
Improving Mitosis Detection via UNet-Based Adversarial Domain
Homogenizer
Tirupati Saketh Chandra*
a
, Sahar Almahfouz Nasser*
b
, Nikhil Cherian Kurian
c
and Amit Sethi
d
Electrical Engineering Department, Indian Institute of Technology Bombay, Mumbai, Maharashtra, India
∗
Indicates Equal Contribution
Keywords:
MIDOG, Domain Generalization, Mitosis Detection, Domain Homogenizer, Auto-Encoder.
Abstract:
The effective counting of mitotic figures in cancer pathology specimen is a critical task for deciding tumor
grade and prognosis. Automated mitosis detection through deep learning-based image analysis often fails
on unseen patient data due to domain shifts in the form of changes in stain appearance, pixel noise, tissue
quality, and magnification. This paper proposes a domain homogenizer for mitosis detection that attempts to
alleviate domain differences in histology images via adversarial reconstruction of input images. The proposed
homogenizer is based on a U-Net architecture and can effectively reduce domain differences commonly seen
with histology imaging data. We demonstrate our domain homogenizer’s effectiveness by showing a reduction
in domain differences between the preprocessed images. Using this homogenizer with a RetinaNet object
detector, we were able to outperform the baselines of the 2021 MIDOG challenge in terms of average precision
of the detected mitotic figures.
1 INTRODUCTION
In many practical applications of machine learning
models domain shift occurs after training, wherein
the characteristics of the test data are different from
the training data. Particularly in the application of
deep neural networks (DNNs) to pathology images,
the test data may have different colors, stain con-
centrations, and magnification compared to what the
DNN was trained on due to changes in scanner, stain-
ing reagents, and sample preparation protocols. MI-
DOG2021 (Aubreville et al., 2021) (organized with
MICCAI 2021) was the first challenge that addressed
the problem of domain shift in pathology – in this
case, the scanner – as it is one of the reasons behind
the failure of machine learning models after training,
including those for mitosis detection.
Domain generalization is the set of techniques that
improve the prediction accuracy of machine learning
models on data from new domains without assuming
access to those data during training. Proposing and
a
https://orcid.org/0000-0002-0325-5821
b
https://orcid.org/0000-0002-5063-9211
c
https://orcid.org/0000-0003-1713-0736
d
https://orcid.org/0000-0002-8634-1804
testing various domain generalization techniques was
the main goal of the MIDOG2021 challenge.
In this paper, we present our work which is an
extension of our proposed method for MIDOG2021
(Almahfouz Nasser et al., 2021). Our contribution is
three-fold and can be summarised as follows. Firstly,
we modified (Almahfouz Nasser et al., 2021) by shift-
ing the domain classifier from the latent space to the
end of the autoencoder, which improved the results
drastically. Secondly, we showed the importance of
perceptual loss in preserving the semantic informa-
tion, which affects the final accuracy of the object de-
tection part. Finally, unlike our previous work, train-
ing the auto-encoder along with the object detection
network end-to-end improved the quality of the ho-
mogenized outputs substantially.
2 RELATED WORK
In this section, we introduce the most significant no-
table solutions for the MIDOG20201 challenge be-
fore introducing our proposed method.
In (Wilm et al., 2021) the authors modified Reti-
naNet network (Algaissi et al., 2020) for mitosis de-
tection by adding a domain classification head and a
52
Chandra, T., Nasser, S., Kurian, N. and Sethi, A.
Improving Mitosis Detection via UNet-Based Adversarial Domain Homogenizer.
