
Efficient Representation of Biochemical Structures for Supervised and
Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
Katrin Sophie Bohnsack
a
, Alexander Engelsberger
b
, Marika Kaden
c
and Thomas Villmann
d
Saxon Institute for Computational Intelligence and Machine Learning, University of Applied Sciences Mittweida,
Technikumplatz 17, 09648 Mittweida, Germany
Keywords:
Machine Learning, Embedding, Dissimilarity Representation, Graph Kernels, Structured Data, Small
Molecules.
Abstract:
We present an approach to efficiently embed complex data objects from the chem- and bioinformatics domain
like graph structures into Euclidean vector spaces such that those data bases can be handled by machine
learning models. The method is denoted as sensoric response principle (SRP). It uses a small subset of objects
serving as so-called sensors. Only for these sensors, the computationally demanding dissimilarity calculations,
e.g. graph kernel computations, have to be executed and the resulting response values are used to generate
the object embedding into an Euclidean representation space. Thus, the SRP avoids to calculate all object
dissimilarities for embedding, which usually is computationally costly due to the complex proximity measures
in use. Particularly, we consider strategies to determine the number of sensors for an appropriate embedding as
well as selection strategies for SRP. Finally, the quality of the embedding is evaluated w.r.t. to the preservation
of the original object relations in the embedding space. The SRP can be used for unsupervised and supervised
machine learning. We demonstrate the ability of the approach for classification learning in context of an
interpretable machine learning classifier.
1 INTRODUCTION
The automatic analysis of databases for biochemical
molecules and structures is a rapidly growing field
in bioinformatics, accelerated by the increased num-
ber of available machine learning tools. Frequently,
this involves the comparison of respective structured
data in the form of graphs, which is computation-
ally demanding. A great diversity of graph com-
parison strategies exists, ranging from exact match-
ing procedures based on graph isomorphism, over
inexact matching schemes like graph edit distances
(Gao et al., 2010) to topological descriptors (Li et al.,
2012) or domain-specific variants like molecular fin-
gerprints in the context of virtual screening (Cereto-
Massagué et al., 2015). Graph kernels have attracted
considerable interest as an alternative during the last
decade (Kriege et al., 2020), especially in the machine
a
https://orcid.org/0000-0002-2361-107X
b
https://orcid.org/0000-0002-8547-2407
c
https://orcid.org/0000-0002-2849-3463
d
https://orcid.org/0000-0001-6725-0141
learning community.
The approaches resulting in a statistical (feature-
based) data representation permit the application of
traditional vector-based machine learning algorithms
but generally fail to capture the rich topological and
semantic information encoded by graphs. In contrast,
kernel approaches and edit distances work directly on
the structural representation and may include domain
knowledge about the data at the same time. Hence,
they are often more appropriate for comparison.
However, these approaches restrict the model
choice for machine learning algorithms to
(dis)similarity-based variants. In the context of
classification tasks, the respective models are e.g.
k-nearest neighbors (Cover and Hart, 1967) or sup-
port vector machines (Schölkopf and Smola, 2002).
These methods suppose the generation of a data
proximity matrix which depends for one thing on the
number of objects N, then again on the complexity
of the dissimilarity calculation K. Particularly,
N · (N − 1)/2 proximity calculations are necessary,
yielding an overall complexity of O(N
2
· K) to obtain
a ready-to-use data proximity representation.
Bohnsack, K., Engelsberger, A., Kaden, M. and Villmann, T.
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric Embeddings.
DOI: 10.5220/0011644000003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 59-69
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
59

While the distance calculation between a pair of
vectors is linear in the number of features, for graphs
it frequently grows exponentially in the number of
nodes. Consequently, the computation load may be
unfeasible for huge data sets or large graphs, let alone
their combination. But precisely such data are often
present in bioinformatics, for example, given by pro-
tein contact (Di Paola et al., 2013) or metabolic net-
works (Jeong et al., 2000).
In the seminal work by (Pekalska and Duin, 2005),
an alternative data representation based on an object
mapping into a proximity space is introduced, which
was resumed and extended to the graph domain by
(Riesen and Bunke, 2010). This work brings together
the two advantages of direct structure-based graph
comparison and a resulting vectorial representation.
However, it is still highly affected by unfavorable
complexities in distance calculation as given by graph
kernels.
1.1 Our Contribution
We present a strategy that draws on this dissimilar-
ity representation of graphs but avoids calculating
all N · (N − 1) /2 distances between the objects. In-
stead, we propose to select n ≪ N objects as refer-
ences and only calculate their distances to all other
objects. Then each object can be represented by a n-
dimensional vector containing the object’s distances
to the references. This realizes a generally nonlinear
embedding into R
n
which now entails only complex-
ity O(N ·K). Our method provides assistance with de-
termining a sufficient amount of reference instances
by means of a geometric stop criterion for successive
reference set generation. Furthermore, we provide a
measure for evaluating how much of the original data
relations remain unchanged when applying this data
embedding while ensuring huge savings in computa-
tion load. The resulting vectorial data representation
may be used in any standard (un-)supervised learning
algorithm.
Although the presented concept seems closely re-
lated to that from (Bohnsack et al., 2022), the basic
ideas should be thoroughly distinguished: Here, we
investigate a data embedding induced by multiple ref-
erences but one proximity measure, while in our pre-
vious work we relied on multiple notions of proximity
but one datum as reference.
1.2 Roadmap
The remainder of this contribution is structured as fol-
lows: Section 2 provides primers on structured data
comparison by graph kernels and data classification
by variants of learning vector quantization. Readers
already familiar with these concepts may join the train
of thoughts in Section 3, where the sensoric response
principle is introduced. In Section 4, the challenges of
suitable sensor (reference) selection are highlighted,
accompanied by conceivable solutions. We demon-
strate the approaches abilities in Section 5 on illustra-
tive classification problems from the biochemical do-
main and put these findings into perspective for future
investigations in Section 6.
2 BACKGROUND
2.1 Graph Comparison by Kernels
Kernels. Informally, a kernel is a function to com-
pare two objects. Mathematically, it corresponds to
an inner product: Let G be a non-empty set of data
points and κ : G × G → R be a function. Then κ is a
kernel on G if there is a Hilbert space H
κ
and a feature
map φ : G → H
κ
such that κ(g
i
,g
k
) = ⟨φ(g
i
),φ(g
k
)⟩
for g
i
,g
k
∈ G, where ⟨·, ·⟩ denotes the inner product
of H
κ
. Such a feature map exists iff the function κ is
positive semi-definite and symmetric. Every real ker-
nel determines a (semi)metric between structures g
i
and g
k
by
δ
κ
(g
i
,g
k
) =
p
κ(g
i
,g
i
) − 2κ(g
i
,g
k
) + κ(g
k
,g
k
) . (1)
Kernels are of interest because they can sometimes
provide a way of efficiently computing inner products
in high-dimensional spaces and may be defined for
any type of data.
