
An Easy-to-Use and Robust Approach for the Differentially Private
De-Identification of Clinical Textual Documents
Yakini Tchouka, Jean-Franc¸ois Couchot and David Laiymani
Femto-ST Institute, Univ. Bourg. Franche-Comt
´
e, CNRS, France
Keywords:
De-Identification, Differential Privacy, Deep Learning, Natural Language Processing, Clinical Textual
Document.
Abstract:
Unstructured textual data is at the heart of healthcare systems. For obvious privacy reasons, these documents
are not accessible to researchers as long as they contain personally identifiable information. One way to share
this data while respecting the legislative framework (notably GDPR or HIPAA) is, within the medical struc-
tures, to de-identify it, i.e. to detect the personal information of a person through a Named Entity Recognition
(NER) system and then replacing it to make it very difficult to associate the document with the person. The
challenge is having reliable NER and substitution tools without compromising confidentiality and consistency
in the document. Most of the conducted research focuses on English medical documents with coarse substitu-
tions by not benefiting from advances in privacy. This paper shows how an efficient and differentially private
de-identification approach can be achieved by strengthening the less robust de-identification method and by
adapting state-of-the-art differentially private mechanisms for substitution purposes. The result is an approach
for de-identifying clinical documents in French language, but also generalizable to other languages and whose
robustness is mathematically proven.
1 INTRODUCTION
Unstructured textual data is at the heart of healthcare
systems. The details included in these documents
allow us to clearly and precisely describe patients’
diseases and medical procedures, and to efficiently
manage and study their pathologies. These textual
documents can be analyzed by Artificial Intelligence,
given the impressive advances in Natural Language
Processing techniques in recent years (Kersloot et al.,
2020; Velupillai et al., 2018).
However, on the one hand, these AI-based tech-
nologies are currently only accessible to computer re-
searchers and not to medical staff, who have access
to medical data. On the other hand and for obvious
privacy reasons, these medical documents are not ac-
cessible to researchers as long as they contain person-
ally identifiable information. Medical managers are
therefore faced with a familiar dilemma: should they
share this medical data and compromise privacy and
medical secrecy to allow patients to benefit from the
latest medical advances available thanks to the artifi-
cial intelligence implemented on this data?
The GDPR does, however, allow researchers to
work on this type of data, provided that it has
been anonymized beforehand (EU, 2016, Recital 26).
GPDR is the European legal framework, on the US
side we have the Health Insurance Portability and Ac-
countability Act (HIPAA) (Cohen and Mello, 2018)
which defines 18 categories of so-called personal in-
formation (PHI) that must be removed from a medical
document before it can be shared. To comply with this
legal framework, it is therefore sufficient for medical
authorities to provide researchers with de-identified
documents. Such a document is a document where the
medical information is present but where all personal
data (names, dates, locations, for example) have been
modified to make any identification very difficult.
Practically, this can be done in two steps. De-
identification consists of first, applying a Named
Entity Recognition (NER) task revealing words
that would allow the document to be re-associated
with a particular person. Then, these entities are
replaced with alternative words making it very
difficult to associate the document with its patient
while preserving the utility of the document. The
challenging aspect in this work is implementing
a system that reliably detects identifying enti-
ties and substitute these recognized ones without
compromising privacy. The following is our guiding
example that will be developed throughout the article.
Thread Example. Consider the following fictional
sentence. It is typical of what can be present in a
94
Tchouka, Y., Couchot, J. and Laiymani, D.
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents.
DOI: 10.5220/0011646600003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 94-104
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

medical text document of a hospital. ”Mr. Durand
born in Dijon, 40 years old, was admitted to the hos-
pital from 12/02/2020 to February 26, 2020 following
a road accident in Dijon”.
To date, the most efficient methods in terms of
Named Entity Recognition are those based on the at-
tention concept, BERT (Devlin et al., 2018) for Bidi-
rectional Encoder Representations from Transformers
and its derivatives (Le et al., 2019; Huang et al., 2019;
Lee et al., 2019). To achieve detection scores where
precision and recall are very high, they require pre-
viously labeled datasets for training. This kind of la-
beled dataset exists in English language (Sun et al.,
2013; Johnson et al., 2016). However, it is severely
lacking in other languages, especially French. This
labeled learning dataset doesn’t need to be perfectly
coherent from a medical point of view. What is im-
portant is the format encountered and the context. It
then seems relevant to make use of an existing de-
identification algorithm, even if imperfect, to provide
this new dataset to be labeled afterward.
This article first shows how the utilization of a
French dataset anonymized using a recent but not
perfect de-identification algorithm followed by its
manual annotation, allowed the implementation of a
transformer-based NER approach. The precision and
recall results exceed all existing approaches in French
and are at the level of those in English.
After this NER phase, the following step is to sub-
stitute the detected sensitive entities. This step is often
neglected in research work because it is not consid-
ered relevant. This is indeed the case for entities such
as phone numbers or email addresses that can be re-
placed by any random number or email address. The
same is not true for dates or locations. Indeed, the
chronology of medical events is essential in detecting
correlations between them for example. Date substi-
tution methods exist but are not satisfactory. It has
indeed been shown (Tchouka et al., 2022) that apply-
ing a uniform shift between dates allows guaranteeing
the chronology but does not protect in any way a re-
identification of the document. One could think of
applying methods based on differential confidential-
ity (Duchi et al., 2013), the only method to date pro-
viding a metric for the level of data leakage. Applied
to temporal elements (date, age), it strongly protects
privacy by making the original date indistinguishable
from a published date. However, it significantly de-
grades the usefulness of the data because of the mag-
nitude of the interval in which the algebraic choice of
the date to be published is made. This article shows
how d-privacy brings a concrete answer to this prob-
lem of amplitude.
Finally, the location elements of textual medical
records must be treated with great care. Randomly
substituting a name of a city with another effectively
protects privacy but this is done at the detriment of the
medical context of the city. There was possibly radon
in this one at the origin of cancers, and pollution at
the origin of respiratory disorders. An approach based
on geo-indistiguishability (Bordenabe et al., 2014) is
not the most relevant since it only takes into account
the geographical position and not the medical and/or
statistical data associated with the city. We present in
this paper an innovative approach based on d-privacy.
The result is a global approach to de-identification
of medical documents dedicated to French textual
documents but which could be generalized to any
other language. This approach is first of all reliable
in the detection of identifying entities. Based on dif-
ferential privacy, substitution is robust to attacks by
definition. Moreover, they are optimized to preserve
data utility in the context of further processing by ma-
chine learning.
Our contributions in this paper can be summarized
as follows:
1. We provide a model identifying sensitive infor-
mation according to HIPAA categories in clinical
textual documents. This one manages to detect
all the categories we want to detect, as well as to
compete with the English detection models.
2. We provide a robust surrogate generation ap-
proach based on advances in differential privacy
that combines security and utility.
3. An open-source implementation of the surrogate
generation approaches proposed in this paper is
available on GitHub
1
.
