
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes
Jonas Schurr
1,2 a
, Andreas Haghofer
1,2 b
, Marian F
¨
ursatz
3,4 c
, Hannah Janout
1,2 d
,
Sylvia N
¨
urnberger
3,4,5 e
and Stephan Winkler
1,2 f
1
Bioinformatics Research Group, University of Applied Sciences Upper Austria, Softwarepark 11–13, Hagenberg, Austria
2
Department of Computer Science, Johannes Kepler University, Altenberger Straße 69, Linz, Austria
3
Department of Orthopedics and Trauma-Surgery, Division of Trauma-Surgery, Medical University of Vienna, Austria
4
Ludwig Boltzmann Institute for Traumatology, The Research Center in Cooperation with the AUVA, Vienna, Austria
5
Austrian Cluster for Tissue Regeneration, Vienna, Austria
Keywords:
Heuristic Optimization, Machine Learning, Image Processing, Spheroids, High Throughput Screening.
Abstract:
Cell Spheroids are of high interest for clinical cell applications and cell screening. To allow the extraction
of early readout parameters a high amount of image data of petri dishes is created. To support automated
analyses of spheroids in petri dish images we present a method for analysing and quantification of spheroids in
its development stages. The algorithm is based on multiple image processing algorithms and neural networks.
With an evolutionary strategy, engraved grid cells on petri dish are extracted and on top a Unet is used for the
segmentation and quantification of different cell compartment states. The measured f1-scores for the different
states are 0.77 for monolayer grid cells, 0.86 for starting formation grid cells and 0.85 for spheroids. As we
describe in this study we can provide thorough analyses of cell spheroid in petri dishes, by automating the
quantification process.
1 INTRODUCTION
Cell spheroids are an important 3D model for in vitro
testing and are gaining interest for their use in clini-
cal applications. Especially for chondrogenic differ-
entiation, this is the golden standard model system
(Zhang et al., 2010; Johnstone et al., 1998). How-
ever, its drawbacks encompass a large consumption
of time and reagents in addition to lacking early read-
out parameters, limiting its usefulness as a screen-
ing system. To remedy this, a petri dish system was
created, allowing for the creation of many spheroids
within one vessel while reducing media consumption
via self-assembly from cell monolayers (F
¨
ursatz et al.,
2021). This was achieved by subdividing the growth
surface of standard cell culture treated petri dishes us-
ing CNC-guided CO2 laser engraving. The engraved
surface yielded anti-adhesive properties confining cell
attachment only to the compartments and leading to
self-assembly over time. This self-assembling pro-
a
https://orcid.org/0000-0001-8536-8354
b
https://orcid.org/0000-0001-6649-5374
c
https://orcid.org/0000-0003-4990-3326
d
https://orcid.org/0000-0002-0294-3585
e
https://orcid.org/0000-0002-5175-5118
f
https://orcid.org/0000-0002-5196-4294
cess can be monitored over time using macroscopic
and microscopic imaging, and the resulting formation
kinetics can be used to assess the effect of treatments
(e.g. anti-inflammatory compounds) due to treatment
depending change in formation behavior. However,
due to the amount of image data and spheroids per
well, automated detection of formation state is nec-
essary to allow for efficient use of data. While the
detection of fully formed spheroids would already
give a good measurement of the formation progress,
the detection of starting and not fully completed
formation refine this data further. To achieve this,
macroscopic images of petri dish plates seeded with
adipose-derived stem cells (1x10
6
cell/plate) were ob-
tained over the time course of the spheroid forma-
tion using an Olympus OM-D E-M1 digital camera
with an Olympus M.Zuiko Digital ED 60mm 1:2.8 46
makro objective, fixed to a macrostrand at a distance
of ∼230 mm. The formation process was separated
into four separate classes to describe the current state
of self-assembly:
• (1) Monolayer describes a compartment with a
completely attached cell layer.
• (2) starting formation describes a compartment
where >50 % of the cell layer remains attached.
• (3) late-stage formation describes a compartment
78
Schurr, J., Haghofer, A., Fürsatz, M., Janout, H., Nürnberger, S. and Winkler, S.
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes.
DOI: 10.5220/0011648700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 2: BIOIMAGING, pages 78-85
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

where <50 % of the cell layer remains attached.
• (4) fully formed describes a completely detached
and condensed spheroid. Due to the detachment,
this class can also translocate to other compart-
ments.
The differentiation between these groups is impor-
tant as it allows a closer insight into the current for-
mation state and, therefore a better characterization
and possible earlier read-out of treatment effects com-
pared to solely detecting fully formed spheroids.
Improvements in cell spheroid in vitro testing se-
tups and screening systems with petri dishes causes
new challenges for subsequent data analysis and fast
data extraction. Improvements in technology enable a
faster generation of image data due to simpler setups
and a higher experiment frequency. To analyse a large
amount of images An algorithm that provides auto-
matic extraction allows more efficient data analyses,
a faster overall experiment execution and increases
objectivity, reproducibility and comparability. Many
essential steps like localization and quantification are
performed manually or with the help of multiple al-
gorithms carried out in different software like ones
of microscope manufacturers or ImageJ. While these
often deliver a wide range of algorithm, many subse-
quent steps still need to be carried out manually or re-
quire expert knowledge. To allow faster and simple
experiment analyses and information extraction, an
easy-to-use end-to-end workflow for automated anal-
yses are of high interest.
In this work, we show a robust method for the au-
tomatic extraction of the grid structure in petri dishes
and the applicability of automated segmentation for
the classification and subsequent quantification of dif-
ferent types of cell compartments in petri dish im-
ages. Multiple steps are needed to enable automatic
information extraction and subsequent quantification.
