
Interpretability and Explainability of Logistic Regression Model for
Breast Cancer Detection
Emina Tahirovi
´
c
1
and Senka Krivi
´
c
2
1
Faculty of Engineering and Natural Sciences, International Burch University, Sarajevo 71000, Bosnia and Herzegovina
2
Faculty of Electrical Engineering, University of Sarajevo, Sarajevo 71000, Bosnia and Herzegovina
Keywords:
Logistic Regression, Explainable AI, Transparency, Healthcare.
Abstract:
Artificial Intelligence techniques are widely used for medical purposes nowadays. One of the crucial applica-
tions is cancer detection. Due to the sensitivity of such applications, medical workers and patients interacting
with the system must get a reliable, transparent, and explainable output. Therefore, this paper examines the
interpretability and explainability of the Logistic Regression Model (LRM) for breast cancer detection. We
analyze the accuracy and transparency of the LRM model. Additionally, we propose an NLP-based interface
with a model interpretability summary and a contrastive explanation for users. Together with textual explana-
tions, we provide a visual aid for medical practitioners to understand the decision-making process better.
1 INTRODUCTION
An accurate cancer diagnosis is essential for planning
the best action and establishing a treatment plan. Over
a hundred risk factors can simultaneously be involved
in estimating a single post-test probability, making
manual prediction incredibly difficult. Machine learn-
ing models can be beneficial for processing large
numbers of variables and thereby bridging the gap be-
tween risk factors and risk estimation. However, legal
and ethical accountability issues make fully indepen-
dent AI medical systems unlikely. An alternative is
the possibility of developing explainable AI systems,
which would aid humans in decision-making while
keeping the simplicity of its background processes.
Among the numerous computer models used for
predicting clinical outcomes can be distinguished two
main subcategories: models built by the statistics
community and models built by the machine learn-
ing community. Logistic Regression (LR) is a sta-
tistical fitting model widely used to model medical
problems, such as estimating disease risk in coronary
heart disease, breast cancer, prostate cancer, postoper-
ative complications, and stroke. Several studies (Ayer
et al., 2010; Aviv et al., 2009) have shown that LR is a
valuable tool in medical diagnosis since the method-
ology is well established.
As ML models penetrate critical and sensitive
areas such as medicine, what becomes increasingly
challenging is the inability of humans to understand
these models (Lipton, 2018). It is of utter importance
that ML models used in the medical domain can be
trusted. Even if a model achieves high accuracy, it is
still desirable that medical practitioners can decide if
the diagnosis makes sense and that the output is inter-
pretable even to a patient. In a hypothetical scenario,
an LR model would output a cancer diagnosis and ex-
plain why this sample was classified as a benign or
malignant tumor. The medical practitioner could con-
clude if the patient needs more invasive tests to be
conducted or if the diagnosis is clear enough as is.
A semantic explanation paired with the visualized aid
that clarifies the deciding factors and features for a
specific case can be used for this purpose, which we
propose in this paper.
We propose an Explainable AI (XAI) system for
breast cancer detection consisting of a classification
model and semantic and visual models providing the
decision of an LR model together with interpretations
and explanations for a medical practitioner interact-
ing with the system. The classification segment is
based on a binary LR model, trained on breast can-
cer characteristics data. The semantic model is an
NLP-based user interface using automated question-
answering models, where the user is prompted to
ask questions about the classification of the tumor.
This way, we produce an output tailored to the user’s
needs. We combine the inherent interpretability of LR
with a contrastive explanations approach. Figure 1
gives a diagram explaining this system.
Tahirovi
´
c, E. and Krivi
´
c, S.
Interpretability and Explainability of Logistic Regression Model for Breast Cancer Detection.