This article is organized as follows. The follow-
ing section summarizes the state of the art regarding
NER as well as the substitution of sensitive elements
for de-identification purposes. Section 3 shows how
the NER task can be strengthened thanks to the con-
struction of an annotated dataset on the one hand, and
thanks to a deep learning-based model taking into ac-
count the context on the other hand. Section 4 finally
shows how to finely substitute temporal (age and date)
and location entities without compromising the confi-
dentiality of the data. Finally, the last section presents
a conclusion and future work.
2 RELATED WORK
This section summarizes the state of the art of de-
identification methods applied to the textual medical
1
https://github.com/healthinf/
Surrogate-generation-Strategies-in-De-identification
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents
95

document. The first section is dedicated to NER step
whereas the second one focuses on the surrogate gen-
eration.
2.1 Named Entity Recognition
For the NER phase (English dataset), several works
have experimented machine learning models such as
SVM, Decision trees, or Condition Random Field
(CRF) (Lafferty et al., 2001). With the emergence
of neural networks, researchers (Dernoncourt et al.,
2016; Liu et al., 2017) have proposed the first neu-
ral network-based model. Recurrent neural networks
(RNNs) of Dernoncourt et al (Dernoncourt et al.,
2016) lead to F
1
-scores of 97.85% and 99.23% on
i2b2 (Sun et al., 2013) and MIMIC (Johnson et al.,
2016) datasets respectively representing the state of
the art in de-identification. Some papers have ob-
tained results almost as accurate as those of Der-
noncourt by combining the machine learning method
(CRF) and the neural recurrent network method
(RNN). Following the recent advance of NLP with
the emergence of transformers (Vaswani et al., 2017)
and BERT (Devlin et al., 2018) which are the state
of the art in a contextualized text representation, it
has been proven that the most accurate models for
NER are those based on transformers. Among the
abundant works in this field, we can cite (Hanslo,
2021) and (Polignano et al., 2021). Research on
the de-identification of French medical documents
is mainly done by C. Grouin (Grouin et al., 2015)
with a machine learning model (CRF reaching 80%
in F
1
-score). A recent work (Bourdois et al., 2021)
is dedicated to de-identification of French emergency
medical records. It is based on a twofold approach.
First, FlauBERT (Le et al., 2019) assigns a label to
documents which require de-identification. Next a
combination of rules-based techniques and LSTM,
via Flair (Akbik et al., 2019) is implemented. Un-
fortunately, there is no dataset like MIMIC or i2b2
in the French language. This forced the authors
in (Tchouka et al., 2022) to combine the machine
learning method (CRF done by C. Grouin) and the
neural network method based on transformers on
WikiNER dataset (Nothman et al., 2013) to integrate
all the attributes to be detected. This hybrid system
reaches 94.7% in F
1
-score which serves as the base-
line in this work.
2.2 Surrogate Generation
The complexity of the substitution phase (Sweeney,
1996) depends on the analysis of the documents. The
most direct way is to delete the detected informa-
tion or replace it with its entity name (Durand by
NAME e.g.). This method protects privacy, but de-
grades the readability of the document and reduces
the usefulness of the data. To preserve the struc-
ture of the document, several authors have tried other
methods. The work of these papers (Douglass et al.,
2004; Levine, 2003; Uzuner et al., 2007; Douglass
et al., 2004; Deleger et al., 2014) has led to the
following strategy: Names are replaced by a ran-
dom name from a pre-established list, alphanumeric
strings are replaced by a randomly generated string,
and for dates, a uniform shift of days is performed
while keeping the format. As for ages, they have
been capped at 89 years, whereas locations are re-
placed randomly from a pre-established list. The most
used system in recent research is the system devel-
oped by Stubbs et al. (Stubbs et al., 2015). This one
combines the strategies of the previously described
work. This system has been used to build the 2014
i2b2 (Kumar et al., 2015) dataset for example. The
Stubbs method (Stubbs et al., 2015) which consists
in making a uniform shift of the dates of a docu-
ment is easily attackable. Since the interval between
the substituted dates remains unchanged, an attacker
only needs to know one date in file to reconstruct the
others. In (Tchouka et al., 2022) the authors have
shown that the system proposed by Stubbs on dates
and ages is easily attackable, thus compromising pri-
vacy in a medical context. Furthermore, for locations,
this random method significantly degrades the level
of information. The goal is to protect privacy while
keeping as much information as possible. To do so,
in (Tchouka et al., 2022) it was proposed to substi-
tute dates and ages through the Local Differential Pri-
vacy (LDP) (Duchi et al., 2013) with the bounded
Laplace mechanism (Dwork et al., 2006) and to sub-
stitute locations by a geo-indistinguishability (Bor-
denabe et al., 2014) algorithm. The problem with
LDP on dates or ages is that we cannot precisely con-
trol the noise added on two distant or close dates,
which sometimes leads to inconsistencies in the doc-
ument (e.g. the duration of a stay). The geo-
indistinguishability method gives a coherent result but
is not relevant in a medical context.
3 STRENGTHENING NAMED
ENTITY RECOGNITION
This section starts with our thread example. The
first section starts with the motivation for the need
to strengthen the NER stage. The second one shows
how we obtained a new labeled medical dataset.
Thirdly, the new machine learning-based NER
HEALTHINF 2023 - 16th International Conference on Health Informatics
96
approach is presented. Its evaluation on a medical
dataset is finally presented in the fourth section.
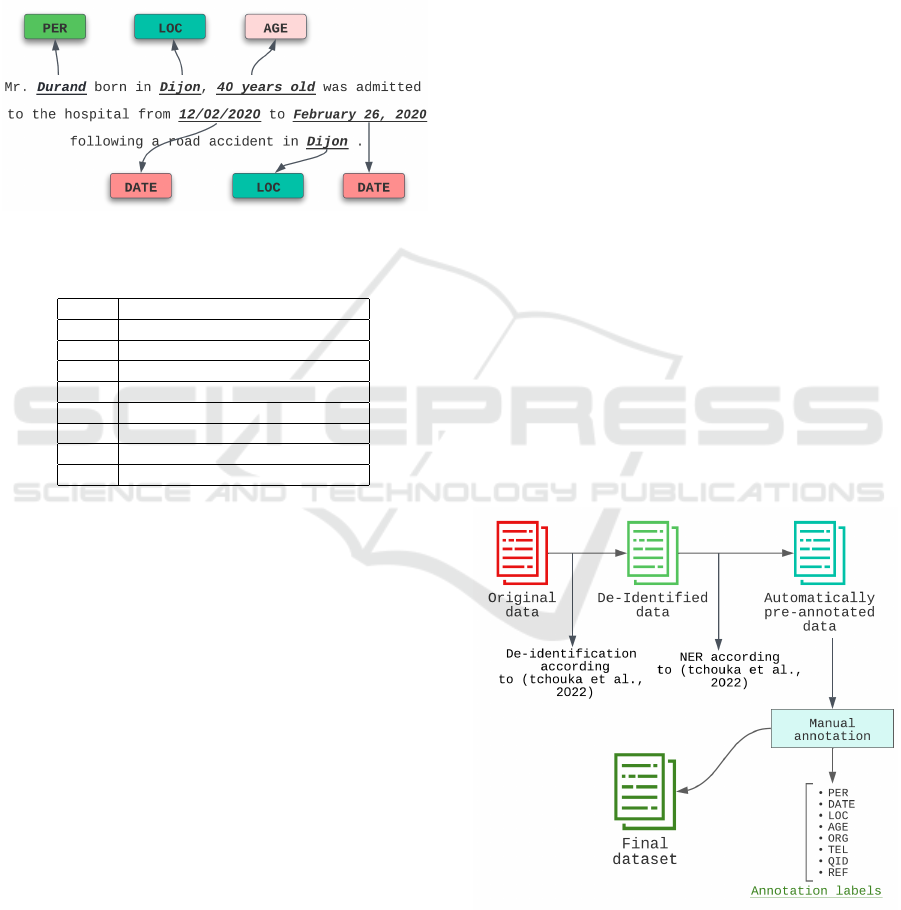
Thread Example. Figure 1 illustrates the result of
a perfect detection (NER) process applied to the
threaded example. HIPAA labels (Cohen and Mello,
2018) with their descriptions are summarized in Ta-
ble 1.