Kernels on Structured Data. Kernels for struc-
tured data such as graphs are usually instances of
so-called convolution kernels (Haussler, 1999). This
concept is based on substructure decomposition. The
graph gets divided into parts, on which base kernel
functions are defined, leading to a new kernel on the
composed object. Kernels may be designed by choos-
ing H and φ, and simply evaluating ⟨φ(g
i
),φ(g
k
)⟩
H
.
This, however, requires operations in H , which might
be computationally demanding such that efficient cal-
culations of κ(g
i
,g
k
) are aspired instead (kernel trick).
Graph kernels differ in the structural properties
they utilise as becomes apparent by considering the
following prominent instances:
• Vertex histogram kernel: Compares the vertex la-
bel histograms by means of a linear or Gaussian
RBF kernel.
• Shortest path: Compares the sequences of vertex
and/or edge labels that are encountered through
traversals through graphs.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
60

• Weisfeiler-Lehman subtree kernel: Compares re-
fined node label histograms, emerged from an iter-
ative relabeling (color refinement) process based
on neighborhood aggregation.
Unfortunately, the flexibility that graph kernels offer
is overshadowed by their mostly prohibitive compu-
tational load if rich structural and label information
is taken into account. For further details, comprehen-
sive surveys can be found in (Kriege et al., 2020) and
(Nikolentzos et al., 2021).
2.2 Classification by Learning Vector
Quantization
The Rise of Interpretable Models. In so-called
black-box models, the rules and insights used to make
predictions frequently remain unclear. However, es-
pecially in life science applications, trustworthiness
and interpretability become more and more impor-
tant for practicioners (Lisboa et al., 2021), forming
the cornerstones of explainable artificial intelligence
(Barredo Arrieta et al., 2020). Prototype-based clas-
sifiers like variants of learning vector quantization
(LVQ) are well-known representatives for models that
are interpretable by design.
Learning Vector Quantization. Generalized
Learning Vector Quantization (GLVQ) as intro-
duced in (Sato and Yamada, 1996) supposes a set
X = {x
x
x
i
}
n
i=1
⊂ R
n
of training data with class labels
c(x
x
x
i
) ∈ C = { 1, . . . ,C }. Further, trainable prototype
vectors w
w
w ∈ W = { w
w
w
j
}
|W |
j=1
⊂ R
n
with class labels
c(w
w
w
j
) ∈ C are required such that each class of C is
represented by at least one prototype. GLVQ aims to
distribute the prototype vectors in the data space such
that the class label of any new input x
x
x /∈ X can be
inferred by means of the nearest prototype principle
given by c(w
w
w
s(x
x
x)
) where s(x
x
x) = argmin
j
d(x
x
x,w
w
w
j
)
and d(x
x
x,w
w
w
j
) is a distance measure usually chosen
as the Euclidean metric. The questions where
and how to place the prototypes are guided by a
dissimilarity-based objective function approximating
the classification error. This objective relates to
the concept of large margin classification ensuring
robust classification (Crammer et al., 2003). An
important conceptual extension of GLVQ is given
by matrix relevance learning (GMLVQ) (Schneider
et al., 2009). This framework addresses the problem
that weighting the input (data) dimensions equally
like in standard GLVQ is an undesirable property
for most practical applications. Only a parametric
form of the dissimilarity measure is fixed in ad-
vance, while its parameters are considered adaptive
quantities that can be optimized in the data-driven
training phase along with the prototypes. Particu-
larly, a semi-metric d
Ω
Ω
Ω
(x
x
x,w
w
w) is considered where
d
2
Ω
Ω
Ω
(x
x
x,w
w
w) = (Ω
Ω
Ω(x
x
x − w
w
w))
2
and Ω
Ω
Ω ∈ R
m×n
, m ≤ n, is a
mapping matrix subject to adaptation during learning.
Yet, GMLVQ remains a robust classifier by means of
implicit margin optimization like GLVQ (Saralajew
et al., 2020).
Inferences on Feature Relevances. After training,
insightful information may be derived by consider-
ing the classification correlation matrix (CCM) Λ
Λ
Λ =
Ω
Ω
Ω
⊺
Ω
Ω
Ω. The entries Λ
jl
reflect the correlations between
the j
th
and l
th
feature, that contribute to a class dis-
crimination. If |Λ
jl
| ≫ 0 the respective feature cor-
relation is important to separate the classes, whereas
|Λ
jl
| ≈ 0 indicates that either the correlation between
those features does not contribute to the decision or
that this information is already contained elsewhere.
The vector λ
λ
λ = (λ
1
,...,λ
n
)
⊺
with λ
k
=
∑
l
|Λ
kl
| pro-
vides the overall importance of the k
th
feature for the
separation of the data set and is denoted as classifi-
cation influence profile (CIP) (Kaden et al., 2022).
Such investigations should be done together with ma-
chine learning experts in order to obtain valid inter-
pretations of feature relevance (Strickert et al., 2013;
Frenay et al., 2014).
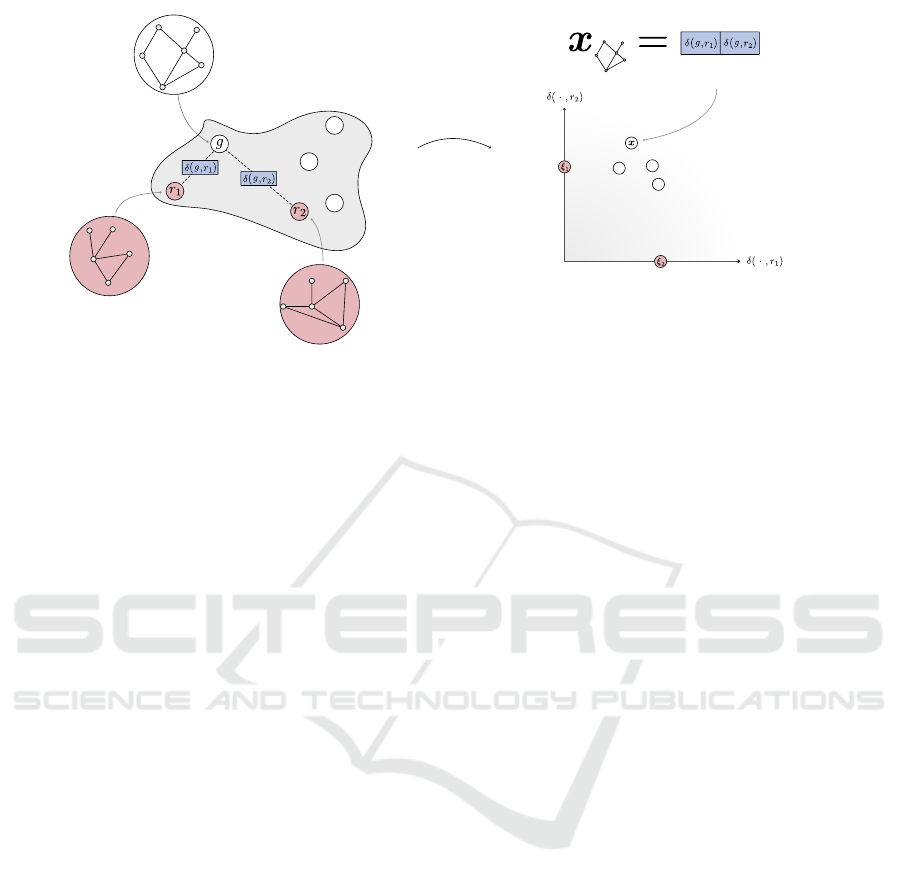
3 OBJECT EMBEDDING BY
MULTI-SENSOR RESPONSES
Let G = {g
i
}
N
i=1
be a finite set of objects, i.e. struc-
tured data like graphs or respective variants such as
trees or sequences. The dissimilarity measure be-
tween elements of G is denoted as δ and may be given
as e.g. graph kernel distance, see Equation (1). Con-
sideration of all pairwise object dissimilarities yields
the matrix ∆
∆
∆ ∈ R
N×N
with entries δ
ik
= δ(g
i
,g
k
). Ob-
viously, determination of ∆
∆
∆ requires N
2
calculations.