DOI: 10.5220/0011629700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 2: BIOIMAGING, pages 52-56
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

gradient reversal layer to encourage domain agnosti-
cism. In this work, they used a pre-trained Resnet18
for the encoder. For their discriminator, it was a
simple sequence of three convolutional blocks and
a fully connected layer. The domain classifier was
placed at the bottleneck of the encoder. Breen et
al (Breen et al., 2021) proposed a U-Net type archi-
tecture that outputs the probability map of the mi-
totic figures. These probabilities get converted into
bounding boxes around the mitotic figures. They used
a neural style transfer (NST) as a domain adapta-
tion technique. This technique casts the style of one
image on the content of another. The method pro-
posed by (Chung et al., 2021) consists of two parts,
a patch selection and a style transfer module. To
learn the styles of images from different scanners,
they used a StarGAN (Choi et al., 2018). A two
steps domain-invariant mitotic detection method was
proposed by (Nateghi and Pourakpour, 2021). This
method is based on Fast RCNN (Girshick, 2015). For
domain generalization purposes they used StainTools
software (Peter Byfield and Gamper, 2022) to aug-
ment the images. StainTools package decomposes the
image into two matrices, a concentration matrix C and
a stain matrix S. By combining the C and S matrices
from different images they produced the augmented
images. A cascaded pipeline of a Mask RCNN (He
et al., 2017) followed by a classification ensemble was
proposed by (Fick et al., 2021) to detect mitotic can-
didates. A Cycle GAN (Zhu et al., 2017) was used
to transfer every scanner domain to every other scan-
ner domain. In (Jahanifar et al., 2021) the authors
used a stain normalization method proposed by (Va-
hadane et al., 2016) as a preprocessing step for the
images. Others like (Dexl et al., 2021) merged hard
negative mining with immense data augmentation for
domain generalization was proposed by (Dexl et al.,
2021). Stain normalization techniques such as (Rein-
hard et al., 2001) and (Vahadane et al., 2015) were
used in (Long et al., 2021) to account for the domain
difference between images. Almahfouz Nasser et al.,
(Almahfouz Nasser et al., 2021) proposed an autoen-
coder trained adversarially on the sources of domain
variations. This autoencoder makes the appearance of
images uniform across different domains.
In the rest of the paper, we describe our proposed
method, the data and experiments. Then, we show
qualitative and quantitative results of our method and
conclude with the take-home meesage from this work.
3 METHODOLOGY
3.1 Notations
In domain generalization there are source (seen)
domains, which are shown to the model during
training, and there are target (unseen) domains,
which are used only during testing. Labelled sam-
ples from the source domains are represented by
D
ls
={(x
ls
i
, y
ls
i
)}
N
ls
i=1
, unlabelled source domains are
represented by D
us
={(x
us
i
)}
N
us
i=1
, and labelled target do-
mains are represented by D
lus
={(x
lus
i
, y
lus
i
)}
N
lus
i=1
. Let
the source images from all subsets be represented by
D
s
=D
ls
∪ D
us
.
3.2 Adversarial End-to-End Trainable
Architecture
Inspired by the work of (Ganin and Lempitsky, 2015),
we have used an encoder-decoder network to trans-
late the patches from different domains (scanners) to a
common space. The translated images are then passed
through RetinaNet for object detection (Algaissi et al.,
2020). The architecture also consists of an adversarial
head with domain classification as an auxiliary task.
This head encourages the encoder-decoder network to
erase all the domain-specific information using a gra-
dient reversal layer. The architecture of our method is
as shown in figure 1.
Figure 1: The pipeline of our proposed method for mitosis
detection.
3.3 Training Objectives
The object detection loss consists of bounding box
loss (L
bb
) and instance classification loss (L
inst
). The
bounding box loss (L
bb
) is computed as smooth L1
loss and the focal loss function (Lin et al., 2017b) is
used for the instance classification (L
inst
). The equa-
tion for the focal loss with p
k
as probability that the
instance belong class k is given by,
FL(p
k
) = (1 − p
k
)
γ
log(p
k
) (1)
Improving Mitosis Detection via UNet-Based Adversarial Domain Homogenizer
53
In order to ensure that the images translated by
encoder-decoder network contains the semantic infor-
mation a perceptual loss (L
percp
) is used. We have
used the perceptual loss based on pretrained VGG-
16, which is proposed in (Johnson et al., 2016). The
perceptual loss is given by equation 2
L
( j)
percp
=
1
C
j
H
j
W
j
||Φ
j
( ˆx) − Φ
j
(x)||
2
2
(2)
where x and ˆx are the input and reconstructed im-
ages respectively. Φ is the pretrained frozen VGG net-
work, j is the level of the feature map (zero-indexed)
of size C
j
× H
j
× W
j
obtained from Φ. At the end
of the adversarial head we have used standard cross
entropy loss (L
CE
) for domain classification.