Figure 1: Perfect NER of PHI entities on thread example.
Table 1: Description of HIPAA labels.
Label Description
PER All names of persons
DATE All date sequences in all formats
LOC All geographical locations and zip codes
ORG Organizational entities
AGE Ages
TEL Phone Numbers
REF All references related to individuals
QID Any ID sequence
3.1 Motivation and Global Overview
Getting near-perfect scores in the NER is an absolute
necessity for successful de-identification. Undetected
sensitive information is a risk for re-identification of
the document. The NER task is a problem-dependent
task. This means that we often don’t have the right
dataset for our problem. The best results of NER
in de-identification in the literature are the ones ob-
tained by implementing English models (consistent
and complete domain-specific English datasets). In
French, such a dataset is rare and, to our knowledge,
does not exist in the medical domain. It is, there-
fore, necessary to build a sufficient dataset adapted to
the context of our application, i.e. a medical corpus,
which includes all the identifying attributes to detect.
With such a dataset, we are then able to apply a Trans-
former based NER method which is the state of the art
in NER.
3.2 Building a Labelled Dataset
As mentioned, the most difficult step is finding a
dataset that is large enough for the implementation
of an accurate model, that includes all the categories
of sensitive information, and finally, that is adapted
to the medical corpus. Such a dataset (in French)
is not available at the moment, at least not accessi-
ble to everyone. In (Tchouka et al., 2022) the au-
thors used WikiNER dataset which includes only a
few tags with a very general vocabulary. This re-
quires them to combine several methods so that all
categories could be integrated into a de-identification
tool. In this current work, as part of our collaboration
with a French public hospital, we have access (on-
site) to a large set of unlabeled medical notes. We
propose that hospital members semi-manually anno-
tate a subset of these notes. As this dataset will be
exported from the hospital afterward, we ask them to
apply the existing de-identification method (Tchouka
et al., 2022) on them to obtain new de-identified docu-
ments. Then we ask them to manually label all the de-
identified documents. To facilitate this manual task,
the NER step of the same tool (Tchouka et al., 2022)
has been used. The process is illustrated in Figure 2.
The obtained dataset, further denoted as to French-
HospitalNER is partially de-identified, according to
the (Tchouka et al., 2022) approach, and labeled. This
annotation step required 25 hours of work for one
person (1 minute per file). The FrenchHospitalNER
Dataset contains 14925 sentences.
Figure 2: FrenchHospitalNER Dataset Construction.
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents
97

3.3 Supervised Learning on a Dedicated
Labelled Dataset
This part starts by the presentation of the architec-
ture of our model. Then we describe how supervised
learning was implemented. Finally, an evaluation of
our model is presented in parallel with existing de-
identification models.
3.3.1 Model Architecture: Transformer Based
Approach
Due to the the availability of FlauBERT which is a
BERT-based pre-trained French models, such a trans-
former is easily accessible This decision is strength-
ened by the fact that it has been shown in (Polignano
et al., 2021) that a language specifically dedicated to
the French language model like FlauBERT, improves
the results compared to multilingual BERT models.
3.3.2 Finetuning Transformer Model
Starting from a pre-trained model such as FlauBERT,
what is left is finetuning it on a smaller and more
specialized dataset. Instead of starting from scratch
to build our text classification or feature detection
model, we will start from the pre-trained BERT and
add a dense layer or a classification layer to build the
model as described in Figure 3.
3.3.3 Training
The learning process has been implemented with the
previously described dataset using a deep learning-
based NLP model.
• the Learning Rate controls the size of the update
steps along the gradient. Usually, a very small
value is set (10
−4
in this work), so that the weights
are less modified at each iteration, which avoids
missing the optimal values of the error function
• the Dropout is a regularization technique for re-
ducing overfitting in neural networks. It is set to
0.1 in this work, which means that 10% of se-
lected neurons are ignored during training
• the Training Batch Size is the number of train-
ing samples to work through before the model’s
internal parameters are updated.
• the Maximum Length defines the maximum
number of words in the sentences
• the Number of Epochs is the number of complete
passes through the training dataset.
The NER is a multi-class classification model
(taking tags as classes). The CrossEntropy er-
ror function is well adapted for this task. As
an optimizer (backpropagation function), we use
the adamW(Loshchilov and Hutter, 2017) algorithm
which is one of the latest evolutions of optimizers
and is proven to be better in neural network learning.
The dataset (FrenchHospitalNER) is randomly split
into training and test sets (90/10). The validation set
is provided by the HNFC hospital and contains over
6000 sentences.
In machine learning, the question remains: how
to select the optimal values of the hyperparameters
to obtain the most accurate results? There are sev-
eral methods of hyper-parameter optimization such
as Grid Search, Random Search, and model-based
Bayesian method. Studies (Bergstra et al., 2013)
on hyper-parameter optimization show that Bayesian
methods give largely more accurate results. In this
paper, for hyper-parameter optimization, we used
the Tree-structured Parzen Estimator (Bergstra et al.,
2011) which is a classical Bayesian optimization al-
gorithm sufficient for a classification model as in our
case. We have experimented with different combina-
tions of parameters according to the Tree-structured
Parzen Estimator algorithm as illustrated in Figure 4.
The optimization during training was performed
on three hyper-parameters: the number of epochs, the
maximum length, and the batch size. At the end of
the training, the model with the highest F
1
score is se-
lected with the following hyper-parameters: number
of epochs = 20, maximum length = 128, batch size =
64.
It is this model that is used in the Evaluation sec-
tion below.