Assuming symmetry and zero-diagonal (given if δ
is a proper metric) still requires
N(N−1)
2
calculations
which becomes computationally unfeasible for huge
N and operations of high time complexity.
Let R = {r
j
}
n
j=1
⊂ G with n ≪ N be a subset
of objects, henceforth denoted as references. Consid-
eration of pairwise dissimilarities between all objects
and references yields the reduced dissimilarity matrix
∆
∆
∆
R
∈ R
N×n
with entries δ
i j
= δ(g
i
,r
j
).
• In fact, row δ
δ
δ
i·
·
·
= (δ(g
i
,r
1
),...,δ(g
i
,r
n
)) charac-
terizes object g
i
in terms of dissimilarities (re-
sponses) to the elements of this reference set (sen-
sors) and can be understood as embedding (see
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
61

Figure 1). Henceforce, this procedure is de-
noted as multiple sensor response principle (SRP).
The embedding of all g
i
∈ G yields the set X =
{x
x
x
i
}
N
i=1
with x
x
x
i
= δ
δ
δ
i·
·
·
∈ R
n
. The subset of embed-
ded references r
j
∈ R yields X
R
= {ξ
ξ
ξ
j
}
n
j=1
.
• Analogously, the column vector δ
δ
δ
·
·
· j
=
(δ(g
1
,r
j
),...,δ(g
N
,r
j
))
⊺
characterizes refer-
ence r
j
in dependence on all objects.
Assuming δ fulfills the metric properties, the mapped
data lie within an n-dimensional (potentially asym-
metric) prism, whose lower bound is given by the hy-
perplane containing the mapped references (P˛ekalska
et al., 2006).
By means of standard vector dissimilarities d such
as the squared Euclidean distance, we can consider
D
D
D ∈ R
N×N
with entries d
ik
= d(x
x
x
i
,x
x
x
k
) denoting the
dissimilarity between embedded objects. Recapitulat-
ing, both δ and d measure object dissimilarities, albeit
in fundamentally different ways: δ in the original (ob-
ject) space and d in the proximity (embedding) space.
4 SENSOR SELECTION
STRATEGIES
The most simple strategies for sensor (reference) se-
lection are i) to consider all available data as refer-
ences or ii) to choose references uniformly at random.
However, the first one requires all pairwise distances,
such that it does not reduce the computational costs,
whereas the latter may introduce noisy or redundant
information into the embedding space. According
to (Riesen and Bunke, 2010) the selection procedure
should avoid too similar references or potential out-
liers as sensors.
Drawbacks of available selection procedures
Given a complete dissimilarity matrix, various
schemes are available to obtain a suitable reference
set: In (Riesen and Bunke, 2010; P˛ekalska et al.,
2006) various geometrically inspired selectors are in-
vestigated. However, all of these procedures inher-
ently rely on determination of the median or marginal
in the graph domain, and thus all pairwise distances.
Alternatively, k-medians (Bradley et al., 1996) or
learning procedures, capable of handling proximity
data, such as Median neural gas (Cottrell et al., 2006)
or Affinity propagation (Frey and Dueck, 2007) may
be considered in order to approximate the data distri-
bution. However, as already emphasised, calculation
of the complete dissimilarity matrix may be computa-
tionally inconvenient. For this reason, also traditional
feature subset selection/reduction algorithms (Guyon
and Elisseeff, 2003), in conjunction with considering
all available data as references, are not an alternative.
The dimensionality of the mapping space is a
crucial parameter of the SRP. However, an adequate
choice is frequently left to the applicant’s judge-
ment or subject to a grid search, involving the train-
ing/consideration of many models for the subsequent
task.
In the following sections, we tackle both chal-
lenges: First, we address strategies for finding refer-
ences (circumventing the calculation of all pairwise
graph distances) and evaluating their quality with re-
spect to the resulting data mapping. And second, we
present a model-independent technique for obtaining
a suitable (sufficient but not excessive) number of ref-
erences for the task at hand.
4.1 Selecting Instances of References
We propose using the following strategies:
• Random selection: Sample references indepen-
dently and uniformly at random from G.
• k-means++ initialization: Samples references
with probability proportional to their squared dis-
tance to the closest already chosen reference
(Arthur and Vassilvitskii, 2007), see Algorithm 1
(Bhattacharya et al., 2020).
Algorithm 1: k-means++ based reference selector.
1: procedure SAMPLE K-MEANS++(G, n)
2: Sample r
1
independently and uniformly at
random from G
3: Let R = {r
1
}
4: while |R | < n do
5: for g
i
∈ G do
6: p(g
i
) :=
min
r
j
∈R
δ(g
i
,r
j
)
2
∑
g
k
∈G
min
r
j
∈R
δ(g
k
,r
j
)
2
7: Sample r
l
from G, where every g
i
∈ G has
probability p(g
i
)
8: Update R = R ∪ {r
l
}
9: return R = { r
1
,...,r
n
}
Further, we investigate the following approach, which
generally is believed to be more inconvenient w.r.t.
the described requirements for reference selectors.
Thus, it is considered as a negative benchmark in this
study:
• Next neighbour strategy: The reference is chosen
as the closest (minimum distance) to the previ-
ously chosen sensor set.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
62
space of
graphs
sensor
graph
sensor
graph
graph
mapping
dissimilarity
space
sensoric response
for two references
Figure 1: Visualization of the mutiple-SRP embedding.
4.2 Selecting the Number of References
If the amount of chosen references is not sufficient, it
may not be possible to display all structural informa-
tion. If, on the other hand, too many are selected,
this information may be hidden in noise, which in
turn may harm the classifier (curse of dimensionality).