The overall loss for the end-to-end training is
given by,
L = E
(x,y)∈D
ls
[L
bb
+ L
inst
] + E
(x,y)∈D
s
[λ
1
L
( j)
percep
+ λ
2
L
CE
]
(3)
4 DATA AND EXPERIMENTS
4.1 Dataset
The experiments were conducted on MIDOG 2021
dataset (Aubreville et al., 2021) which consists of
50 whole slide images of breast cancer from four
scanners namely Hamamatsu XR NanoZoomer 2.0,
Hamamatsu S360, Aperio ScanScope CS2, and Le-
ica GT450 forming four domains. Two classes of ob-
jects are to be detected namely mitotic figures and
hard negatives. The whole slide images from scan-
ners other than the Leica GT450 areis labelled. Small
patches of size 512 x 512 are mined for supervised
end-to-end training such that the cells belonging to at
least one of the mitotic figures or hard negatives are
present in the patch.
The seen and unseen domains i.e., the scanners are
D
ls
={Hamamatsu XR NanoZoomer 2.0, the Hama-
matsu S360}, D
us
={Leica GT450}, D
lus
={Aperio
ScanScope CS2} (refer 3.1 for notations.)
4.2 Implementation Details
The model is implemented using Pytorch (Paszke
et al., 2019) library. For supervised end-to-end train-
ing a batch size of 12 is used with equal number of
patches being included from each scanner. Here the
model is trained using FastAI (Howard et al., 2018)
library default settings with an initial learning rate of
1e
−4
. In the equation 3 we have set the values of hy-
perparameters as j=1, λ
1
=10 and λ
2
=25. These values
are chosen by grid search over a range of values. Fur-
ther tuning of these values can yield better results.
Our code is available on Github (Almah-
fouz Nasser et al., ).
4.3 Results
Two classes of objects – hard negatives, and mitotic
figures – are detected. The models are evaluated on
D
ls
∪ D
lus
. One of the standard metric for object de-
tection Average precision (AP) at intersection over
union (IoU) threshold of 0.5, which is introduced in
PASCAL VOC challenge (Everingham et al., ), is
used as metric for evaluation. It represents the aver-
age of precision values obtained at various bounding
box confidence thresholds.
End-to-end training with (AEC RetinaNet
+ Pecp) and without using perceptual loss
(AEC RetinaNet) were tried. The results are com-
pared with the reference algorithm DA RetinaNet
(Wilm et al., 2021), RetinaNet (Lin et al., 2017a)
with and without data augmentation. The results
obtained are as shown in table 1.
The count of mitotic figures is an important clini-
cal goal. So, the performance on the class of mitotic
figures was our focus. The results in the table 1 show
that the newly designed end-to-end training architec-
tures performs better than the reference algorithm and
the basic RetinaNet based algorithms. The improve-
ment is in terms of detection performance for the class
mitotic figures,.