DOI: 10.5220/0011627600003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 3, pages 161-168
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
161
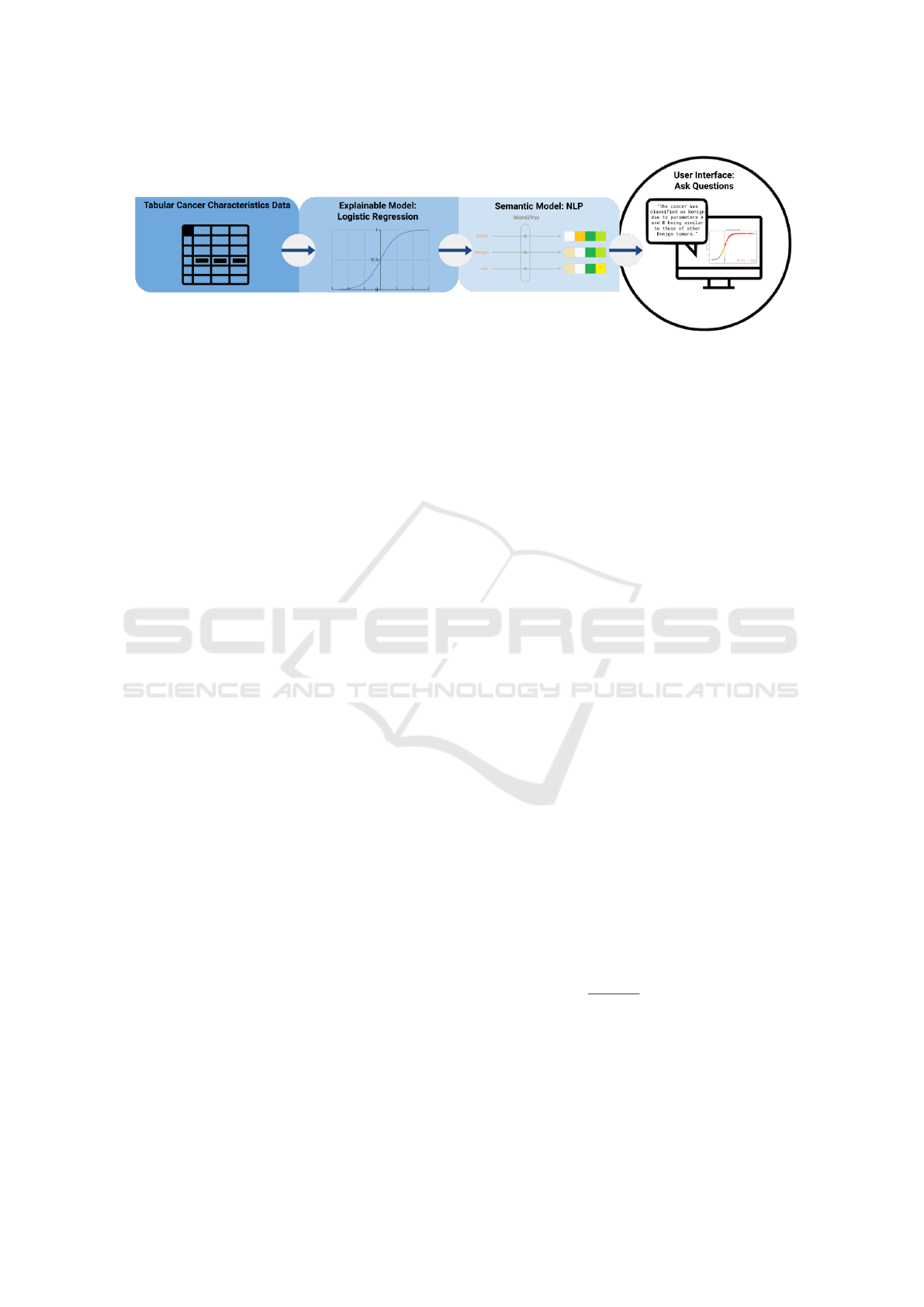
Figure 1: Diagram of the proposed XAI system for Breast Cancer Detection.
This paper is structured as follows. Section 2 de-
scribes related work. In section 3, we explain LR and
its interpretability and explainability. In section 4, we
start with a brief introduction to the dataset, move on
to explaining our LR model, and finally present the
NLP semantic user interface. Section 5 presents re-
sults and section 6 concludes the paper.
2 RELATED WORK
The development of machine learning methods made
many breakthroughs in challenging clinical tasks such
as assisted diagnosis, automatic image analysis, and
prediction (Zhao et al., 2020). Logistic Regression
is successfully used for several medical prediction
tasks (Naji et al., 2021; Anisha et al., 2021). The
LR model for binary data is probably the most widely
used in medical research (Hastie et al., 2009). Ayer
et al. (Ayer et al., 2010) compared the performance
of LR and Artificial Neural Networks (ANNs) on the
Breast Cancer Wisconsin (BCW) dataset, which we
also use in this research. They note that in terms of
clinical interpretation, LR models have better clini-
cal references than ANNs. Sultana and Jilani (Sul-
tana and Jilani, 2018) used Simple Logistic Regres-
sion. They trained the classifier on the BCW dataset.