The cell compartments are segmented and classified
for the extraction of the classes described above. For
the quantification of monolayer (type 1) and forma-
tion (types 2 and 3), each grid cell of the petri dish
is assigned to cell compartment type. Therefore, an
automated grid extraction to extract single grid cells
is crucial in addition to the segmentation step.
1.1 Data
As shown in Figure 1, the datasets consist of petri dish
images from a digital single-lens reflex camera using
a 60mm F2.8 macro lens. All images provide a color
depth of 24 bits and a dimension of 4608 x 3456 pix-
els. All shown annotations are created using the slide
runner labeling tool (Aubreville et al., 2018)
Figure 1: Representation of an example image out of the
used dataset, including the manually labeled objects to be
segmented and classified. The blue marks represent the ini-
tial monolayer class. The yellow and gray marks are used
for the early and late formation of future spheroids. The
green color indicate the final spheroids.
Figure 2: Example of the four stages from the initial mono-
layer (1) to the formation stages of early (2) and late (3)
formations until the final spheroid form (4).
Figure 3: Simplified representation of the quantification
workflow, including the three independent detection steps.
As shown by the individual cells in Figure 2, ini-
tially a monolayer (1) forms within each compartment
of the petri dish. During the ongoing formation pro-
cess, this monolayer transforms into the final spheroid
form (4). The states in between are separated into
early (2) and late (3) formations, which are combined
into one formation class for the workflow in this pa-
per. The whole image dataset contains 20 images
which are divided into three separate sets for train-
ing the segmentation models (12 images), validation
(2 images) and testing (6 images).
2 METHODS
Our presented workflow has a modular design and is
based on three detection steps as shown in Figure 3.
This design decision allows future adjustments of the
used image processing algorithms as well as replace-
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes
79
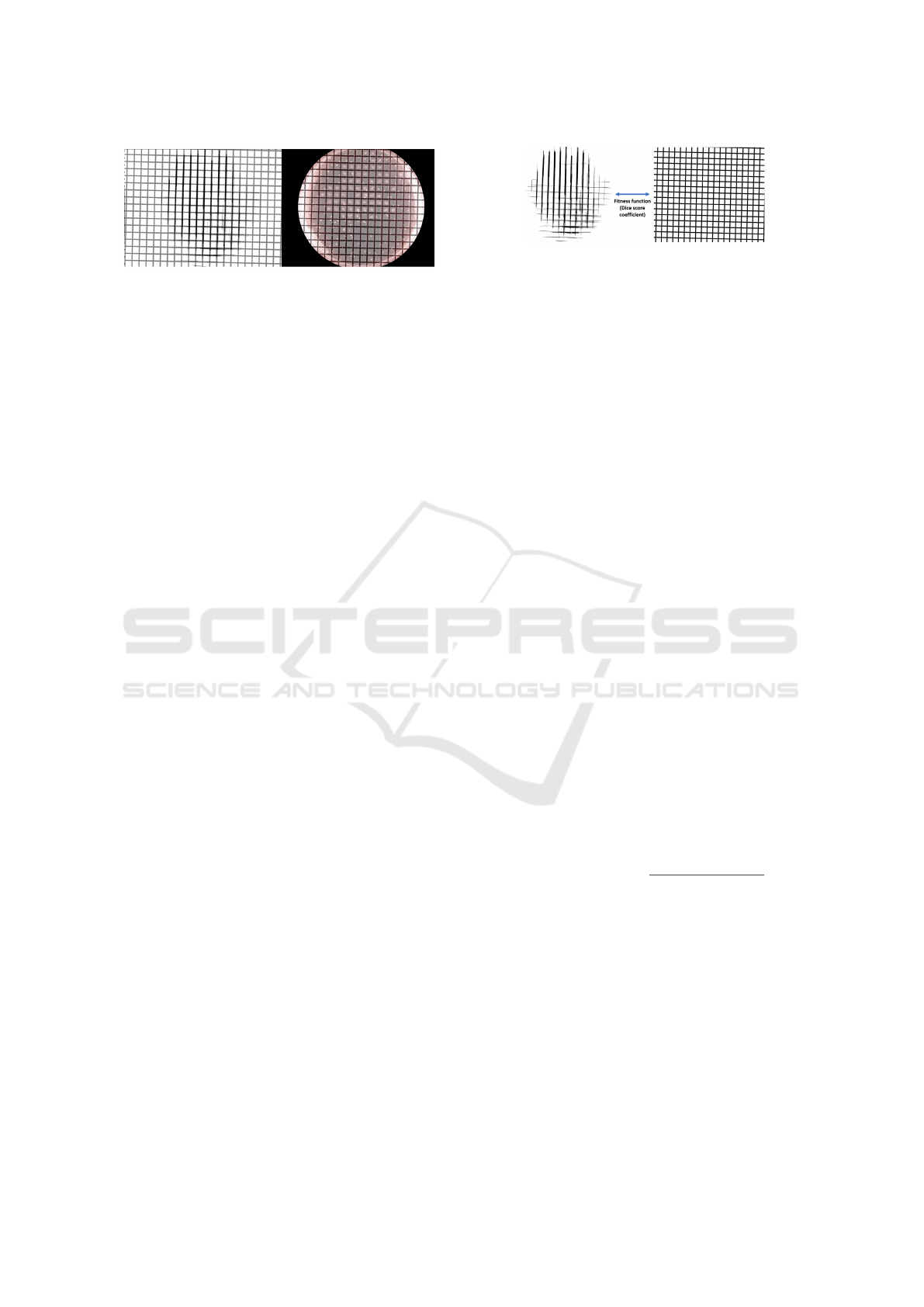
Figure 4: Examples of the grid identification with the
mapped grid in binary (left) used for the fitness function
and the mapped grid in the RGB image (right).
ments of the current segmentation neural networks.