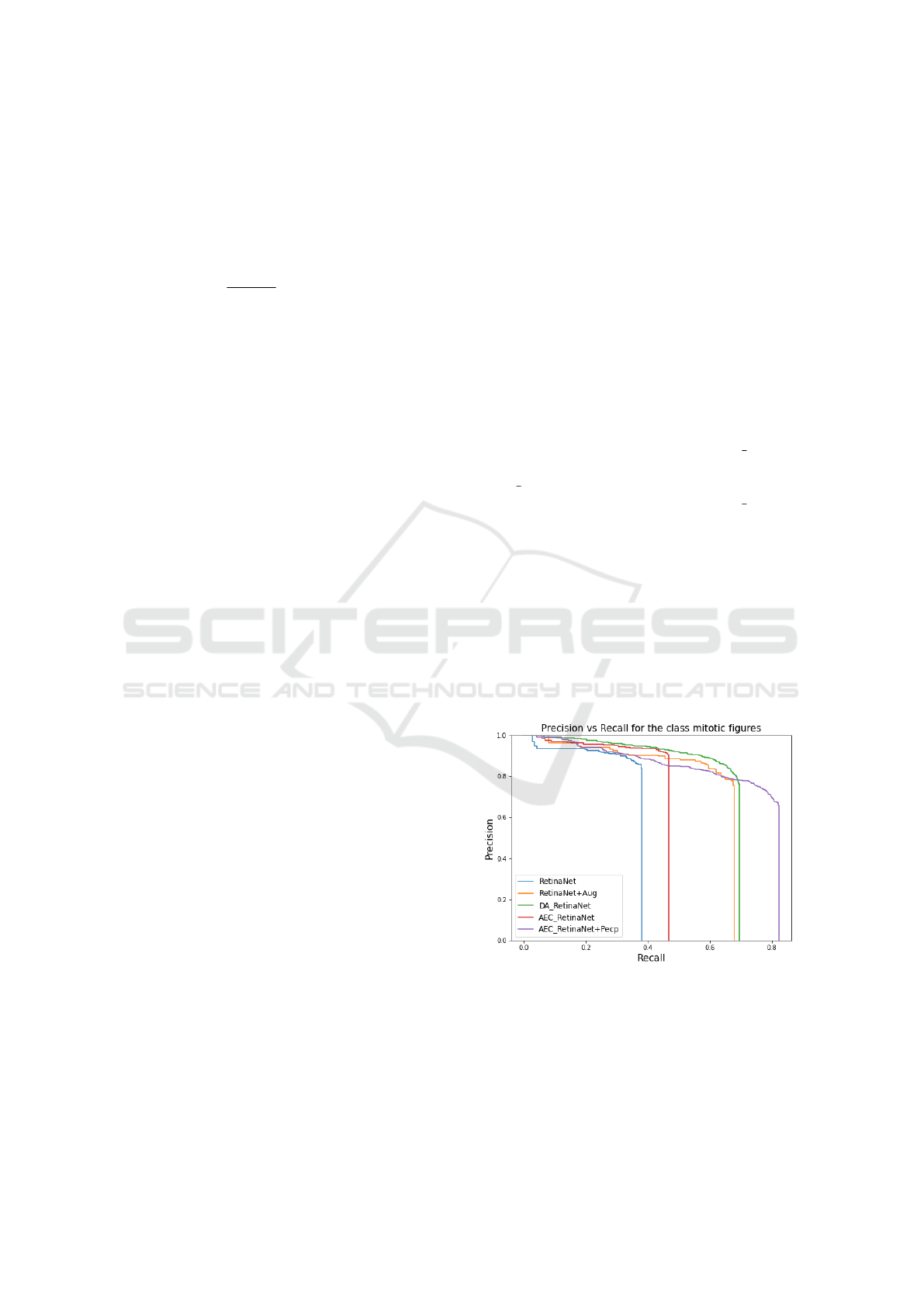
Figure 2: The precision vs recall plot, which represents
the values of precision and recall at various IoU thresholds,
shows that the newly designed end-to-end model performs
better than the baselines in terms of recall values without
compromising on the precision.