Furthermore, due to the highly complex graph com-
parison measures considered, we want to keep dis-
tance calculations to a minimum. For this, we present
multiple strategies of finding a suitable amount by
means of forward selection, i.e., incremental refer-
ence set construction:
• Estimate the intrinsic (Hausdorff) dimension of
the data in the mapping space, e.g. by using cor-
relation integrals according to (Grassberger and
Procaccia, 1983) and iteratively add references
until the values reach a saturation point. Then this
dimension corresponds to the number of variables
needed in a minimal representation of the data, i.e.
the number of dominating parameters to describe
the data manifold.
• Given a metric δ in object space, the triangle in-
equality and its reverse can be utilized to deter-
mine the uncertainty of unseen entries in the dis-
tance matrix.
Yet, estimating the intrinsic dimension of data by
correlation integrals is noise sensitive and requires
a huge number of data (Camastra and Vinciarelli,
2001), which is obviously not valid for the reference
set. Therefore we focus on the second of the above
options.
The Triangle Span as a Stopping Criterion.
Given a subset of objects (references) with known dis-
tance values between them, the triangle inequality can
be used to estimate an interval covering the range of
an unknown (non-calculated) distance between each
pair of objects of the full data set. Let r
j
∈ R be a
single reference, g
i
,g
k
∈ G are objects and δ is the
given distance measure in G. Then the inequality
l
r
j
(g
i
,g
k
) ≤ δ(g
i
,g
k
) ≤ u
r
j
(g
i
,g
k
)
holds with u
r
j
(g
i
,g
k
) = δ(g
i
,r
j
) + δ(r
j
,g
k
) being an
upper bound and l
r
j
(g
i
,g
k
) = |δ(g
i
,r
j
) − δ(r
j
,g
k
)| is
a lower bound. We consider the triangle span
T S
r
j
(g
i
,g
k
) = u
r
j
(g
i
,g
k
) − l
r
j
(g
i
,g
k
).
If r
j
∈ {g
i
,g
k
}, i.e. r
j
= g
i
or r
j
= g
k
is valid both
bounds are equal and, hence, the span becomes zero.
In case of multiple references R = { r
j
}
n
j=1
the corresponding span T S
R
(g
i
,g
k
) is calculated
according to the modified bounds: u
R
(g
i
,g
k
) =
min
r
j
∈R
u
r
j
(g
i
,g
k
) and, analogously, l
R
(g
i
,g
k
) =
max
r
j
∈R
l
r
j
(g
i
,g
k
).
One can easily show for an extended reference
set R
′
⊃ R the inequality T S
R
(g
i
,g
k
) ≥ T S
R
′
(g
i
,g
k
)
is valid, the triangle span is monotonically de-
creasing with increasing reference set converging to
zero. Hence, the mean of all triangle span values
T S
R
(g
i
,g
k
) can be used as a stopping criterion for
reference set expansion by thresholding.
4.3 An Evaluation Measure for the
Reference Induced Embedding
Ideally, the SRP mapping preserves the original rela-
tions according to δ between the objects w.r.t. an ap-
propriate distance measure d in the embedding space.
We can compare the corresponding dissimilarity ma-
trices ∆
∆
∆ and D
D
D by means of the Normalized Rank
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
63

Equivalence (NRE) measure (Nebel et al., 2017): We
consider the dissimilarity rank matrix P
P
P
δ
w.r.t. dis-
similarity measure δ. The entries
p
δ
ik
=
N
∑
l=1
H(δ(g
i
,g
k
) − δ(g
i
,g
l
))
denote the number of objects from G which have a
higher similarity to object g
i
than g
k
. Analogously, we
take P
P
P
d
w.r.t. the dissimilarity measure d for elements
of the embedding space X . Then the absolute rank-
equivalence measure is given as
ϒ
G,X
(δ,d) =
N
∑
i=1
N
∑
k=1
|p
δ
ik
− p
d
ik
|
This quantity can be normalized by the constant
c =
(
N
N
2
2
if N is even
N
(N−1)(N+1)
2
if N is odd
to enable comparability between different data set car-
dinalities. The rank-equivalence measure is close to
zero for mostly perfect embedding, preserving the
topological relations. Hence, it can serve for evalu-
ation of the embedding.
Note that we use this evaluation measure which
requires full ∆
∆
∆ solely to highlight the possibilities
and limitations of the proposed mapping to proxim-
ity space. In order to be able to make estimates re-
garding the rank-equivalence in real applications, we
recommend considering the subsets ∆
∆
∆
T
⊂ ∆
∆
∆ ∈ R
n×n
with δ
jl
= δ(r
j
,r
l
) and D
D
D
T
⊂ D
D
D ∈ R
n×n
with d
jl
=
d(ξ
ξ
ξ
j
,ξ
ξ
ξ
l
), which have to be calculated for the mapping
anyway.
5 EXPERIMENTS
This section aims at empirically evaluating the multi-
sensor embedding of graphs.
5.1 Data Set Description and
Experimental Setup
Data Sets. The experiments were conducted on the
TUDataset benchmark collection of data sets for su-
pervised learning with graphs (Morris et al., 2020).
Particularly, the following biochemically-motivated
classification tasks on small molecule and protein
graphs were considered.
Small molecules are modelled as graphs, where
vertices represent atoms and edges represent covalent
bonds. Their labels correspond to the atom type and
the bonding order (valence of the linkage), respec-
tively. Explicit hydrogen atoms are omitted.
• AIDS: For these compounds obtained from the
AIDS Antiviral Screen Database the task is to pre-
dict whether or not they are active against HIV
(Riesen and Bunke, 2008).
• MUTAG: It is to predict whether or not the con-
tained (hetero)aromatic nitro compounds have a
mutagenic effect on the Gram-negative bacterium
Salmonella typhimurium (Debnath et al., 1991).
• PTC-MR: For organic compounds from the pre-
dictive toxicology challenge (Helma et al., 2001)
their carcinogenic effect on rodents, particularly
male rats is to be predicted.
Furthermore, a graph data set about protein structures
was considered:
• ENZYMES: Graphs are modelled from enzymes
obtained from the BRENDA database (Schom-
burg, 2004). Secondary structure elements (SSE)
are considered as vertices, annotated by their type,
i.e. sheet, turn or helix. Edges are drawn between
vertices if they are either neighbours in the amino
acid sequence or among the 3 nearest neighbours
in 3D space. They are annotated with their type,
i.e. sequential or structural. The task is to as-
sign them to one of six top-level Enzyme Com-
mision (EC) classes, indicating the chemical re-
actions they catalyse (Borgwardt et al., 2005).
Node attributes available for some of the data sets
were neglegted.
Implementation Details. The kernel calculation
was conducted via the GraphKernels library by
(Sugiyama et al., 2018), which implements the ker-
nels described in Section 2.1. Classification bases on
the GMLVQ, see Section 2.2, with one prototype per
class and 10-fold cross-validation.