2.1 Heuristic Grid Identification
Grid mapping is performed to analyse single grid cells
in the image of a petri dish. A generated grid mask
maps the underlying grid. This grid mask defines sin-
gle grid cells and is used to extract the single grids
correctly. The grid is optimized with image process-
ing algorithms and an evolutionary strategy.
2.1.1 Preprocessing
Multiple preprocessing steps are performed before the
final grid optimization can be executed. To avoid
possible noise outside of the petri dish and to fur-
ther on enable the identification of grid cells within
it, the petri dish itself is extracted. This is achieved
by hough circles (Barabas et al., 2013; Forero et al.,
2020; Duda and Hart, 1972). The parameters for
blurring, canny edge detection, and the accumulator
threshold of the hough transformation were identified
manually on 14 images. It is presumed that the cen-
ter of the petri dish is within a window of 30% image
length in the middle of the image. As fitness func-
tion of the evolutionary strategy, a binary mask is ex-
tracted automatically from the original RGB image.
This processed binary mask is used as a ground truth
image in the fitness function, which is defined by the
inverted Dice coefficient, whereas the prediction is
the solution candidate representing a mask counting
a predicted grid (as shown in 5. To extract the binary
mask, after CLAHE, to increase the contrast, borders
are extracted with hough lines representing the grid
on the petri dish image (Zuiderveld, 1994; Duda and
Hart, 1972; Hansard et al., 2014). To allow the extrac-
tion of hough lines, a binary image with Roberts edge
detection is created based on the equalized RGB im-
age (Roberts et al., 1965; Bhardwaj and Mittal, 2012).
The connection of multiple extracted hough lines for
one line, is realized by morphological filtering espe-
cially closing. Kernel size is reflected by the size
of the border and is extracted in relation to the im-
age size. The final extracted mask, consists of white
Figure 5: Representative images for Dice coefficient be-
tween preprocessing image used as ground truth (left) and
predicted grid (right).
background and black lines. A too-low minimum line
length increases the amount of noise within the image,
whereas a too high number of minimum line length
misses too many lines. In our evaluations, a mini-
mum length given with 10% of the image height pro-
vides robust results. In the experimental setup, light is
coming from the sides of the images, which increases
the contrast of vertical borders and allows an easier
extraction. Therefore, a lower threshold can be used
optionally for horizontal lines to avoid overlooking
them. With the described steps, a sufficient amount
of horizontal and vertical borders are extracted within
the petri dish, which is important as ground truth for
a proper evaluation of the solution candidates.
2.1.2 Grid Optimization by Evolution strategy
Due to low contrast, shadows and disturbing effects,
not all borders in the image are visible or can be ex-
tracted from the image. Therefore, an evolution strat-
egy is used for the approximation of the full grid and
the prediction of missing borders in the petri dish
(Borgmann et al., 2012). The goal is to use the ex-
tracted lines in the preprocessing step as ground truth
and fit a predicted full grid onto the extracted ground
truth image based on the fitness function. The error of
the fitness function is defined by the Dice coefficient
between the extracted binary images of the prepro-
cessing step containing the extracted lines and a pre-
dicted grid image represented by a solution candidate,
as shown in equation 1.
DiceC oe f f icient =
2 ∗ TP
2 ∗ T P + FP + FN
(1)
Defined by: true positive (TP), false positive (FP) and
false negative (FN). Each solution candidate defines
a grid (see 6). Four adjustable parameters leaned on
affine transformation parameters are used to represent
a grid:
• Rotation: Rotation of the grid lines to the origin.
• Scaling: Width of a single grid cell.
• Horizontal and Vertical offset: Position and trans-
lation of the grid.
For each solution candidate, an image containing the
grid using the parameters is created to evaluate the fit-
ness of the solution candidate. An applicable amount
BIOIMAGING 2023 - 10th International Conference on Bioimaging
80
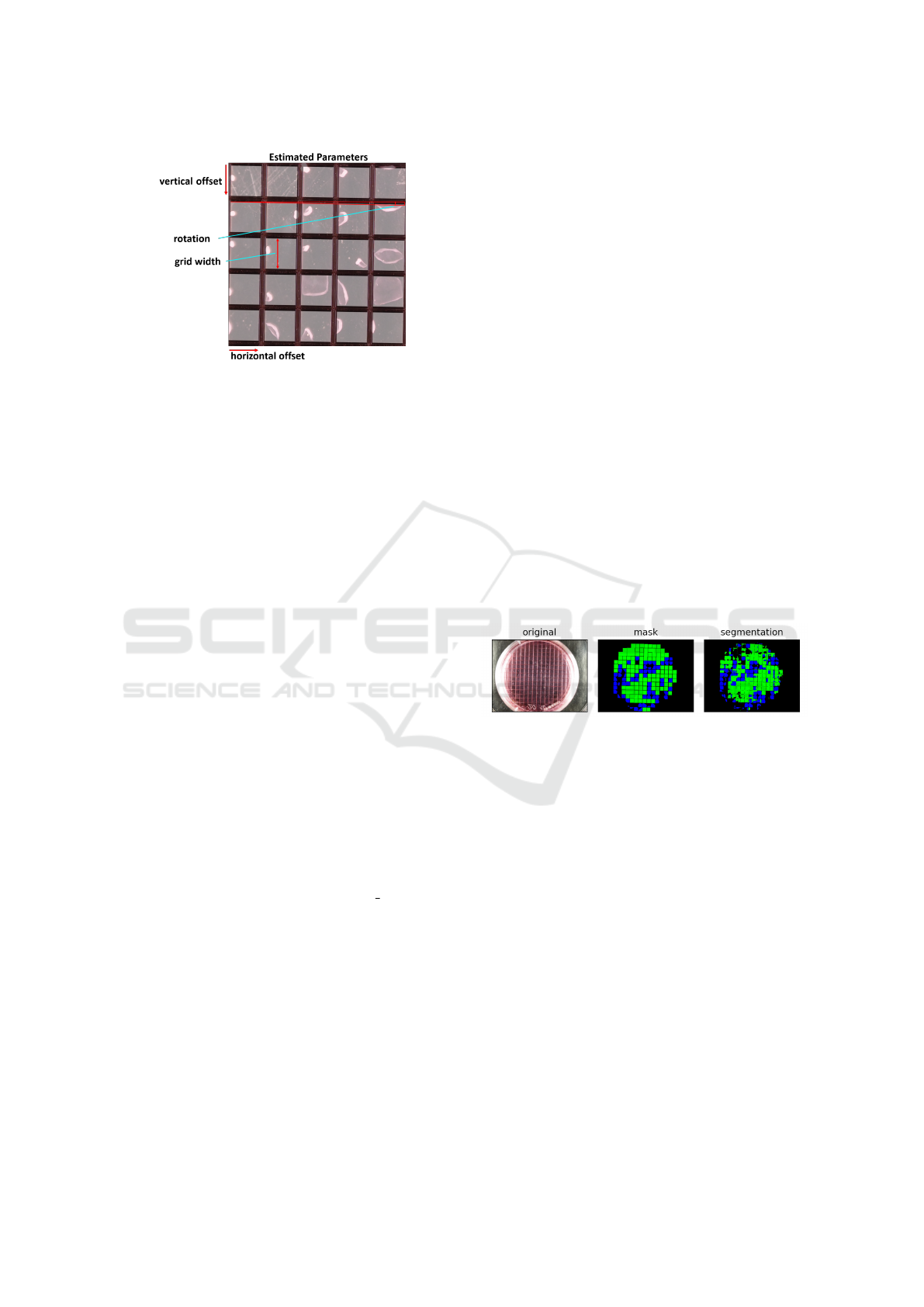
Figure 6: Explanation of solution candidate parameters.
for the population and starting sigma of the evolution-
ary strategy to allow a robust convergence was evalu-
ated. Elitism was used for the creation of new genera-