From the precision-recall plot shown in figure 2
represents the values of precision recall at various IoU
thresholds. Our method is able to achieve a better re-
BIOIMAGING 2023 - 10th International Conference on Bioimaging
54
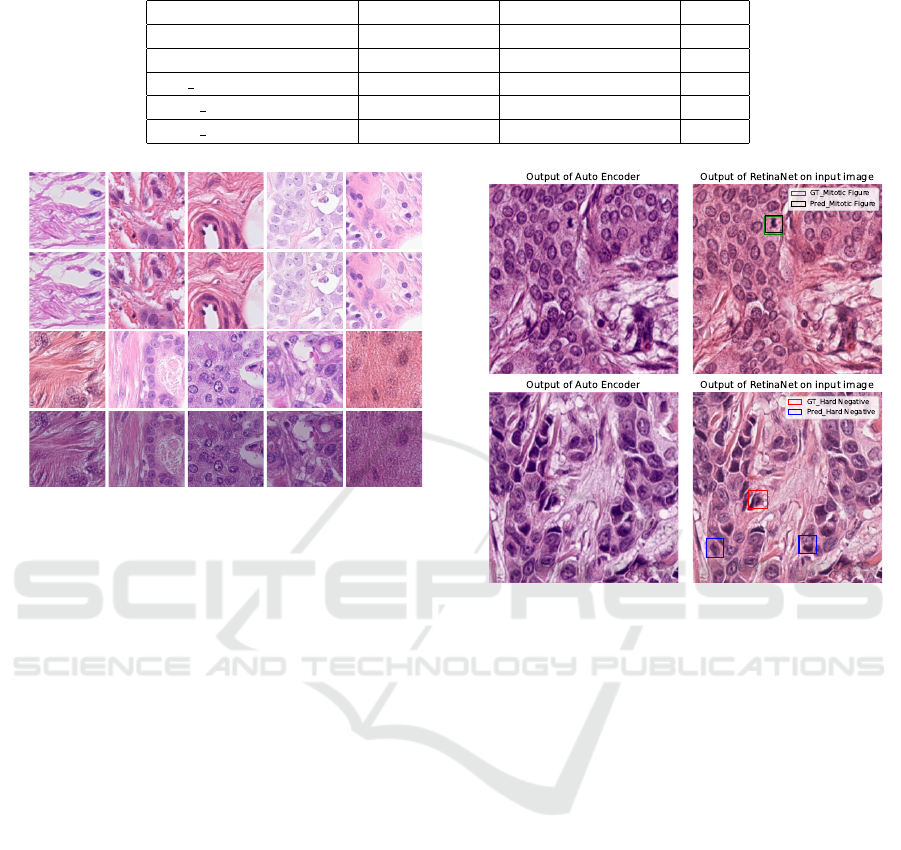
Table 1: Results obtained using end-to-end training of models.
Model AP-Hard Neg AP-Mitotic figures mAP
RetinaNet 0.196 0.352 0.274
RetinaNet + Aug 0.238 0.619 0.429
DA RetinaNet 0.347 0.655 0.501
AEC RetinaNet 0.289 0.448 0.369
AEC RetinaNet + Pecp 0.248 0.72 0.484
Input Image
Output of Domain
Homogenizer
Input Image
Output of Modified
Domain Homogenizer
Figure 3: A visual comparison of the performances of the
domain homogenizer and the modified domain homoge-
nizer (proposed method) on a randomly sampled set of
patches. The modified version is able to transform the im-
ages from various scanners to a common space and the
translated images cannot be visually distinguished on the
basis of scanner whereas the original domain homogenizer
produced the exact same input images.
call without compromising on the precision.
The perceptual loss added at the output of the
decoder helps in retaining the semantic information.
This information which helps in better object detec-
tion. This is also validated by higher AP score ob-
tained when perceptual loss component is added.
As shown in figure 3 the modified domain homog-
enizer produced much more plausible images than
the original domain homogenizer (Almahfouz Nasser
et al., 2021).Besides, figure 4 shows the detection ac-
curacy of our proposed method.