5.2 Results and Discussion
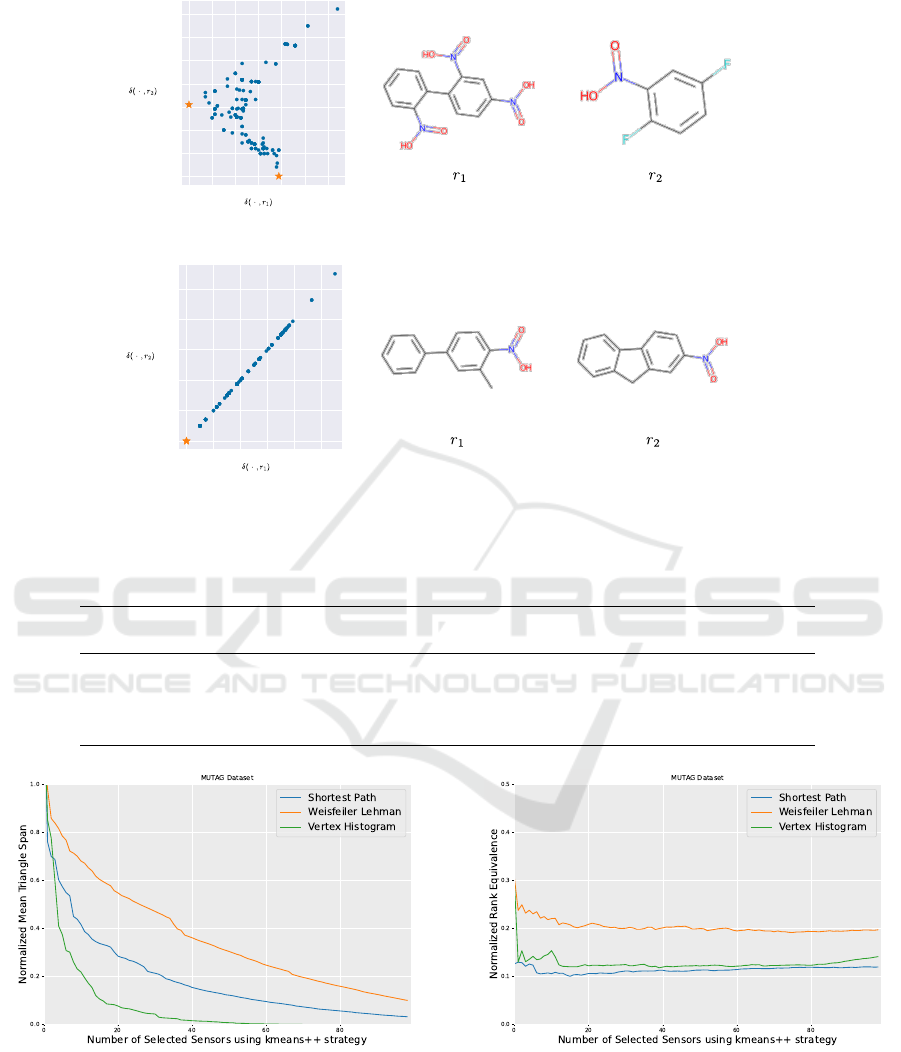
Reference Selection. A quick success of the de-
scribed embedding strongly depends on a favourable
choice of the underlying references. This can be
understood by considering the minimal example in
Figure 2: Depicted are the mapping spaces δ
δ
δ
i·
·
·
=
(δ(g
i
,r
1
),δ(g
i
,r
2
)) of MUTAG induced by two refer-
ences chosen via the k-means++ and next-neighbour
strategy based on the Vertex Histogram kernel, as well
as the reference’s chemical structures. While for k-
means++ the respective data manifold actually is in-
trinsically 2-dimensional, it remains 1-dimensional
for the next-neighbour approach. This implies that
for the latter case consideration of the second refer-
ence graph did not provide/capture new information
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
64

or properties not already represented before. This is
obvious as the molecules display high structural simi-
larity, therefore kernel distances w.r.t. them are highly
correlated. We might need more iterations to capture
the relevant information and thereby unnecessary in-
crease the data dimensionality for downstream appli-
cations.
It has been proven that the k-means++ initializa-
tion for k-means leads in probability to an optimal
distribution of its prototypes (here references) in the
sense of information optimum coding (Arthur and
Vassilvitskii, 2007). Thus, it overcomes the problem
of initialization sensitive behavior (stucking in local
optima) of the standard k-means. In the context of the
problem at hand, particularly, the probabilistic model
for prototype initialization in k-means++ has to be
emphasized. Here it is used to determine the reference
vectors (see Algorithm 1). In fact, it prevents an unfa-
vorable selection of molecule/graph outliers, which is
unsuitable as discussed in (Riesen and Bunke, 2010).
So far, purely mathematical criteria have been
considered for reference selection. However, if task-
driven prior domain knowledge is available, this can,
and should, be integrated into the selection scheme.
Obviously, this would contribute to a better inter-
pretability. For example, domain knowledge of bio-
chemists regarding specific properties of molecules or
molecule groups could be used to select references
that represent certain classes of molecules. Other-
wise, heuristic selection strategies may complicate
later interpretations but could be unavoidable for spe-
cific problems.
Stop Criterion. In general, the original data in use
are Euclidean embeddable only under certain condi-
tions, which can be formulated in terms of the full dis-
similarity matrix (Pekalska and Duin, 2005). If these
conditions are not fulfilled and an Euclidean embed-
ding is forced, topological distortions, i.e. disconti-
nuities in the mapping occur. These distortions may
be captured by the NRE by values greater than 0 (see
Figure 4).
In this sense, the SRP based on the presented sen-
sor selection strategies defines a surrogate or approx-
imation model for such an information-optimal em-
bedding. Theoretically, there is a sensor configura-
tion with minimum approximation error, i.e. mini-
mum rank equivalence between the data in the origi-
nal and the embedding space. But, because it is nec-
essary to know the complete kernel matrix in advance
to calculate this measure, it is not feasible for huge
data sets.
The MTS (see Figure 3), as explained before, sup-
ports the decision making, whether adding another
sensor probably can improve our knowledge about the
properties of the original (full) but unknown dissim-
ilarity matrix. If the MTS is small, it is possible to
estimate the whole matrix with only small deviations
and, hence, few rank swaps. Consequently, although
the MTS is not directly connected to the NRE of the
embedding, it gives strong insights into the informa-
tion retrieval of the selection.
In our experiments we used a fixed threshold for
the mean triangle span as stopping point. Thresholds
dependent on the chosen kernel or the data set size, or
finding the stopping point by taking the shape of the
function into account (e.g. finding saturation points)
could lead to even better results.
More sophisticated selection schemes based on
the prediction of the whole matrix can be considered.