In their research, this accuracy was higher than any
other classifier, some of which were Nearest Neigh-
bor Classifier, Random Forest, and Decision Tree.
Limitations on the use of deep and ensemble
learning models in the medical domain are reflected
in their lack of interpretability (Chakrobartty and El-
Gayar, 2021). Gunning and Aha (Gunning and Aha,
2019) define XAI as ”AI systems that can explain
their rationale to a human user, characterize their
strengths and weaknesses, and convey an understand-
ing of how they will behave in the future”.
The measures and models involved in decision-
making and solutions to explain them explicitly
had been inspected by Yang et al. (Yang et al.,
2022). They demonstrated the research trends toward
trustable AI and showcased promising XAI results for
the two most widely investigated classification and
segmentation problems in medical image analysis.
The fundamental conceptual differences of Ex-
plainable Artificial Intelligence (XAI) for regression
and classification tasks were analyzed by Letzgus et
al. (Letzgus et al., 2021). They focus on ’post-hoc’
explanation where they try to attribute the prediction
for each data sample to its input features in a mean-
ingful manner. They cite contextual utility and feature
importance as the bases of early approaches toward
understanding the decision processes of ML models.
They concluded that explanation methods are favor-
able in the regression scenario and that XAI cannot
be transferred between different types of ML prob-
lems without adaptation.
One point of view that has the potential to broaden
the scope of XAI explanations is contrastive ques-
tions. Cashmore et al. (Cashmore et al., 2019) give
an example of this type of question: ”Why A rather
than B?”. When answering a contrastive explanation,
one must consider a situation where scenario B might
be better suited than scenario A. In other words, one
must consider why scenario B would be more appro-
priate by arguing why scenario A would be less so.
This type of explanation is called a contrastive expla-
nation Krarup et al. (Krarup et al., 2021) argue that
users of explainable user interfaces must be able to
ask explicit contrastive questions.
Natural Language Processing (NLP) may offer a
helping hand in a quest to make medical domain re-
gression tasks more explainable. Krieger (Krieger,
2016) identifies the medical NLP statements used by
medical professionals as gradation words. E.g. ”X-
ray technician A suspects that patient B suffers from
breast cancer”. Modeling such language in an NLP
semantic output could lead to more trust in medical
domain regression tasks. Therefore, we use such lan-
guage to leave an approachable and professional im-
pression.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
162

3 METHODS
This section describes the proposed approach of an
XAI system consisting of an LR classifier, inter-
pretability, and explainability modules.
3.1 Logistic Regression
Logistic Regression determines the relationship be-
tween a binary outcome (dependent variable) and pre-
dictors (independent variables). It estimates the prob-
ability of an event’s occurrence. It is widely used
in biostatistical applications where binary responses
(two classes) occur quite frequently (Hastie et al.,
2009). In the medical problem, we address the two
classes which make the binary response 1 for malig-
nant tumors and 0 for benign. Here, the diagnosis is
the dependent variable, and the parameters which de-
scribe the cancer cells are the independent variables.
Let y denote the presence of the disease, where
y = 0 or y = 1, and let x denote the vector of predic-
tors (features). Given that p denotes the probability
of breast cancer (the probability that y = 1), we can
define Logistic Regression with
P(y = 0, 1|x, w) =
1
1 + e
−(w
T
x+b)
(1)
Here, (w, b) are weights, with b being a constant,
w the regression coefficients to their respective pre-
dictor variables x, and y the class label. These coeffi-
cients are estimated from the available data.
If the training instances are from the dataset
M = (x
1
, y
1
), ..., (x
n
, y
n
) with the labels y
i
∈ {0, 1},
one estimates (w, b) by minimizing the negative log-
likelihood: min
w,b
∑
n
i=1
log(1 + e
−(w
T
x
i
+b)
) (2).
3.2 Interpretability of Logistic
Regression
An oft-made claim is that linear models are preferable
to deep neural networks because of their interpretabil-
ity. The high accuracy of complex models comes at
the expense of interpretability. Hence, even the con-
tributions of individual features to the predictions of
such a model become challenging to understand (Lou
et al., 2012). Interpretability can be seen as a reflec-
tion of several different ideas than a monolithic con-
cept (Lipton, 2018). Another view of interpretability
is that it represents the degree to which a human can
understand the cause of a decision (Miller, 2019).