tions to keep the best candidate until this point. More-
over, the initial search space is reduced by problem-
specific constraints. The maximum width of a grid
cell also defines the maximum offset. Additionally,
the grid cells can not exceed 10% of the image’s
height. The rotation is limited to a maximum of 5
degrees. The evolutionary strategy is used for the op-
timization of the grid prediction and, therefore, the
accurate extraction of every single grid cell.
2.2 Object Detection
2.2.1 Segmentation Models
Descendent from the Unet architecture (Ronneberger
et al., 2015) which represents a specialized artifi-
cial neural network for image segmentation, the used
Unet ++ (Zhou et al., 2018) represents the base ar-
chitecture for our segmentation models. Provided
by the segmentation models package (Iakubovskii,
2019) for the used Pytorch framework these models
were trained with the support of the Pytorch-lightning
framework (Falcon et al., 2019). Out of the several
provided backbones for the Unet ++ , our segmen-
tation models are built with the regnety 120 back-
bone (Radosavovic et al., 2020) which delivered the
best results for our use case. Due to limitations of
the available sample images, our modelling process
also included the use of data augmentation techniques
(Shorten and Khoshgoftaar, 2019) which represents
a commonly used approach for artificially extending
the available amount of image data. In our case, it
was necessary to use image cropping, which was re-
alised by randomly selecting small parts of the images
instead of using the whole image for the input of the
neural network. This technique was used on the train-
ing as well as the validation data. For the training
data, color and contrast adjustments combined with
image distortion, transposing, scaling, and rotation
added some variation in the available image proper-
ties. All in all, it was possible to extend our training
and validation data by a factor of 30. The separated
test data was not changed using any of the mentioned
algorithms. The original dataset without augmenta-
tion included 12 training images, 2 validation images
and 6 test images with a resolution of 4608x3456
pixel. The concept of using two models instead of
one is built up on the idea that the first stages of a
forming spheroid can not move within the petri dish.
2.2.2 Monolayer and Forming Spheroids
Classification
As Figure 7 shows, our segmentation model separates
the monolayers (blue) as well as the currently forming
spheroids (green) from the background represented
by the rest of the petri dish. This segmentation neu-
ral network classifies the corresponding stage for each
pixel without the possibility for the same pixel to be
part of more than one class. This design decision was
made due to the property that these objects can not
overlay each other, which led to the use of a softmax
activation within the final layer.
Figure 7: Example image of the segmentation result (right)
based on the raw RGB image (left) in comparison to the
mask (middle) representing the to be achieved separation
of the two classes. Green represents the already forming
spheroids out of the blue monolayer.
To further improve the accuracy of the prediction
used for a final quantification of monolayer cells and
currently forming spheroid cells, a prediction for ev-
ery single grid cell is performed. The prediction is
based on the segmentation results. Since a predic-
tion of both classes can be within a single grid cell
of the grid a final decision to correctly classify such a
grid cell has to be made. The quantification of each
cell type is allowed by the assignment of the class to
each valid grid cell. Only grid cells within the petri
dish are considered. Therefore, grid cells that are not
fully within the petri dish and grid cells with a high
percentage of background are removed for the eval-
uation since they cannot contain any cell or the cell
is cut and not fully in the image. The estimated grid
defines each grid cell. For each grid cell, a predic-
tion of either one class is performed. If a grid cell
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes
81

contains a prediction for a monolayer, it is assigned
as a monolayer and vice versa for the second class of
forming spheroids. A prediction within a grid cell is
extracted by connected components. If both classes
are predicted within a single grid cell, the monolayer
class is taken if the predicted overall area is larger or
if the confidence of the model is higher in comparison
to the second class. This is suitable since monolayers
are larger by definition.