Classification. Table 1 gives an overview of the
achieved classification accuracies by GMLVQ on the
embedded data for different graph kernels side-by-
side with benchmark results of an SVM classifier
in the original graph domain by (Nikolentzos et al.,
2021). The corresponding number of selected sensors
is given in Table 2. In general, it can be observed that
our results are comparable under the premise of enor-
mous savings in computation time. Due to the chosen
criterion, always less than 5% percent of the data set
were used as sensors. This scales the problem down
by a complexity (time) factor of at least 20.
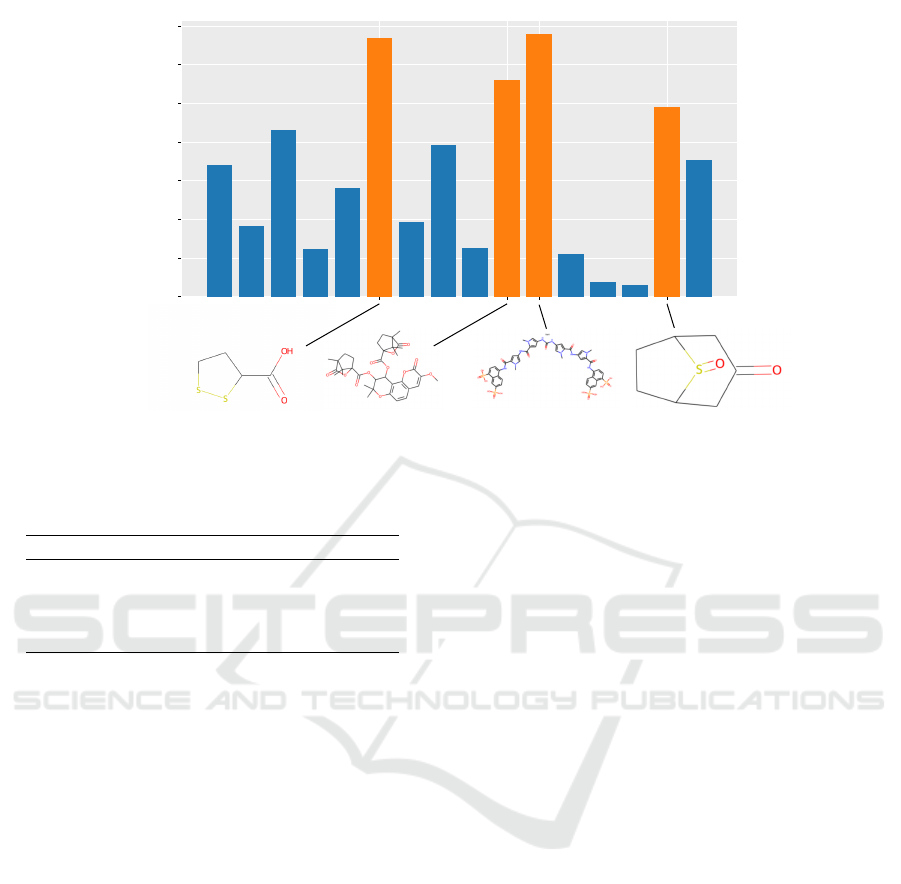
Figure 5 highlights the features, i.e. refer-
ences/sensors with high values in GMLVQ’s CIP for
the AIDS data set and the shortest path kernel. The
spatial relations (distance values) w.r.t. the depicted
chemical structures in orange have great significance
in terms of the classification decision and, hence, may
give valuable information regarding the sensitivity of
the embedding with respect to given molecule struc-
tures.
At this point it should be reflected that also the
insights provided by interpretable models have their
limitations. In the presented approach, the reduction
of the problem to the dissimilarity space by sensors
introduces an information bottleneck. Since only dis-
tances in terms of certain graph kernels are consid-
ered, inferences about relevant properties and features
of underlying graph structures are challenging to say
the least.
6 CONCLUSIONS
In this contribution, we propose a multi-sensoric re-
sponse principle for efficient embedding of graph ob-
jects into an Euclidean feature vector space based on
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
65
0 2 4 6 8 10 12
0. 0
2. 5
5. 0
7. 5
10 . 0
12 . 5
15 . 0
17 . 5
Dissimilarity Space
(a) kmeans++
0 2 4 6 8 10
0
2
4
6
8
10
Dissimilarity Space
(b) nearest neighbour
Figure 2: Dissimilarity space visualization for the kmeans++ selection (a) and the nearest neighbor selection (b).
Table 1: Comparison of GMLVQ using accuracy and standard deviation with kmeans++ selected sensors and a fixed threshold
for mean triangle span compared to SVM results from (Nikolentzos et al., 2021).
VH WL-VH SP
GMLVQ SVM GMLVQ SVM GMLVQ SVM
AIDS 96.1 (±1.4) 80.0 (±2.3) 98.8 (±0.4) 98.3 (±0.8) 97.4 (±1.5) 99.3 (±0.4)
ENZYMES 17.8 (±4.5) 20.0 (±4.8) 25.3 (±3.2) 50.7 (±7.3) 23.8 (±4.5) 37.3 (±8.7)
MUTAG 85.1 (±7.1) 69.1 (±4.1) 73.4 (±12.5) 86.7 (±7.3) 81.4 (±7.5) 82.4 (±5.5)
PTC-MR 60.2 (±6.0) 57.1 (±9.6) 57.6 (±7.5) 64.9 (±6.4) 61.2 (±7.1) 60.2 (±9.4)
Figure 3: Course of the Mean triangle span as a function of
the number of sensors/references.
their proximities obtained by graph kernels. The re-
sulting embedding representation can be used in both
supervised and unsupervised machine learning. For
this purpose, only a small subset of all available ob-
jects is selected to serve as references/sensors. Only
Figure 4: Course of the Normalized Rank Equivalence as a
function of the number of sensors/references.
for these references the proximities to all available
objects have to be calculated, which avoids the de-
termination of the complete proximity/kernel matrix
as usual. For the cardinality of the reference set, a
good tradeoff between the maintenance of a sufficient
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
66
0.0 0
0.0 2
0.0 4
0.0 6
0.0 8
0.1 0
0.1 2
0.1 4
Influence Profile - AIDS - Shortest Path
Figure 5: Example for an influence profile determined by GMLVQ for AIDS: Most relevant sensors are highlighted and the
corresponding molecules are depicted.
Table 2: Selected number of sensors in our experiments and
their proportion of the complete dataset.
VH WL-VH SP
AIDS 3 (0.2%) 43 (2.2%) 17 (0.9%)
ENZYMES 2 (0.3%) 22 (3.7%) 7 (1.2%)
MUTAG 3 (1.6%) 7 (3.7%) 2 (1.1%)
PTC-MR 5 (1.5%) 16 (4.7%) 7 (2.0%)
amount of relations from the original graph domain
and the computational complexity is striven as well as
an appropriate selection scheme, especially for poten-
tial real-world applications. For both problems, fea-
sible solutions are provided. Results in molecule and
structure classification serve as proof of concept.