3.4 Evaluation
To evaluate our model we used the classical metrics
Precision (P), Recall (R), and F
1
-score. To get a sense
of the overall performance of the system, we use the
micro-average of Accuracy of the labeling process is
evaluated across precision, recall, and F
1
-score met-
rics.
Table 2: NER results on the evaluation dataset.
Methods (Tchouka et al., 2022) PROPOSAL (Dernoncourt et al., 2016)
Dataset HNFC i2b2
Metrics P R F
1
P R F
1
P R F
1
PER 96.3 99.8 98 97.2 98.9 98 98.2 99.1 98.6
ORG 41.1 57.3 47.8 90 51 65.6 92.9 71.4 80.7
LOC 88.4 95.8 92 99.4 94.4 96.9 95.9 95.7 95.8
DATE 97.7 86.7 91.9 99.2 95.7 97.4 99 99.5 99.2
AGE 91.5 66.9 77.3 98.2 91.8 95 98.9 97.6 98.2
TEL 99.5 97.9 98.7 99.4 99.8 99.6 98.7 99.7 99.2
REF - 96.1 79.5 87 -
QID - 77.2 32 45.3 99.2 98.7 99
Overall 94.6 94.9 94.7 98.5 96.4 97.4 98.3 98.53 98.4
HEALTHINF 2023 - 16th International Conference on Health Informatics
98
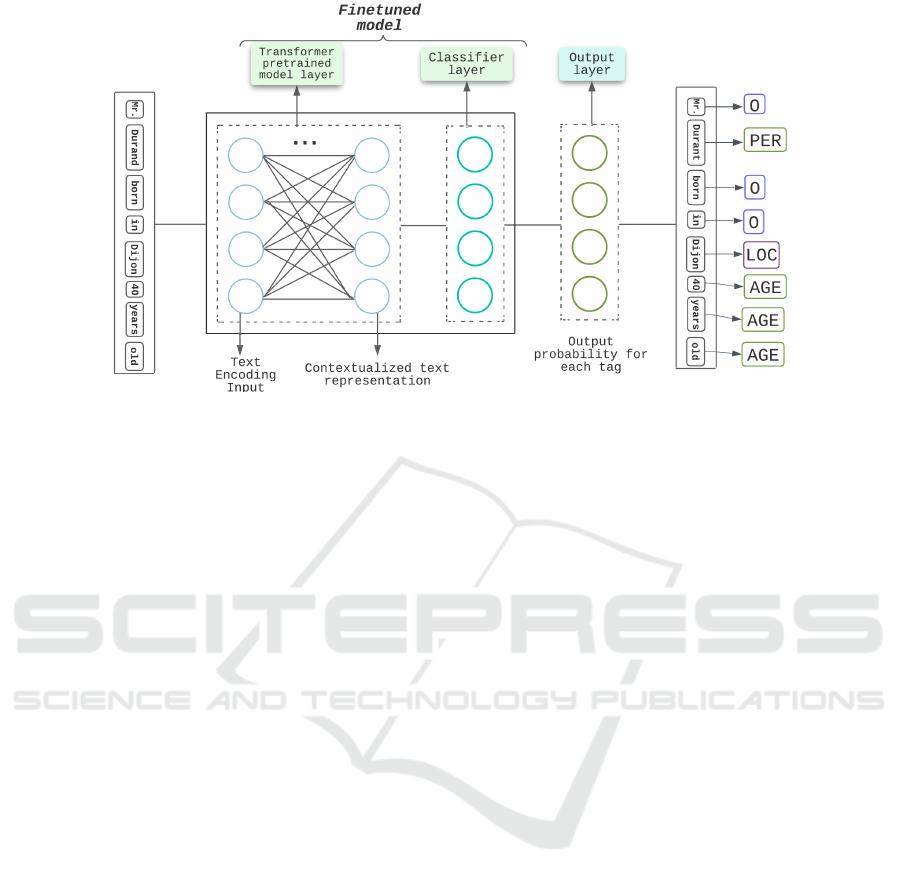
Figure 3: Deep Learning Model Architecture for NER.
To be fair with (Tchouka et al., 2022), we asked
the HNFC hospital to evaluate the NER step of this
approach on their HNFC-dataset. The results are de-
tailed in Table 2. This proposal largely surpasses re-
sults obtained in (Tchouka et al., 2022) in several cat-
egories (DATE, AGE. . . ). This is due to the BERT-
based layer which allows us to have a precise con-
textualization of the sequence. Our low score on
the organization level compared to the i2b2 model
is explained by the fact that they are structured in-
formally in the medical documents (abbreviation, iso-
lated word. . . ). Increasing the dataset will help solve
these types of problems and generally improve the
scores in the different categories.
The next step is substituting the detected entities,
as described in the next section.
4 SURROGATE GENERATION
STRATEGIES
The challenge here is to substitute personal informa-
tion detected by NER with relevant surrogates regard-
ing medical content whilst preserving privacy.
As argued in (Tchouka et al., 2022; Stubbs et al.,
2015), not all entities have the same level of criticality
or importance. A random strategy may be chosen for
instance for replacing names, phone numbers. . .
Moreover, to avoid averaging attacks and for
consistency in the document, memoization has been
implemented as in (Erlingsson et al., 2019; Arcolezi
et al., 2022). This consists in using the same substi-
tute for given sensitive information in the document.
Thread Example. In the thread example, Durand
could be replaced by any name, Julien for instance.
In contrast, temporal and location data inherently
carries information that is both medically important
and highly identifiable. In (Tchouka et al., 2022), for
temporal entities, the authors opted for local differen-
tial privacy with bounds in time categories (recalled
hereafter) to calibrate the added noise. About geo-
graphical locations, geo-indistinguishability (Borden-
abe et al., 2014) was retained as a direct mechanism to
provide a location close to the original one and whose
privacy leak is measured by ε − d privacy. These two
approaches allow the aforementioned method to re-
spect privacy. However, the relevance of substitutions
in this specific context of medical data has some lim-
its which will be detailed in following two sections.
4.1 Date & Age: Substitution Strategy
Beyond the fact that a date is identifying in a medical
document, we can not afford to randomly substitute
them. Providing an algorithm that respects privacy
means accepting that only the patient is allowed to
modify his or her data in such a way that given two
sanitized data of two patients, it is difficult (from
a probabilistic point of view) to reassign one to the
first and the other to the second. Local Differential
Privacy (Duchi et al., 2013) (LDP) formalizes the
algorithm robustness and its definition is recalled
hereafter.