While the output of the LR model is binary, the
precedent of this output is a probability output in the
range from 0 to 1. The probability output is trans-
formed into the binary output by using a cutoff value
of 0.5. A positive classification (= 1) is the result
of a probability of ≥ 0.5, and a negative classifica-
tion (= 0) is the result of a probability of < 0.5. We
use this probability of each classification as an inter-
pretability approach, essentially utilizing an aspect of
LR’s inherent interpretability. By analyzing Eq.(1) we
get
P(y = 1|x, w)
1 − P(y = 1|x, w)
=
P(y = 1|x, w)
P(y = 0|x, w)
= e
w
T
x+b
(2)
We can now examine how the feature weights and
values affect the probability. Let us say that x is a
vector of x
1
, ..., x
n
∈ N variables, and feature values of
x
i
changes from x
k
= a to x
k
= c where c−a = 1. Now
we can examine how the change of the feature from a
to c impacted the model’s outcome by observing the
ratio the probability gets scaled by a factor of e
w
k
as
we can deduce from:
e
(b+w
1
x
1
+...+cw
k
+...+w
n
x
n
)
e
(b+w
1
x
1
+...+aw
k
+...+w
n
x
n
)
= ... = e
(a+1)w
k
−aw
k
= e
w
k
(3)
To highlight this impact for a user and provide the
interpretability of the decision, we use the following
plots:
1. Sigmoid plot, to clarify to the user where the cur-
rently observed cancer sample lies on the plot,
2. value counts plots to clarify the dominance of fea-
ture values for each classification, and
3. classification shift at the classification border with
changing values of x
k
.
3.3 Explainability of Logistic
Regression
If we look at LR as a classification model, there is im-
plicit knowledge associated with the class. This can
serve as a piece of a potential explanation of LR (Let-
zgus et al., 2021). The explainability of a model de-
pends on the level of known information about how
the parameters of a model impact the decision pro-
cess. In this respect, the coefficients of an LR model
make it directly explainable. Hence, as valuable infor-
mation for the user, we select visualization of feature
importance.
Another important aspect that needs to be kept in
mind when developing XAI models is the recipient
of the information. The information must be tailored
to the end user. As our interface is meant to be used
by medical personnel, we tailor our explanations in a
way that will help relate their pre-existing knowledge
on the topic, while helping them understand how our
model makes predictions. The explainability of our
model relies on three different methods:
Interpretability and Explainability of Logistic Regression Model for Breast Cancer Detection
163

• identification of a relevant subset of features,
• interpretation of feature importance scores,
• explainability by contrastive example.
We present feature importance plots, parallel
plots, classification shift plots, box plots, and heat
maps to help the end user understand what kind of
data leads to a certain prediction.
Contrastive Explanation: is a specific type of
explanation which answers a question of type: ”Why
A rather than B ?”. It includes a contrastive exam-
ple with the goal of presenting the difference in the
decision-making to the user. It is commonly used in
various AI fields (Krarup et al., 2021). In our medical
XAI system case, we propose presenting two different
feature vectors x
ce
and x
c f
as contrastive examples of
feature vectors to the user. For the current input vec-
tor x
i
and the decision of an LR classifier y
i
, we define
these contrastive vectors as follows. x
ce
is the value
of a feature vector with minimal Euclidean distance
from the input vector x
i
which results in the opposite
class result ¬y
i
. x
c f
is a vector in which variation in
a single feature value results in the prediction class
change and becomes ¬y
i
.
4 IMPLEMENTATION DETAILS
XAI MEDICAL SYSTEM
This section explains the dataset used for this research
and the implementation process. In the first subsec-
tion, we go through the nature of the dataset and its
features. In the following subsections, we explain the
implementation of the LR model and the NLP seman-
tic user interface.
4.1 Dataset Description
The dataset used for this research is the well-known
Wisconsin Breast Cancer Dataset obtained from the
UC Irvine Machine Learning Repository (Mangasar-
ian et al., 1995). The nine predictor variables ana-
lyzed for this research are visually assessed cytologi-
cal characteristics of a Fine Needle Aspiration (FNA)
sample. These variables take integer values between
1 and 10. Wolberg and Mangasarian (Wolberg and
Mangasarian, 1990) chose the nine features based on
statistical analysis, which showed a significant dif-
ference in values for benign and malignant samples.