This work offers versatile starting points for fu-
ture investigations: Regarding the reference set selec-
tion from the data, density-based approaches may be
considered. Moreover, methods for low-rank approxi-
mations of kernel matrices via Nyström like the ridge
Leverage score (Alaoui and Mahoney, 2015) or an-
chor nets (Cai et al., 2022) may be adapted for the
selection process. The sensors/references are known
as landmark points in this context. Alternatively,
reference-graphs may be given data-independent by
taking artificial graphs such as graphlets. Other stop
criteria for the incremental reference set construction
may be defined: Foremost, measures based on in-
formation gain are considered to reflect the essen-
tial properties w.r.t. reference induced redundancies
in the data representation. Evaluating the quality of
the induced embedding may be refined. Particularly,
other quality scores such as (Lee and Verleysen, 2009;
Mokbel et al., 2013) may be considered. Finally,
other time-consuming graph proximity measures than
graph kernels may underlie the principle, e.g. graph
edit distances (Gao et al., 2010). But the principle
can even be generalized to other data structures that
involve high distance computation loads as e.g. se-
quence data with respective costly edit (alignment)
distances (Smith and Waterman, 1981; Needleman
and Wunsch, 1970). This becomes especially inter-
esting for the guide tree construction step in MSAs
(Blackshields et al., 2010).
Future investigations may combine the presented
approach with the multi-proximity response princi-
ple introduced in (Bohnsack et al., 2022), which is
closely related to the concept of multiple kernel learn-
ing (Donini et al., 2017). Following this methodology,
the SRP may become a promising and efficient alter-
native to standard approaches for handling heteroge-
neous data in machine learning, which have complex
structures requiring computational intensive proxim-
ity calculations.
ACKNOWLEDGEMENTS
This work has partially been supported by the
project “MaLeKITA” funded by the European Social
Fund (ESF), the project “AIMS” (subprojects “IAI-
XPRESS” and “DAIMLER”) funded by the German
Space Agency (DLR) and the project “PAL” funded
by the German Federal Ministry of Education and Re-
search (BMBF).
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
67

AUTHOR’S CONTRIBUTION
K.S.B. and A.E. contributed equally.
REFERENCES
Alaoui, A. and Mahoney, M. W. (2015). Fast randomized
kernel ridge regression with statistical guarantees. In
Cortes, C., Lawrence, N., Lee, D., Sugiyama, M., and
Garnett, R., editors, Advances in Neural Information
Processing Systems, volume 28. Curran Associates,
Inc.
Arthur, D. and Vassilvitskii, S. (2007). K-means++: The
Advantages of Careful Seeding. In Proceedings of the
Eighteenth Annual ACM-SIAM Symposium on Dis-
crete Algorithms, pages 1027–1035, PA, USA. Soci-
ety for Industrial and Applied Mathematics Philadel-
phia.
Barredo Arrieta, A., Díaz-Rodríguez, N., Del Ser, J., Ben-
netot, A., Tabik, S., Barbado, A., Garcia, S., Gil-
Lopez, S., Molina, D., Benjamins, R., Chatila, R., and
Herrera, F. (2020). Explainable Artificial Intelligence
(XAI): Concepts, taxonomies, opportunities and chal-
lenges toward responsible AI. Information Fusion,
58:82–115.
Bhattacharya, A., Eube, J., Röglin, H., and Schmidt,
M. (2020). Noisy, greedy and not so greedy k-
Means++. In Grandoni, F., Herman, G., and
Sanders, P., editors, 28th Annual European Sym-
posium on Algorithms (ESA 2020), volume 173
of Leibniz International Proceedings in Informat-
ics (LIPIcs), pages 18:1–18:21, Dagstuhl, Germany.
Schloss Dagstuhl–Leibniz-Zentrum für Informatik.
Blackshields, G., Sievers, F., Shi, W., Wilm, A., and Hig-
gins, D. G. (2010). Sequence embedding for fast con-
struction of guide trees for multiple sequence align-
ment. Algorithms for Molecular Biology, 5(1):21.
Bohnsack, K. S., Kaden, M., Voigt, J., and Villmann, T.
(2022). Efficient classification learning of biochem-
ical structured data by means of relevance weighting
for sensoric response features. In ESANN 2022 Pro-
ceedings, page 6.
Borgwardt, K. M., Ong, C. S., Schonauer, S., Vish-
wanathan, S. V. N., Smola, A. J., and Kriegel, H.-P.
(2005). Protein function prediction via graph kernels.
Bioinformatics, 21(Suppl 1):i47–i56.
Bradley, P., Mangasarian, O., and Street, W. (1996). Clus-
tering via concave minimization. In Mozer, M., Jor-
dan, M., and Petsche, T., editors, Advances in Neu-
ral Information Processing Systems, volume 9. MIT
Press.
Cai, D., Nagy, J., and Xi, Y. (2022). Fast Deterministic Ap-
proximation of Symmetric Indefinite Kernel Matrices
with High Dimensional Datasets. SIAM Journal on
Matrix Analysis and Applications, 43(2):1003–1028.
Camastra, F. and Vinciarelli, A. (2001). Intrinsic Dimen-
sion Estimation of Data: An Approach Based on
Grassberger–Procaccia’s Algorithm. Neural Process-
ing Letters, 14(1):27–34.
Cereto-Massagué, A., Ojeda, M. J., Valls, C., Mulero, M.,
Garcia-Vallvé, S., and Pujadas, G. (2015). Molecu-
lar fingerprint similarity search in virtual screening.
Methods, 71:58–63.
Cottrell, M., Hammer, B., Hasenfuß, A., and Villmann, T.
(2006). Batch and median neural gas. Neural Net-
works, 19(6-7):762–771.
Cover, T. and Hart, P. (1967). Nearest neighbor pattern clas-
sification. IEEE Transactions on Information Theory,
13(1):21–27.
Crammer, K., Gilad-Bachrach, R., Navot, A., and A.Tishby
(2003). Margin analysis of the LVQ algorithm. In
Becker, S., Thrun, S., and Obermayer, K., editors, Ad-
vances in Neural Information Processing (Proc. NIPS
2002), volume 15, pages 462–469, Cambridge, MA.
MIT Press.
Debnath, A. K., Lopez de Compadre, R. L., Debnath,
G., Shusterman, A. J., and Hansch, C. (1991).
Structure-activity relationship of mutagenic aromatic
and heteroaromatic nitro compounds. Correlation with
molecular orbital energies and hydrophobicity. Jour-
nal of Medicinal Chemistry, 34(2):786–797.
Di Paola, L., De Ruvo, M., Paci, P., Santoni, D., and
Giuliani, A. (2013). Protein Contact Networks: An
Emerging Paradigm in Chemistry. Chemical Reviews,
113(3):1598–1613.
Donini, M., Navarin, N., Lauriola, I., Aiolli, F., and Costa,
F. (2017). Fast hyperparameter selection for graph
kernels via subsampling and multiple kernel learning.