2.2.3 Spheroid Localization
In contrast to the monolayer and forming spheroid
stages, the final spheroid objects can move around and
overlay other objects. This property could lead to a
pixel with more than one class, which is possible by
using a second model specifically for this class. Nor-
mally, this would be realized by a multiclass segmen-
tation model for all stages at once, but this decision
would inhibit the ability of the workflow to only allow
spheroid objects to overlap other classes. As shown in
Figure 8: Example of the test dataset with an overlay of
the segmentation on the original RGB image. All detected
spheroids are represented by the white dots.
Figure 8 this second neural network only separates the
possible spheroid objects from the background with-
out any knowledge of the other stages. Therefore,
all the other stages are considered as background.
Based on this segmentation result, the actual local-
ization of the individual objects is realized using the
connected components algorithm from the Open CV
Python framework (OpenCV, 2015). After a filtering
step that removes every object smaller than 70 percent
of the average object size, the resulting objects are lo-
cated by their center pixel as assigned to the spheroid
class for the final quantification step. This size filter-
ing has to be done individually for each image to be
segmented by the fact that the object size also relies
on the camera parameters and the used lens. There-
fore, it would not be possible to set a specific size limit
for all now-arriving objects. This 70 percent of the av-
erage size limit was set based on the current dataset.
It still allows some wrong detected objects, but it in-
Figure 9: Comparison of a sufficient and insufficient result
for grid alignment on an image of a petri dish.
hibits nearly all artifacts that are smaller than all valid
spheroids, like wrongly as spheroids detected reflec-
tions or air bubbles. Based on the used imaging setup,
this limit can be adjusted by the user for future appli-
cations. Due to the possibility of the final spheroid
stage merging together with other spheroids, an up-
per size limit is not considered to have any positive
impact on the segmentation quality.
3 RESULTS
3.1 Monolayer & Forming Spheroid
Quantification
3.1.1 Parameter evaluation of Grid Estimation
In Figure 9 the applicability of the evolutionary strat-
egy and its used fitness function is shown. Two differ-
ent results on the same image with its corresponding
fitness score histories can be seen. In the first results
(A.1 and A.2) the fitness is improved over multiple
generations down to a value of 0.034. The low value
is also connected to a good result (as shown in sub-
image A.2). Whereas in the second result (B.1 and
B.2) the error (fitness score) can be reduced slightly
in the first generations but there is no further improve-
ment also after multiple generations. This states a
false optimization in a local optimum and is reflected
in a higher value of 0.093. The higher error is also re-
flected in the corresponding sub-image (B.2), as it can
clearly be seen that the grid is not aligned and even
divides the actual grid cells on the petri dish into two
parts. The second result was generated by an ES with
an insufficient size for the population. The aligned
grid and the converging error over multiple genera-
tions in sub-images A.1 and A.2 show the applicabil-
ity of the used fitness function and also the overall
approach.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
82
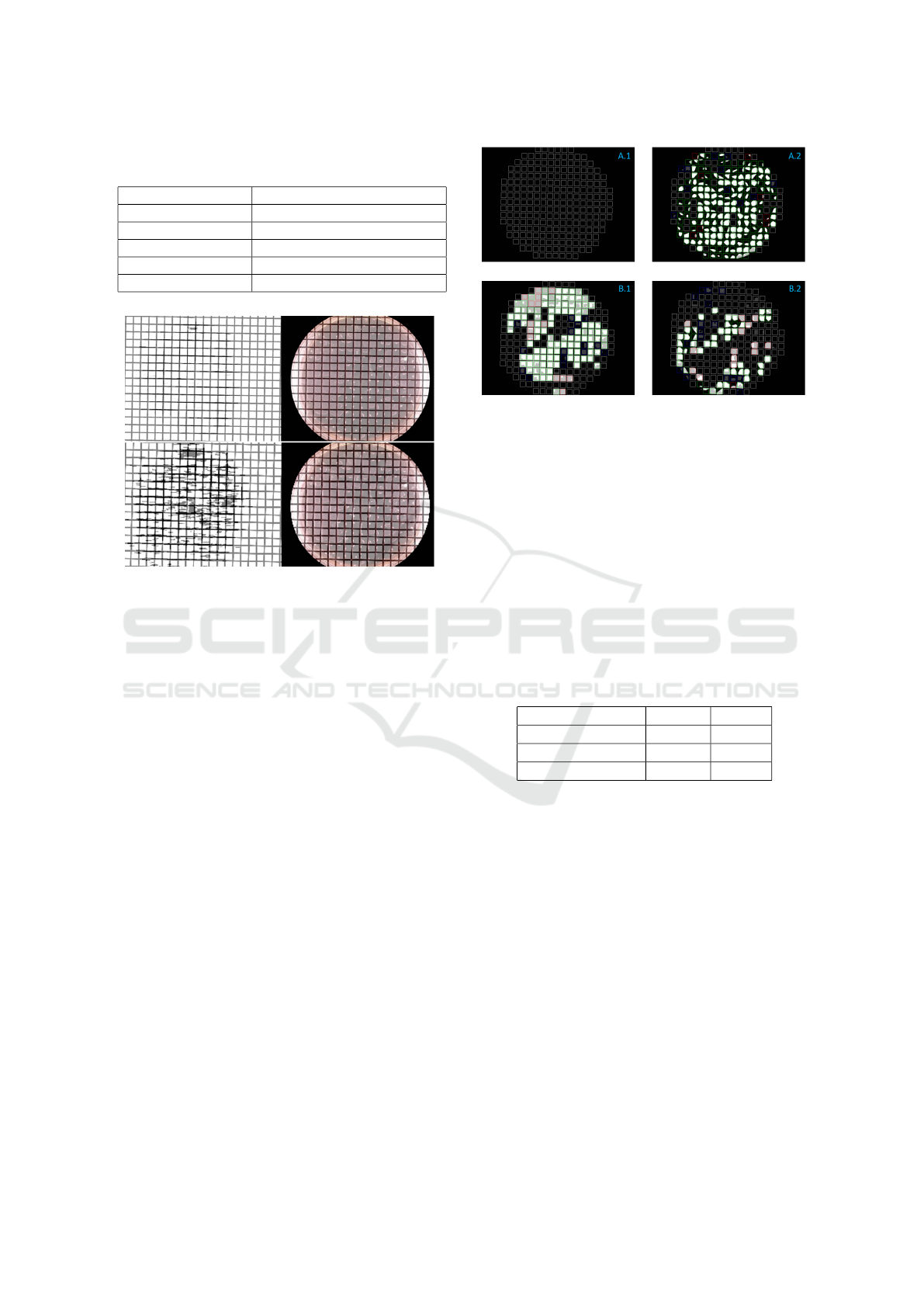
Table 1: Fitness comparison of multiple runs with different
settings for the evolutionary strategy.