Definition 1 (ε-local differential privacy). A random
mechanism A satisfies ε-local differential privacy if,
for any pair of input values v
1
, v
2
∈ Domaine(A ) and
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents
99
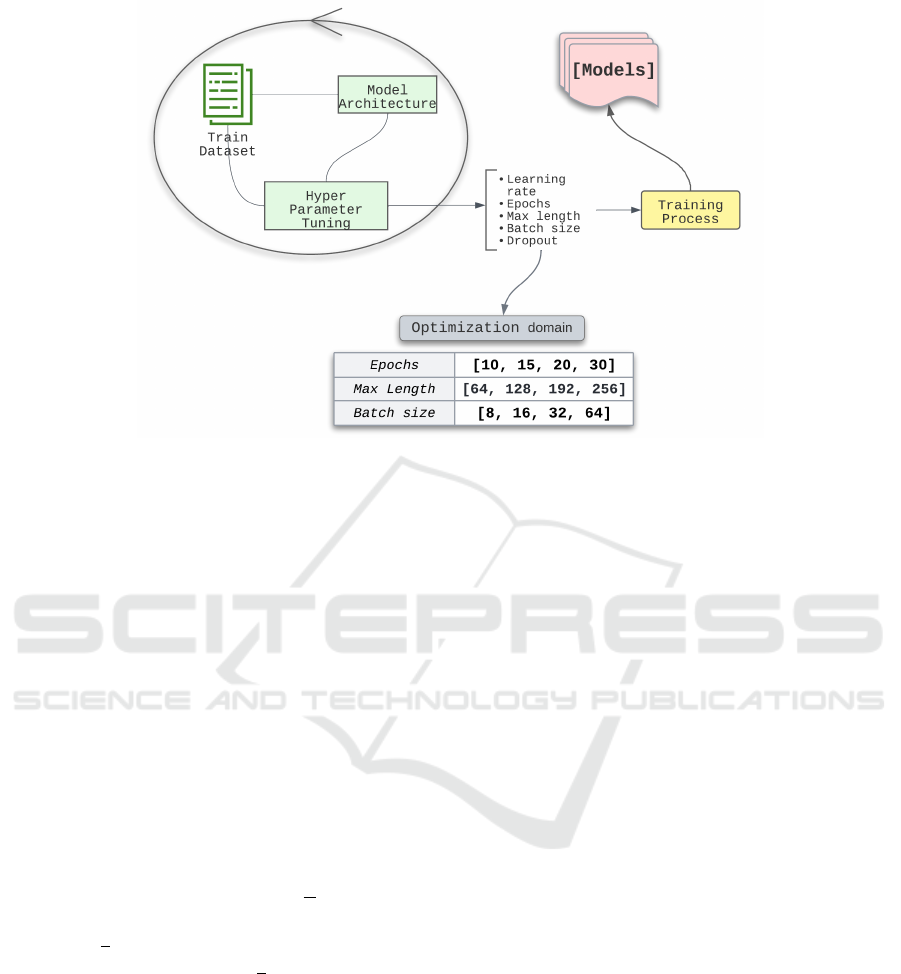
Figure 4: FineTuning Process.
any possible output y of A:
Pr[A(v
1
) = y] ≤ e
ε
· Pr[A(v
2
) = y]. (1)
LDP mechanisms (Holohan et al., 2017) are
tuned with respect to the data types they handle (real,
integer, . . . ), and to their usefulness. Here, the focus
is on temporal data. As each of them can be seen as a
number (number of days elapsed between the date to
be cleaned and the current date), in (Tchouka et al.,
2022) the authors have focused on the Laplacian
mechanism recalled below.
Definition 2 (Laplacian mechanism in an interval of
amplitude ∆). In the Laplacian mechanism, a nu-
merical value v is sanitized into a numerical value
M
Lap
(v, ∆, ε) with
M
Lap
(v, ∆, ε) = v + Lap
∆
ε
(2)
where Lap
∆
ε
is the Laplace distribution centered in
0 and whose scale parameter is
∆
ε
.
In (Tchouka et al., 2022), the authors reached the
conclusion that segmenting a set of dates into 3 cat-
egories (less than 2 months, less than 2 years, more
than two years) was necessary to minimize ∆, i.e. the
introduced noise. Indeed, within these intervals (of
range ∆), the generated dates did not allow us to infer
what their preimages were. However, in the larger cat-
egory, the introduced noise is still too important since
it is necessary to make indistinguishable the cleaning
of two dates like 3 years and 80 years.
It is thus necessary to further segment the space
much more or, equivalently, allow distinguishing
certain dates from others. Two dates that are initially
far apart should not necessarily be made identical by
a differential privacy mechanism. The underlying
idea is therefore privacy depending on the distance
between the values of the elements to be protected.
We find here the notion of ε.d-privacy (Alvim et al.,
2018) recalled below.
Definition 3 (ε.d-privacy). A randomized algorithm
(A satisfies the epsilon.d-privacy if, for any possible
output y of A and for any pair of input values v
1
, v
2
∈
Domain(A), domain with a metric d.
Pr[A(v
1
) = y] ≤ e
ε.d(v
1
,v
2
)
· Pr[A(v
2
) = y]. (3)
Intuitively, the ε.d-privacy protects the precision
of the secret: if we add a metric in the date space, it al-
lows us to distinguish between an old date (of birth for
example) and a recent date (of operation last week).
On the other hand, it guarantees that two very recent
dates (v
1
and v
2
) at a very small distance will generate
the same output y with a very high probability.
Without going into further details, this version
of ε.d-privacy generalizes both the ε-local differen-
tial privacy and ε-differential privacy and has the
same properties in terms of composition and post-
processing (Fernandes, 2021).
The question then arises of implementing a mech-
anism that guarantees this ε.d-privacy property for
temporal events.
In (Tchouka et al., 2022), to respect the chronol-
ogy of events each temporal event d (a date, an
age,. . . ) is converted into a duration v in days between
the current date and d. The input domain is thus R
+
HEALTHINF 2023 - 16th International Conference on Health Informatics
100
with the absolute value as distance. Easy to imple-
ment, it would be detrimental to privacy. Indeed, in
the context of ε-LDP, the ∆-amplitude of this mecha-
nism would be equal to 1 day instead of the amplitude
of each category (100 × 365 days for the largest cat-
egory). A precise date such as birth or intervention
will probably be modified. On the other hand, an age
of a few decades will very probably not be modified,
which is not satisfactory.
Moreover, in a medical document which contains
”10 years ago”, what is actually meant is ”about 10
years ago”, and not ”the same day, 10 years be-
fore”. This approximation is also found when tem-
poral events are expressed in months or weeks. The
metric that we will consider will be unit dependent.
It will be in years (in months, weeks . . . resp.) for
events expressed in years (in months, weeks . . . resp.).
With this adaptation, an age (in years) will probably
be modified by a few years, for example.
The Laplacian mechanism (recalled in defini-
tion 2) adds noise following a Laplace-centered distri-
bution of parameter ε
−1
. It is not difficult to demon-
strate (Fernandes, 2021) that this mechanism has the
ε.d-privacy property given by the equation (3).
Notice that, as in a classical differential privacy
approach, the privacy global ε budget is shared
between all the elements to be substituted. This
sharing here can be uniform or not. Without any a
priori, we consider that it is here.