In ESANN 2017 Proceedings, pages 287–292, Bruges,
Belgium.
Frenay, B., Hofmann, D., Schulz, A., Biehl, M., and Ham-
mer, B. (2014). Valid interpretation of feature rele-
vance for linear data mappings. In 2014 IEEE Sympo-
sium on Computational Intelligence and Data Mining
(CIDM), pages 149–156, Orlando, FL, USA. IEEE.
Frey, B. J. and Dueck, D. (2007). Clustering by
Passing Messages Between Data Points. Science,
315(5814):972–976.
Gao, X., Xiao, B., Tao, D., and Li, X. (2010). A survey
of graph edit distance. Pattern Analysis and Applica-
tions, 13(1):113–129.
Grassberger, P. and Procaccia, I. (1983). Characteriza-
tion of Strange Attractors. Physical Review Letters,
50(5):346–349.
Guyon, I. and Elisseeff, A. (2003). An introduction to vari-
able and feature selection. Journal of machine learn-
ing research, 3(Mar):1157–1182.
Haussler, D. (1999). Convolution kernels on discrete struc-
tures. Technical Report.
Helma, C., King, R. D., Kramer, S., and Srinivasan, A.
(2001). The Predictive Toxicology Challenge 2000-
2001. Bioinformatics, 17(1):107–108.
Jeong, H., Tombor, B., Albert, R., and Oltvai, Z. N. (2000).
The large-scale organization of metabolic networks.
Nature, 407:4.
Kaden, M., Bohnsack, K. S., Weber, M., Kudła, M.,
Gutowska, K., Blazewicz, J., and Villmann, T. (2022).
Learning vector quantization as an interpretable clas-
sifier for the detection of SARS-CoV-2 types based on
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
68

their RNA sequences. Neural Computing and Appli-
cations, 34(1):67–78.
Kriege, N. M., Johansson, F. D., and Morris, C. (2020). A
survey on graph kernels. Applied Network Science,
5(1):6.
Lee, J. A. and Verleysen, M. (2009). Quality assessment of
dimensionality reduction: Rank-based criteria. Neu-
rocomputing, 72(7-9):1431–1443.
Li, G., Semerci, M., Yener, B., and Zaki, M. J. (2012). Ef-
fective graph classification based on topological and
label attributes. Statistical Analysis and Data Mining,
5(4):265–283.
Lisboa, P., Saralajew, S., Vellido, A., and Villmann, T.
(2021). The coming of age of interpretable and ex-
plainable machine learning models. In Verleysen, M.,
editor, Proceedings of the 29th European Symposium
on Artificial Neural Networks, Computational Intelli-
gence and Machine Learning (ESANN’2021), Bruges
(Belgium), pages 547–556, Louvain-La-Neuve, Bel-
gium. i6doc.com.
Mokbel, B., Lueks, W., Gisbrecht, A., and Hammer, B.
(2013). Visualizing the quality of dimensionality re-
duction. Neurocomputing, 112:109–123.
Morris, C., Kriege, N. M., Bause, F., Kersting, K., Mutzel,
P., and Neumann, M. (2020). TUDataset: A collec-
tion of benchmark datasets for learning with graphs.
In ICML 2020 Workshop on Graph Representation
Learning and Beyond (GRL+ 2020).
Nebel, D., Kaden, M., Villmann, A., and Villmann, T.
(2017). Types of (dis-)similarities and adaptive mix-
tures thereof for improved classification learning.
Neurocomputing, 268:42–54.
Needleman, S. B. and Wunsch, C. D. (1970). A gen-
eral method applicable to the search for similarities
in the amino acid sequence of two proteins. Journal
of Molecular Biology, 48(3):443–453.
Nikolentzos, G., Siglidis, G., and Vazirgiannis, M. (2021).
Graph Kernels: A Survey. Journal of Artificial Intel-
ligence Research, 72:943–1027.
P˛ekalska, E., Duin, R. P., and Paclík, P. (2006). Prototype
selection for dissimilarity-based classifiers. Pattern
Recognition, 39(2):189–208.
Pekalska, E. and Duin, R. P. W. (2005). The Dissimilar-
ity Representation for Pattern Recognition: Founda-
tions and Applications, volume 64 of Series in Ma-
chine Perception and Artificial Intelligence. WORLD
SCIENTIFIC.
Riesen, K. and Bunke, H. (2008). IAM graph database
repository for graph based pattern recognition and ma-
chine learning. In da Vitoria Lobo, N., Kasparis, T.,
Roli, F., Kwok, J. T., Georgiopoulos, M., Anagnos-
topoulos, G. C., and Loog, M., editors, Structural,
Syntactic, and Statistical Pattern Recognition, pages
287–297, Berlin, Heidelberg. Springer Berlin Heidel-
berg.
Riesen, K. and Bunke, H. (2010). Graph Classification and
Clustering Based on Vector Space Embedding, vol-
ume 77 of Series in Machine Perception and Artificial
Intelligence. WORLD SCIENTIFIC.
Saralajew, S., Holdijk, L., and Villmann, T. (2020). Fast ad-
versarial robustness certification of nearest prototype
classifiers for arbitrary seminorms. In Larochelle, H.,
Ranzato, M., Hadsell, R., Balcan, M., and Lin, H., ed-
itors, Proceedings of the 34th Conference on Neural
Information Processing Systems (NeurIPS 2020), vol-
ume 33, pages 13635–13650. Curran Associates, Inc.
Sato, A. and Yamada, K. (1996). Generalized learning vec-
tor quantization. In Touretzky DS, Mozer MC, H. M.,
editor, Advances in Neural Information Processing
Systems, volume 8, pages 423–429. MIT Press, Cam-
bridge.
Schneider, P., Biehl, M., and Hammer, B. (2009). Adaptive
Relevance Matrices in Learning Vector Quantization.
Neural Computation, 21(12):3532–3561.
Schölkopf, B. and Smola, A. (2002). Learning with Kernels.
MIT Press, Cambridge.
Schomburg, I. (2004). BRENDA, the enzyme database:
Updates and major new developments. Nucleic Acids
Research, 32(90001):431D–433.
Smith, T. F. and Waterman, M. S. (1981). Identification of
common molecular subsequences. Journal of Molec-
ular Biology, 147(1):195–197.
Strickert, M., Hammer, B., Villmann, T., and Biehl, M.
(2013). Regularization and improved interpretation
of linear data mappings and adaptive distance mea-
sures. In 2013 IEEE Symposium on Computational
Intelligence and Data Mining (CIDM), pages 10–17,
Singapore, Singapore. IEEE.
Sugiyama, M., Ghisu, M. E., Llinares-López, F., and Borg-
wardt, K. (2018). Graphkernels: R and Python
packages for graph comparison. Bioinformatics,
34(3):530–532.
Efficient Representation of Biochemical Structures for Supervised and Unsupervised Machine Learning Models Using Multi-Sensoric
Embeddings
69