Population sigma mean ± std [min - max]
1,50 0.05 0.086 ± 0.003 [0.063 - 0.229]
10,50 0.05 0.063 ± 0.005 [0.052 - 0.229]
20,100 0.05 0.061 ± 0.005 [0.05 - 0.231]
20,100 0.1 0.062 ± 0.01 [0.048 - 0.229]
20,100 0.001 0.125 ± 0.048 [ 0.044 - 0.25]
Figure 10: Example results of the mapped grid on an image
with different resolutions (row 1: factor 1.25, row 3: Factor
0.25). On the left the overlap of the grid and the binary
image can be seen. on the right the fitted grid on the rgb
images is shown.
As shown in Table 1, multiple runs for a differ-
ent setting for the population of the ES and the ini-
tial sigma were performed. In it, the average error
over 20 images is shown. The used setting with 20
parents and 100 children and an initial sigma of 0.05
provides the best and most robust results with the low-
est average error of 0.061 and a standard deviation
of 0.005. Whereas other runs with a higher starting
sigma (0.01) and a lower amount for the population
(10,50) provide similar results, lower values for the
population, especially with only one parent or a lower
starting value of sigma for faster convergence is in-
sufficient and provides clearly worse results reflected
in a higher error of 0.086 and 0.125.
As it is important that the grid detection also
works size independently, example images of differ-
ent sizes can be seen in Figure 10. The shown res-
olutions are factor 1.25 and 0.25. In a both results
(rows) the grid is aligned correctly. For resize fac-
tor 1.25 in all tested images, the grid was identified
correctly. The grid lies perfectly on the preprocessed
image. The preprocessed image also contains a very
low amount of noise and a large amount of extracted
hough lines. With smaller images, the amount of
noise increases. The result in row 2 shows a worst
Figure 11: Final grid cell prediction based on two example
images (A and B) for the prediction results of both classes
(monolayer and starting formation). The Figure shows the
final evaluation of the algorithm with its assignments per
grid cell (white: true negative, green: true positive, blue:
false positive, red: false negative).
case scenario with a high amount of noise, but still
shows a correct gird. In almost all cases, the evo-
lutionary strategy is robust enough to allow a cor-
rect alignment also in smaller images or images with
higher noise.
3.1.2 Segmentation of Monolayer & Starting
Formation
Table 2: Monolayer and forming spheroids segmentation
quality measurements on the test dataset.
Class Jaccard F1
Empty 0.8739 0.9671
Monolayer 0.5995 0.3674
Forming spheroid 0.7969 0.7754
As shown in Table 2, our segmentation neural net-
work can identify empty cells and already forming
spheroids with an F1 score of over 0.9671 for the
empty ones and a slightly lower score of 0.7754 for
the forming spheroid class. The main struggle for our
network is the identification of the monolayer class,
which led us to the presented post-processing steps
for an increase of the initial F1 score as low as 0.3674.
3.1.3 Classification Monolayer and Starting
Formation
In the final object detection step the quantification for
each relevant type of cell is fulfilled. In Figure 11 the
classification and quantification of monolayers (class
1) and starting formation cells (class 2) can is shown.