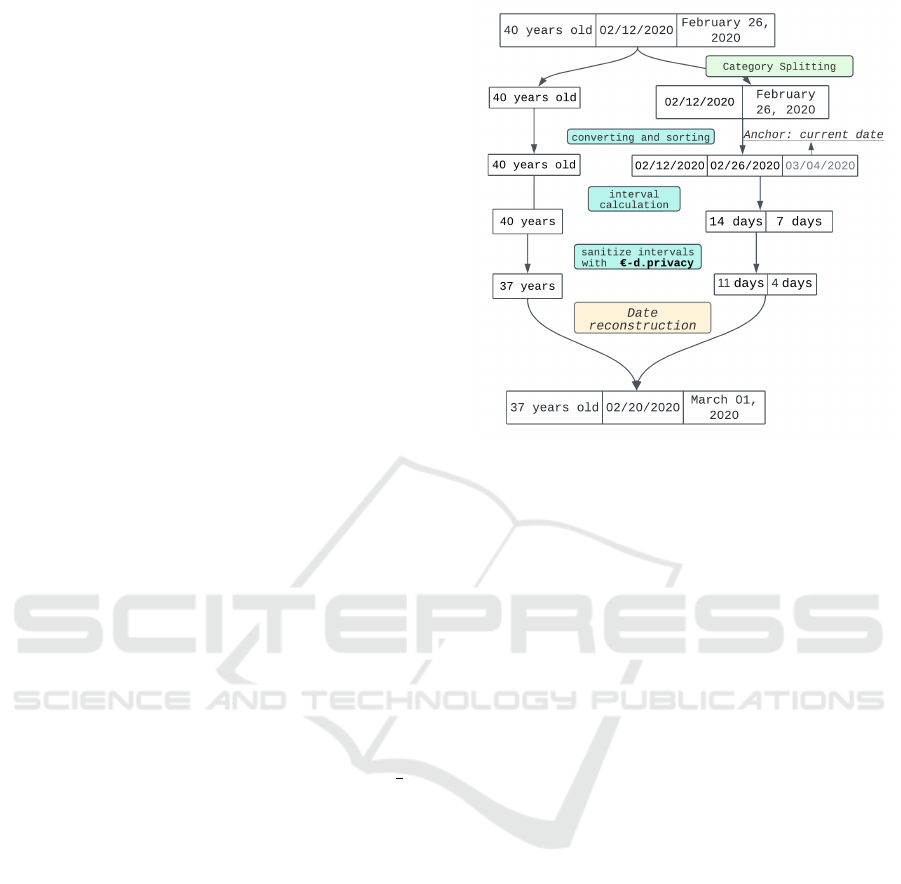
Thread Example. In our thread example, there are 2
detected dates (expressed in days), 1 age expressed in
years, and 1 location (2 times duplicated), i.e. 4 ele-
ments to substitute. Each element will consume
ε
4
of
the privacy budget, the last quarter is dedicated to san-
itizing location. The date substitution process for this
example is detailed in Figure 5. Thus, 40 years be-
comes 37 years, 02/12/2020 and February 26, 2020
respectively lead to 02/20/2020 and March 01, 2020.
4.2 Geographic Locations: Substitution
Strategy
Geo-indistinguishability (Bordenabe et al., 2014) has
been accepted de facto as the gold standard to pre-
serve location privacy (Xiao and Xiong, 2015; Fawaz
and Shin, 2014; Bordenabe et al., 2014). This mech-
anism instantiates ε.d-privacy (as recalled in defini-
tion 3) in the context of locations which are (x, y)
coordinates inside R
2
. In the de-identification con-
text, the authors of (Tchouka et al., 2022) use geo-
indistinguishability to randomly add noise to the co-
ordinates (x, y) of the location to be sanitized leading
to the new tuple (x
′
, y
′
). Thus, they re-associate the
Figure 5: Example of date substitution process on the thread
example.
location which is the closest one to this new tuple.
This method effectively protects privacy and provides
a consistent substitute in the document. However, we
argue that it does not effectively answer the question
of the document’s utility in a medical context. Indeed,
two places relatively close to each other geographi-
cally can be far from each other from a health point
of view. Our motivation here is to have a system that
integrates not only the distance but also some health
criteria that can impact the health of the population.
In this substitution of locations, it seems desirable
to choose randomly among locations that are close not
only in a geographical sense but also in a statistical
(e.g., number of inhabitants) and medical (e.g., the in-
cidence rate of all cancers, the number of strokes, air
pollution, radon level. . . ) sense. Everything depends
on the fact that we can express a distance between
locations that would integrate statistical and medical
characteristics. Many institutional websites freely of-
fer this local information. Figure 6 gives an extract
for some cities of the Bourgogne Franche-comt
´
e, a re-
gion of France. With such features for each location,
it is not hard to compute the distance between them
(for instance the euclidean one) and to apply any LDP
mechanism capable of capturing this distance.
For instance, let us consider a public database
of N locations where each location i is a vector
(x
i
, y
i
, c
1
i
, . . . , c
n
i
) where (x
i
, y
i
) is the geographical lo-
cation and (c
1
i
, . . . , c
n
i
) the features, further considered
to be normalized, i.e. in [0, 1]. Let d
ji
be the vector of
feature differences between locations j and i.
Let v
j
= [(1, d
j1
), (2, d
j2
), . . . ( j, 0), . . . , (N, d
jN
)]
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents
101

Figure 6: Example of exponential mechanism applied on sanitizing Dijon city.
be the sequence of all distances between j and oth-
ers. In a practical situation, this sequence can be re-
duced to the location distances (i, d
ji
) s.t. both the
geographical distance between i and j is lower than
a given threshold and to the k smallest values of the
distances, and where values are sorted in ascending
order according to d. v
′
j
is the result and consti-
tutes the possible substitutes of the city j. This leads
to v
′
j
= [(i
1
, d
ji
1
), . . . (i
k
, d
ji
k
)], with (i
1
, d
ji
1
) = ( j, 0)
since the smallest distance is 0 between j and j. The
score function U may be defined by U( j, i) = 1 − d
ji
for each i ∈ {i
1
, . . . , i
k
} and −∞ elsewhere. This func-
tion is public and is not based on any private data. The
probability distribution function is thus as follows:
P
j
= [a.e
εU( j,i
1
)
, . . . , a.e
εU( j,i
k
)
, 0, . . . , 0] (4)
where a =
∑
k
i=1
e
εU( j,i
1
−1
is the normalization fac-
tor. Notice this mechanism is an adaptation of the cen-
tralized exponential mechanism with public data, i.e.,
without sensitivity. Cities can thus be sanitized ac-
cording to the mechanism given in algorithm 1. This
mechanism is based on a distance. The next section
shows it verifies ε.d-privacy.
Algorithm 1: Local exponential mechanism applied to the
city j.