Therefore, they are prominent candidates for predic-
tor variables for our LR model. These predictor vari-
ables are uniformity of cell shape, uniformity of cell
size, clump thickness, marginal adhesion, single ep-
ithelial cell, bare nuclei, bland chromatin, normal nu-
cleoli, and mitosis. The feature class is the predicted
or the dependent variable in this dataset.
4.2 Logistic Regression Model
Our LR model is implemented from scratch, in
Python, on the Jupyter Notebook platform. It is a bi-
nary LR model with an added bias term. The moti-
vation behind the from-scratch implementation is the
interpretability of each step of LR, where one can
choose to print out different values and interpret them
separately, as opposed to a black-box approach to LR
with a library-provided model.
The original model is trained on all 9 features and
is evaluated using stratified 5-fold cross-validation.
These preliminary scores are 97.10% for accuracy,
and 95.65% for F
1
-score. Models trained on differ-
ent subsets of features are cross-validated individu-
ally. Each subset consists of n most important fea-
tures, as determined by feature importance scores (see
subsection 5.2). The results are recorded and the best-
performing model is selected. The optimal subset size
was determined to be 7 features, which are, in order
of importance: bare nuclei, clump thickness, mitosis,
uniformity of cell shape, marginal adhesion, bland
chromatin, and normal nucleoli. This model is our
finished LR model.
4.3 NLP Models and User Interface
The output of our framework is based on an NLP ex-
plainable user interface, which produces a combina-
tion of textual and pictorial explanations to provide
aid for medical practitioners to better understand the
decision-making process of the LRM. In the follow-
ing subsections, we explain the models used to imple-
ment this user interface.
Word2Vec: is a word embedding model. It is an
unsupervised learning algorithm that generates low-
dimensional vector representations of words. The
variation used in this research is Skip-gram, which
predicts the context, given the word.
GloVe: or global vectors is an unsupervised learn-
ing algorithm that obtains low-dimensional vector
representations for words and performs dimensional-
ity reduction on the word context matrix. It thereby
keeps track of the frequencies of word co-occurrences
in a text, with an additional weight parameter depen-
dent on the distance between the words.
Question Database: consists of twenty-two ques-
tions and their respective answers. Some of these an-
swers are static, while others are assembled with the
help of the information obtained from the model. The
database is shown in table 1. Answers to questions
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
164
13-22 are explained in section 5.2.
Automated Question Answering: The next level
of interpretability is reached through an NLP auto-
mated question-answering model based on Word2Vec
or GloVe embeddings. We use cosine similarity to es-
timate how similar the user-asked question is to the