5 CONCLUSIONS
In this paper, we proposed a modified version of our
previous domain homogenizer proposed by us and
tested it on the data from for the MIDOG 2021 chal-
lenge 2021. We showed that the position of the do-
main classifier has a significant impact on the perfor-
mance of the homogenizer. Shifting the adversarial
head from the latent space to the output of the auto-
encoder helps in erasing all the domain-specific in-
Figure 4: Two examples explaining the results of our pro-
posed method from table 1 i.e., our method is able to detect
the mitotic figures accurately but not the hard negatives.
formation as the inverse of the domain loss will flow
back throughout the decoder and the encoder unlike
the situation in the previous arrangement. Addition-
ally, our experiments revealed that training the ho-
mogenizer along with the object detection network
end-to-end improves the detection accuracy by a sig-
nificant margin. Finally, we showed that our method
substantially improves upon the baseline of the MI-
DOG challenge in terms of mitotic figures detection.
REFERENCES
Algaissi, A., Alfaleh, M. A., Hala, S., Abujamel, T. S.,
Alamri, S. S., Almahboub, S. A., Alluhaybi, K. A.,
Hobani, H. I., Alsulaiman, R. M., AlHarbi, R. H.,
et al. (2020). Sars-cov-2 s1 and n-based serological
assays reveal rapid seroconversion and induction of
specific antibody response in covid-19 patients. Sci-
entific reports, 10(1):1–10.
Almahfouz Nasser, S., Chandra, T., Kurian, N.,
and Sethi. Improving Mitosis Detection Via
UNet-based Adversarial Domain Homogenizer.
https://github.com/MEDAL-IITB/MIDOG.git.
Almahfouz Nasser, S., Kurian, N. C., and Sethi, A. (2021).
Improving Mitosis Detection via UNet-Based Adversarial Domain Homogenizer
55

Domain generalisation for mitosis detection exploting
preprocessing homogenizers. In International Con-
ference on Medical Image Computing and Computer-
Assisted Intervention, pages 77–80. Springer.
Aubreville, M., Bertram, C., Veta, M., Klopfleisch, R.,
Stathonikos, N., Breininger, K., ter Hoeve, N.,
Ciompi, F., and Maier, A. (2021). Mitosis domain
generalization challenge. In 24th International Con-
ference on Medical Image Computing and Computer
Assisted Intervention (MICCAI 2021), pages 1–15.
Breen, J., Zucker, K., Orsi, N. M., and Ravikumar, N.
(2021). Assessing domain adaptation techniques
for mitosis detection in multi-scanner breast cancer
histopathology images. In International Conference
on Medical Image Computing and Computer-Assisted
Intervention, pages 14–22. Springer.
Choi, Y., Choi, M., Kim, M., Ha, J.-W., Kim, S., and Choo,
J. (2018). Stargan: Unified generative adversarial net-
works for multi-domain image-to-image translation.
In Proceedings of the IEEE conference on computer
vision and pattern recognition, pages 8789–8797.
Chung, Y., Cho, J., and Park, J. (2021). Domain-robust
mitotic figure detection with style transfer. In In-
ternational Conference on Medical Image Comput-
ing and Computer-Assisted Intervention, pages 23–
31. Springer.
Dexl, J., Benz, M., Bruns, V., Kuritcyn, P., and Wittenberg,
T. (2021). Mitodet: Simple and robust mitosis de-
tection. In International Conference on Medical Im-
age Computing and Computer-Assisted Intervention,
pages 53–57. Springer.
Everingham, M., Van Gool, L., Williams, C.
K. I., Winn, J., and Zisserman, A. The
PASCAL Visual Object Classes Challenge
2012 (VOC2012) Results. http://www.pascal-
network.org/challenges/VOC/voc2012/workshop/index.html.
Fick, R. H., Moshayedi, A., Roy, G., Dedieu, J., Petit,
S., and Hadj, S. B. (2021). Domain-specific cycle-
gan augmentation improves domain generalizability
for mitosis detection. In International Conference on
Medical Image Computing and Computer-Assisted In-
tervention, pages 40–47. Springer.
