ican Medical Informatics Association, 25(10):1351–
1358.
Depression in Obstructive Sleep Apnea Patients: Is Using Complex Deep Learning Structures Worth It?
211