➔ Let the probability distribution P
j
defined as in (4)
➔ Y
j
= [y
1
, . . . , y
k
] the k possible output cities
➔ the substitute l of the city j is l = Random[Y
j
]
P
j
with Random[Y
j
]
P
j
a
random draw according to the distribution P
j
Property 1. The mechanism defined in Algorithm 1
verifies ε.d-privacy.
Proof. According to the definition 3, for any y whose
probability distribution definition is not null we suc-
cessively have
Pr[A(v
1
)=y]
Pr[A (v
2
)=y]
=
ae
εU(v
1
,y)
ae
εU(v
2
,y)
=
e
ε(1−d(v1,y))
e
ε(1−d(v2,y))
= e
ε(d(v
2
,y)−d(v
1
,y))
≤ e
ε.d(v
1
,v
2
)
Clearly, the features to be integrated for this step
should be defined upstream in concert between the
medical teams (who know the data) and the technical
teams.
Thread Example. Using our example with the loca-
tion ”Dijon”. Considering the features: overall pop-
ulation, cancer incidence rate, and strokes, shown in
blue in Figure 6. The columns (’distance’ & ’scores’)
represent respectively the vector distance (Euclidean
distance with normalized features) and the results of
the score function defined in Algorithm 1, from Dijon
to k = 10 ’nearby’ cities (according to features). After
applying the probability distribution function previ-
ously detailed, we obtain the normalized distribution
illustrated in orange in Figure 6. The random draw
thus follows this distribution.
According to the memoization, all occurrences of
the location Dijon can be replaced by Besanc¸on and
the final result of the substitution step would be: ”Mr.
Julien born in Besanc¸on, 37 years old, was admitted
to the hospital from 02/20/2020 to March 01, 2020
following a road accident in Besanc¸on.”
5 CONCLUSION
This paper detailed a complete accurate differen-
tially private de-identification method. Regarding the
NER step, an existing comprehensive but flawed de-
identification approach was taken to internally build
a new and substantial medical dataset that was then
labeled by hand. Using this new labeled and large
dataset, deep learning was implemented taking into
account the context. NER results we obtained in
French are equivalent to the most accurate results in
the English language, filling the gap between these
two languages. Regarding substitutions of sensi-
tive data, we pointed out the limitations of exist-
ing approaches, especially for temporal and location
data. We believe we have provided the most privacy-
HEALTHINF 2023 - 16th International Conference on Health Informatics
102

friendly method to date and location (since it is based
on differential privacy) that retains sufficient medical
information for further processing. For the NER part,
our future works will focus on the use of multilingual
models such as XLM-RoBERTa and their ability to
enable zero-shot cross-lingual transfer. We will fur-
thermore study how the introduction of a translation
step and the associated English language NER task
can improve our NER approach as in (Sch
¨
afer et al.,
2022) The idea is to tune a such model on an English
NER medical dataset and to study its behavior on a
French evaluation dataset. With the same goal of an-
alyzing medical documents while preserving privacy,
in addition to the anonymization method detailed in
this paper, we will try the methods of perturbing the
training dataset in the word embedding vocabulary
(BERT-based model) by metric-based differential pri-
vacy (Feyisetan et al., 2020; Zhao and Chen, 2022).
ACKNOWLEDGEMENTS
This work was partially supported by the EIPHI-
BFC Graduate School (contract ”ANR-17-EURE-
0002”). All computations have been performed on
the ”M
´
esocentre de Calcul de Franche-Comt
´
e”.
REFERENCES
Akbik, A., Bergmann, T., Blythe, D. A. J., Rasul, K.,
Schweter, S., and Vollgraf, R. (2019). Flair: An easy-
to-use framework for state-of-the-art nlp. In NAACL.
Alvim, M. S., Chatzikokolakis, K., Palamidessi, C., and
Pazii, A. (2018). Metric-based local differential
privacy for statistical applications. arXiv preprint
arXiv:1805.01456.
Arcolezi, H. H., Couchot, J.-F., Al Bouna, B., and Xiao,
X. (2022). Improving the utility of locally differen-
tially private protocols for longitudinal and multidi-
mensional frequency estimates. Digital Communica-
tions and Networks.
Bergstra, J., Bardenet, R., Bengio, Y., and K
´
egl, B. (2011).
Algorithms for hyper-parameter optimization. Ad-
vances in neural information processing systems, 24.
Bergstra, J., Yamins, D., and Cox, D. (2013). Making a sci-
ence of model search: Hyperparameter optimization
in hundreds of dimensions for vision architectures. In
International conference on machine learning, pages
115–123. PMLR.
Bordenabe, N. E., Chatzikokolakis, K., and Palamidessi, C.
(2014). Optimal geo-indistinguishable mechanisms
for location privacy. In Proceedings of the 2014 ACM
SIGSAC conference on computer and communications
security, pages 251–262.
Bourdois, L., Avalos, M., Chenais, G., Thiessard, F.,
Revel, P., Gil-Jardin
´
e, C., and Lagarde, E. (2021).
De-identification of Emergency Medical Records in
French: Survey and Comparison of State-of-the-Art
Automated Systems, volume 34. LibraryPress@UF.
Cohen, I. G. and Mello, M. M. (2018). Hipaa and pro-
tecting health information in the 21st century. Jama,
320(3):231–232.
Deleger, L., Lingren, T., Ni, Y., Kaiser, M., Stoutenbor-
ough, L., Marsolo, K., Kouril, M., Molnar, K., and
Solti, I. (2014). Preparing an annotated gold standard
corpus to share with extramural investigators for de-
identification research. Journal of biomedical infor-
matics, 50:173–183.
Dernoncourt, F., Lee, J., Uzuner, O., and Szolovits, P.
(2016). De-identification of patient notes with recur-
rent neural networks. Journal of the American Medi-
cal Informatics Association : JAMIA, 24.
Devlin, J., Chang, M., Lee, K., and Toutanova, K. (2018).
BERT: pre-training of deep bidirectional transformers
for language understanding. CoRR, abs/1810.04805.
Douglass, M., Clifford, G. D., Reisner, A., Moody, G. B.,
and Mark, R. G. (2004). Computer-assisted de-
identification of free text in the mimic ii database.
In Computers in Cardiology, 2004, pages 341–344.
IEEE.
Duchi, J. C., Jordan, M. I., and Wainwright, M. J. (2013).
Local privacy and statistical minimax rates. In 2013
IEEE 54th Annual Symposium on Foundations of
Computer Science, pages 429–438. IEEE.
Dwork, C., McSherry, F., Nissim, K., and Smith, A. (2006).
Calibrating noise to sensitivity in private data analy-
sis. In Theory of cryptography conference, pages 265–
284. Springer.
Erlingsson,
´
U., Feldman, V., Mironov, I., Raghunathan, A.,
Talwar, K., and Thakurta, A. (2019). Amplification
by shuffling: From local to central differential privacy
via anonymity. In Proceedings of the Thirtieth An-
nual ACM-SIAM Symposium on Discrete Algorithms,
pages 2468–2479. SIAM.