In the figure the results for both classes on two exam-
ple images are shown (Image A, and Image B). Image
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes
83
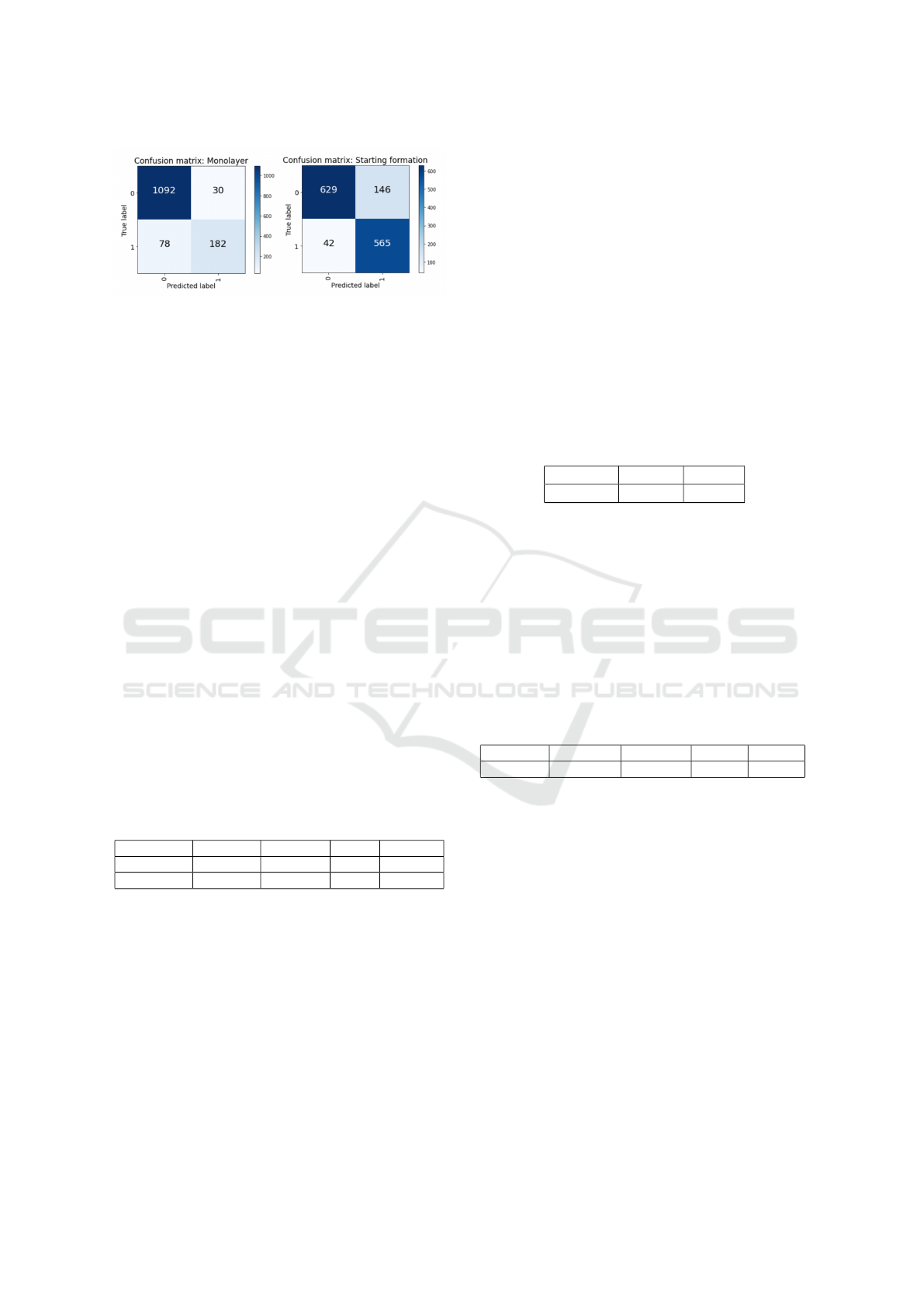
Figure 12: Confusion matrix of grid cell classification for
the classes monolayer & starting formation).
A shows a result without any monolayer cells in the
ground truth. The predictions for the monolayer class
in sub-image A.1 are all correctly true negative. The
model can therefore predict correctly even if there is
no cell in that class. In the prediction of the monolay-
ers (image B.1) in image B a high amount of mono-
layers can be seen. It can be seen that the models’
prediction and the subsequent class assignment to a
grid cell do indicate good results. Despite some false
positives (blue colored grid cells) and false negatives
(red colored grid cells) the prediction result can be
considered as good. Most errors in class 1 appear in
monolayer cells that span the whole grid cell with bad
lighting or starting formations cells that just started
and are still covering almost a full grid cell. Another
problem is the mentioned division of a single cell into
two different classes, which can lead to false nega-
tives in one class and false positives in the other cells.
These problems can also be seen in sub-images B.1
and B2, where most false negatives are starting for-
mation cells with almost full coverage of the grid cell.
In sub-image A.2 these problems appear much less
therefore, a higher performance can be reached. All
in all, the approach shows minor problems with bor-
der cases but a good overall performance.
Table 3: Metrics of grid cell classification for the classes
monolayer and starting formation.
class accuracy precision recall f1-score
monolayer 0.92 0.86 0.70 0.77
formation 0.86 0.80 0.93 0.86
The general performance on all test images is
shown in Table 3 and Figure 12. The result shows a
high accuracy of 0.92 for class 1 and 0.86 for class 2.
Especially the predictions for the monolayer cells do
have high accuracy. This is in comparison to the class
with cells that started the formation due to a very good
prediction of true negatives (also shown in sub-image
A.1 in Figure 11). Overall, the prediction of cells with
starting formation provides slightly better results, as
shown in the f1-score. With scores of 0.77 and 0.86
the error, especially for cells of the starting forma-
tion class, is low. The lower f1-score in the first class
can be explained by a lower recall resulting in more
missed cells, whereas the precision is higher. For the
class of starting formations, most cells are detected
with slightly lower precision. Both have a consider-
ably high performance. With a f1-score of 0.77 and
0.86, the final quantification was increased in com-
parison to the performance of the segmentation itself
and subsequently allows a good automatized quantifi-
cation.
3.2 Spheroid localisation
As shown in Table 4, the raw segmentation mask can
not provide the demanded quality for accurate quan-
tification of the spheroids within an image.
Table 4: Spheroids segmentation quality measured on the
test dataset.
Class Jaccard F1
Spheroid 0.7642 0.6889
The exceptionally low F1/Dice Score is signifi-
cantly increased during the post-processing, including
the actual location of the floating objects, as shown
in Table 5. As explained in section 2.2.3, the local-
ization of each spheroid is represented by its center
pixel. Based on these coordinates, the quality mea-
sures of Table 5 where realized by the comparison
with the label masks. The mentioned filtering, as well
as the added localisation workflow increased the final
F1 Score from 0.6889 to 0.8457.