Ganin, Y. and Lempitsky, V. (2015). Unsupervised do-
main adaptation by backpropagation. In International
conference on machine learning, pages 1180–1189.
PMLR.
Girshick, R. (2015). Fast r-cnn. In Proceedings of the IEEE
international conference on computer vision, pages
1440–1448.
He, K., Gkioxari, G., Doll
´
ar, P., and Girshick, R. (2017).
Mask r-cnn. In Proceedings of the IEEE international
conference on computer vision, pages 2961–2969.
Howard, J., Thomas, R., and Gugger, S. (2018). fastai. Aval-
ableat: https://github. com/fastai/fastai.
Jahanifar, M., Shepard, A., Zamanitajeddin, N., Bashir, R.,
Bilal, M., Khurram, S. A., Minhas, F., and Rajpoot,
N. (2021). Stain-robust mitotic figure detection for
the mitosis domain generalization challenge. In In-
ternational Conference on Medical Image Comput-
ing and Computer-Assisted Intervention, pages 48–
52. Springer.
Johnson, J., Alahi, A., and Fei-Fei, L. (2016). Perceptual
losses for real-time style transfer and super-resolution.
In European conference on computer vision, pages
694–711. Springer.
Lin, T., Goyal, P., Girshick, R., He, K., and Dollar,
P. (2017a). Focal loss for dense object detection
2017 ieee international conference on computer vision
(iccv).
Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Doll
´
ar, P.
(2017b). Focal loss for dense object detection. In 2017
IEEE International Conference on Computer Vision
(ICCV), pages 2999–3007.
Long, X., Cheng, Y., Mu, X., Liu, L., and Liu, J.
(2021). Domain adaptive cascade r-cnn for mito-
sis domain generalization (midog) challenge. In In-
ternational Conference on Medical Image Comput-
ing and Computer-Assisted Intervention, pages 73–
76. Springer.
Nateghi, R. and Pourakpour, F. (2021). Two-step domain
adaptation for mitotic cell detection in histopathology
images. In International Conference on Medical Im-
age Computing and Computer-Assisted Intervention,
pages 32–39. Springer.
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J.,
Chanan, G., Killeen, T., Lin, Z., Gimelshein, N.,
Antiga, L., et al. (2019). Pytorch: An imperative style,
high-performance deep learning library. Advances in
neural information processing systems, 32.
Peter Byfield, a. T. G. and Gamper, J. (Accessed
in 03 Aug 2022). Staintools homepage. In
https://github.com/Peter554/StainTools.
Reinhard, E., Adhikhmin, M., Gooch, B., and Shirley, P.
(2001). Color transfer between images. IEEE Com-
puter graphics and applications, 21(5):34–41.
Vahadane, A., Peng, T., Albarqouni, S., Baust, M., Steiger,
K., Schlitter, A. M., Sethi, A., Esposito, I., and Navab,
N. (2015). Structure-preserved color normalization
for histological images. In 2015 IEEE 12th Inter-
national Symposium on Biomedical Imaging (ISBI),
pages 1012–1015. IEEE.
Vahadane, A., Peng, T., Sethi, A., Albarqouni, S., Wang,
L., Baust, M., Steiger, K., Schlitter, A. M., Esposito,
I., and Navab, N. (2016). Structure-preserving color
normalization and sparse stain separation for histolog-
ical images. IEEE transactions on medical imaging,
35(8):1962–1971.
Wilm, F., Marzahl, C., Breininger, K., and Aubreville, M.
(2021). Domain adversarial retinanet as a reference
algorithm for the mitosis domain generalization chal-
lenge. In International Conference on Medical Im-
age Computing and Computer-Assisted Intervention,
pages 5–13. Springer.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Proceedings of
the IEEE international conference on computer vi-
sion, pages 2223–2232.
BIOIMAGING 2023 - 10th International Conference on Bioimaging
56