EU (2016). Regulation (eu) 2016/679 of the european par-
liament and of the council of 27 april 2016 on the
protection of natural persons with regard to the pro-
cessing of personal data and on the free movement of
such data, and repealing directive 95/46/ec (general
data protection regulation) (text with eea relevance).
Fawaz, K. and Shin, K. G. (2014). Location privacy protec-
tion for smartphone users. In Proceedings of the 2014
ACM SIGSAC Conference on Computer and Commu-
nications Security, pages 239–250.
Fernandes, N. (2021). Differential privacy for metric
spaces: information-theoretic models for privacy and
utility with new applications to metric domains. PhD
thesis,
´
Ecole Polytechnique Paris; Macquarie Univer-
sity.
Feyisetan, O., Balle, B., Drake, T., and Diethe, T. (2020).
Privacy-and utility-preserving textual analysis via cal-
ibrated multivariate perturbations. In Proceedings of
the 13th International Conference on Web Search and
Data Mining, pages 178–186.
Grouin, C., Griffon, N., and N
´
ev
´
eol, A. (2015). Is it pos-
sible to recover personal health information from an
An Easy-to-Use and Robust Approach for the Differentially Private De-Identification of Clinical Textual Documents
103

automatically de-identified corpus of french ehrs? In
Proceedings of the Sixth International Workshop on
Health Text Mining and Information Analysis, pages
31–39.
Hanslo, R. (2021). Deep learning transformer archi-
tecture for named entity recognition on low re-
sourced languages: State of the art results. CoRR,
abs/2111.00830.
Holohan, N., Leith, D. J., and Mason, O. (2017). Optimal
differentially private mechanisms for randomised re-
sponse. IEEE Transactions on Information Forensics
and Security, 12(11):2726–2735.
Huang, K., Altosaar, J., and Ranganath, R. (2019). Clinical-
bert: Modeling clinical notes and predicting hospital
readmission. arXiv preprint arXiv:1904.05342.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). Mimic-
iii, a freely accessible critical care database. Scientific
data, 3(1):1–9.
Kersloot, M. G., van Putten, F. J., Abu-Hanna, A., Cornet,
R., and Arts, D. L. (2020). Natural language pro-
cessing algorithms for mapping clinical text fragments
onto ontology concepts: a systematic review and rec-
ommendations for future studies. Journal of biomedi-
cal semantics, 11(1):1–21.
Kumar, V., Stubbs, A., Shaw, S., and Uzuner,
¨
O. (2015).
Creation of a new longitudinal corpus of clinical nar-
ratives. Journal of biomedical informatics, 58:S6–
S10.
Lafferty, J. D., McCallum, A., and Pereira, F. C. N. (2001).
Conditional random fields: Probabilistic models for
segmenting and labeling sequence data. In Proceed-
ings of the Eighteenth International Conference on
Machine Learning, ICML ’01, pages 282–289, San
Francisco, CA, USA. Morgan Kaufmann Publishers
Inc.
Le, H., Vial, L., Frej, J., Segonne, V., Coavoux, M., Lecou-
teux, B., Allauzen, A., Crabb
´
e, B., Besacier, L., and
Schwab, D. (2019). Flaubert: Unsupervised lan-
guage model pre-training for french. arXiv preprint
arXiv:1912.05372.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2019). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics.
Levine, J. M. (2003). De-identification of ICU patient
records. PhD thesis, Massachusetts Institute of Tech-
nology.
Liu, Z., Tang, B., Wang, X., and Chen, Q. (2017). De-
identification of clinical notes via recurrent neural
network and conditional random field. Journal of
Biomedical Informatics, 75.
Loshchilov, I. and Hutter, F. (2017). Decoupled weight de-
cay regularization. arXiv preprint arXiv:1711.05101.
Nothman, J., Ringland, N., Radford, W., Murphy, T.,
and Curran, J. R. (2013). Learning multilingual
named entity recognition from wikipedia. Artificial
Intelligence, 194:151–175. Artificial Intelligence,
Wikipedia and Semi-Structured Resources.
Polignano, M., de Gemmis, M., and Semeraro, G. (2021).
Comparing transformer-based NER approaches for
analysing textual medical diagnoses. In Faggioli,
G., Ferro, N., Joly, A., Maistro, M., and Piroi, F.,
editors, Proceedings of the Working Notes of CLEF
2021 - Conference and Labs of the Evaluation Forum,
Bucharest, Romania, September 21st - to - 24th, 2021,
volume 2936 of CEUR Workshop Proceedings, pages
818–833. CEUR-WS.org.
Sch
¨
afer, H., Idrissi-Yaghir, A., Horn, P., and Friedrich, C.
(2022). Cross-language transfer of high-quality anno-
tations: Combining neural machine translation with
cross-linguistic span alignment to apply NER to clini-
cal texts in a low-resource language. In Proceedings of
the 4th Clinical Natural Language Processing Work-
shop, pages 53–62, Seattle, WA. Association for Com-
putational Linguistics.
Stubbs, A., Kotfila, C., and Uzuner,
¨
O. (2015). Auto-
mated systems for the de-identification of longitudinal
clinical narratives: Overview of 2014 i2b2/uthealth
shared task track 1. Journal of biomedical informatics,
58:S11–S19.
Sun, W., Rumshisky, A., and Uzuner, O. (2013). Evaluat-
ing temporal relations in clinical text: 2012 i2b2 chal-
lenge. Journal of the American Medical Informatics
Association, 20(5):806–813.
Sweeney, L. (1996). Replacing personally-identifying in-
formation in medical records, the scrub system. In
Proceedings of the AMIA annual fall symposium, page
333. American Medical Informatics Association.
Tchouka, Y., Couchot, J., Coulmeau, M., Laiymani, D.,
Selles, P., Rahmani, A., and Guyeux, C. (2022). De-
identification of french unstructured clinical notes for
machine learning tasks. CoRR, abs/2209.09631.
Uzuner,
¨
O., Luo, Y., and Szolovits, P. (2007). Evaluating the
state-of-the-art in automatic de-identification. Jour-
nal of the American Medical Informatics Association,
14(5):550–563.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
Velupillai, S., Suominen, H., Liakata, M., Roberts, A.,
Shah, A. D., Morley, K., Osborn, D., Hayes, J., Stew-
art, R., Downs, J., et al. (2018). Using clinical natu-
ral language processing for health outcomes research:
overview and actionable suggestions for future ad-
vances. Journal of biomedical informatics, 88:11–19.
Xiao, Y. and Xiong, L. (2015). Protecting locations with
differential privacy under temporal correlations. In
Proceedings of the 22nd ACM SIGSAC Conference
on Computer and Communications Security, pages
1298–1309.
Zhao, Y. and Chen, J. (2022). A survey on differential pri-
vacy for unstructured data content. ACM Computing
Surveys (CSUR).
HEALTHINF 2023 - 16th International Conference on Health Informatics
104