Table 5: Spheroids localisation quality measured on the test
dataset.
Class Accuracy Precision Recall F1
Spheroid 0.7327 0.8962 0.8007 0.8457
As shown in the confusion matrix of Figure 13,
most of the actual spheroid objects are segmented
and classified correctly. Despite the high amount of
falsely as background detected objects, the number of
false positives could be kept on a significantly lower
level, which is realized by the mentioned size filter.
Without this filter, the number of false positives would
increase significantly by a factor of 6.5 due to the in-
creased amount of noise detected as objects.
4 DISCUSSION AND OUTLOOK
Regarding the presented results, our workflow already
provides the functionality for the automated quantifi-
cation of cell compartments on petri dishes by form-
ing spheroids based on RGB images. Simply im-
aged using a normal DSLR camera, these images are
BIOIMAGING 2023 - 10th International Conference on Bioimaging
84

Figure 13: Confusion matrix of the spheroid localisation
showing result measures on the test dataset.
processed using a combination of machine learning
algorithms, heuristic optimization, and computer vi-
sion. This combination of state-of-the-art algorithms
allows our workflow to quantify different stages of
forming spheroids which will be used for statistical
analysis in future work, including the influence of dif-
ferent drugs measured over a period of time. De-
spite the low initial segmentation quality, our pre-
sented post-processing and quantification algorithms
increases the classification performance of our work-
flow significantly, which still allows reliable quan-
tification of the different stages. In future different
segmentation methods will be further evaluated to in-
crease the overall performance. Regarding these first
results, the amount of data used for the modeling steps
will be increased significantly in future work, which
should further increase the final quality of the quan-
tification.
ACKNOWLEDGEMENTS
This work was supported by the Center of Excellence
for Technical Innovation in Medicine (TIMed), the
Dissertation Programme of the University of Applied
Sciences Upper Austria and the Austrian Research
Promotion Agency (FFG, project no. 881547 and In-
dustrienahe Dissertation no 867720).
REFERENCES
Aubreville, M., Bertram, C., Klopfleisch, R., and Maier, A.
(2018). SlideRunner - A Tool for Massive Cell An-
notations in Whole Slide Images. In Bildverarbeitung
f
¨
ur die Medizin 2018, pages 309–314. Springer Berlin
Heidelberg.
Barabas, J., Radil, R., and Gombarska, D. (2013). Image
processing and feature extraction of circular objects
from biological images. In 2013 36th International
Conference on Telecommunications and Signal Pro-
cessing (TSP), pages 612–615. IEEE.
Bhardwaj, S. and Mittal, A. (2012). A survey on vari-
ous edge detector techniques. Procedia Technology,
4:220–226.
Borgmann, D., Weghuber, J., Schaller, S., Jacak, J., and
Winkler, S. (2012). Identification of patterns in mi-
croscopy images of biological samples using evolu-
tion strategies. In Proceedings of the 24th European
Modeling and Simulation Symposium, pages 19–21.
Duda, R. O. and Hart, P. E. (1972). Use of the hough trans-
formation to detect lines and curves in pictures. Com-
munications of the ACM, 15(1):11–15.
Falcon, W. et al. (2019). Pytorch lightning. GitHub.
Note: https://github.com/PyTorchLightning/pytorch-
lightning, 3(6).
Forero, M. G., Medina, L. A., Hern
´
andez, N. C., and Mor-
era, C. M. (2020). Evaluation of the hough and ransac
methods for the detection of circles in biological tests.
In Applications of Digital Image Processing XLIII,
volume 11510, pages 387–395. SPIE.
F
¨
ursatz, M., Gerges, P., Wolbank, S., and N
¨
urnberger,
S. (2021). Autonomous spheroid formation by
culture plate compartmentation. Biofabrication,
13(3):035018.
Hansard, M., Horaud, R., Amat, M., and Evangelidis, G.
(2014). Automatic detection of calibration grids in
time-of-flight images. Computer Vision and Image
Understanding, 121:108–118.
Iakubovskii, P. (2019). Segmentation models pytorch. https:
//github.com/qubvel/segmentation models.pytorch.
Johnstone, B., Hering, T. M., Caplan, A. I., Goldberg,
V. M., and Yoo, J. U. (1998). In vitrochondrogene-
sis of bone marrow-derived mesenchymal progenitor
cells. Experimental cell research, 238(1):265–272.
OpenCV (2015). Open source computer vision library.
Radosavovic, I., Kosaraju, R. P., Girshick, R., He, K., and
Doll
´
ar, P. (2020). Designing network design spaces.
Roberts, L. et al. (1965). Machine perception of three-
dimensional solids, optical and electro-optical infor-
mation processing. MIT Press, Cambridge, MA, pages
159–197.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. In Lecture Notes in Computer Science.
Shorten, C. and Khoshgoftaar, T. M. (2019). A survey on
image data augmentation for deep learning. Journal
of Big Data, 6:60.
Zhang, L., Su, P., Xu, C., Yang, J., Yu, W., and Huang, D.
(2010). Chondrogenic differentiation of human mes-
enchymal stem cells: a comparison between micro-
mass and pellet culture systems. Biotechnology let-
ters, 32(9):1339–1346.
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang,
J. (2018). Unet++: A nested u-net architecture for
medical image segmentation. volume 11045 LNCS.
Zuiderveld, K. (1994). Contrast limited adaptive histogram
equalization. Graphics gems, pages 474–485.
In Vitro Quantification of Cellular Spheroids in Patterned Petri Dishes
85