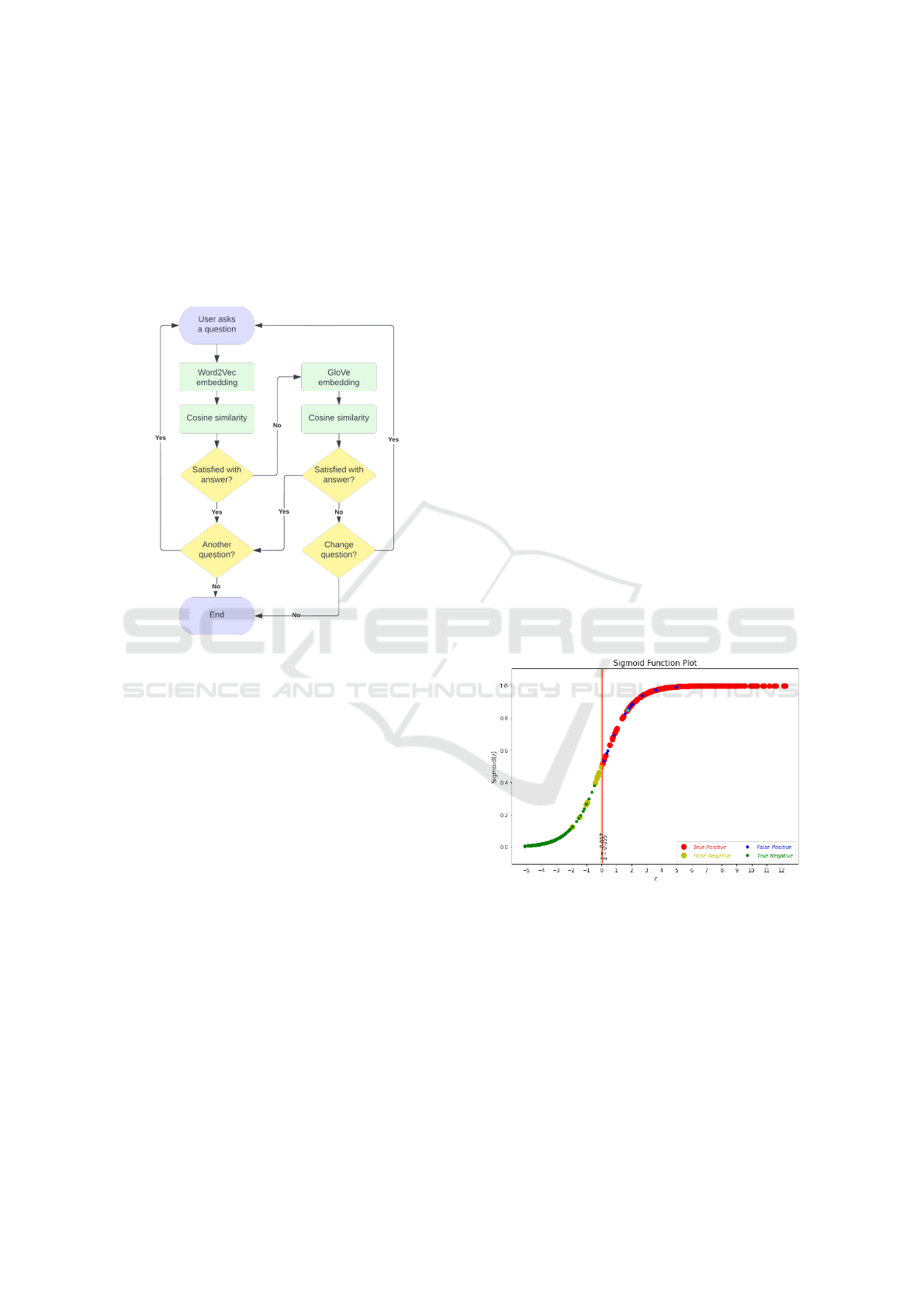
questions in the database. The process is illustrated
from start to finish in figure 2.
Figure 2: User interface state diagram.
5 EXPERIMENTAL RESULTS
This section presents the prediction, interpretability,
and explainability results of our models. We provide
textual, as well as pictorial explanations of our results.
5.1 LR Prediction Evaluation
We present the prediction results of our model in
Table 2 for the test subset and the original dataset.
For the performance evaluation, we compare the CV
score, accuracy, precision, recall, and F1 score. Ta-
ble 3 shows the confusion matrices for the test set
and complete dataset. These results indicate that our
model performs well, as the rates of falsely classified
data are low. The ratio of each portion (true positives,
false positives, false negatives, and true negatives) is
well-preserved in the test set, as evident from compar-
ing the test and entire dataset confusion matrices.
5.2 Interpretability Results
When the user begins the interaction with our inter-
face, they are presented with some basic information
about how their tumor is seen by our model. They are
told what their classification is (benign or malignant),
and how probable it is that the tumor is malignant.
Figure 3 shows the Sigmoid plot for the entire
dataset, where the colors of the dots are clarified by
the legend. The two vertical bars represent the right-
most negative and the left-most positive classification,
along with their respective Z-values. The currently
observed cancer sample is represented by a cyan dot.
This figure is presented along with the answer to
question no. 13 in the semantic output (see Table 1).
Its purpose is to clarify to the user where the currently
observed cancer sample lies on the plot and to get a vi-
sual sense of how similar this sample is to other such
classifications.
In this way, we maintain a level of transparency
as to how our model classifies data. This plot is
presented in combination with a textual output stat-
ing the density of falsely classified samples in the
0.5-neighborhood of the current cancer sample (along
the Z-axis), the values of each of the 7 relevant fea-
tures, and the space between the left-most positive
and right-most negative classification. These state-
ments aim to help the user interpret the classification.
Figure 3: Sigmoid plot for the entire dataset.
Figure 4 shows the percentages of value counts for
bare nuclei, for benign and malignant samples. Anal-
ogous plots are generated for all other features. These
plots are used in questions 14 and 15 to clarify the dif-
ferences in the values that the individual features typ-
ically take for different classifications. This way, the
user can clearly see that for example bare nuclei=1 is
most often seen in benign samples.
Figure 5 shows the shift of the classification prob-
ability of the observed tumor sample with the change
of a single feature, in this case, bare nuclei. This plot
Interpretability and Explainability of Logistic Regression Model for Breast Cancer Detection
165
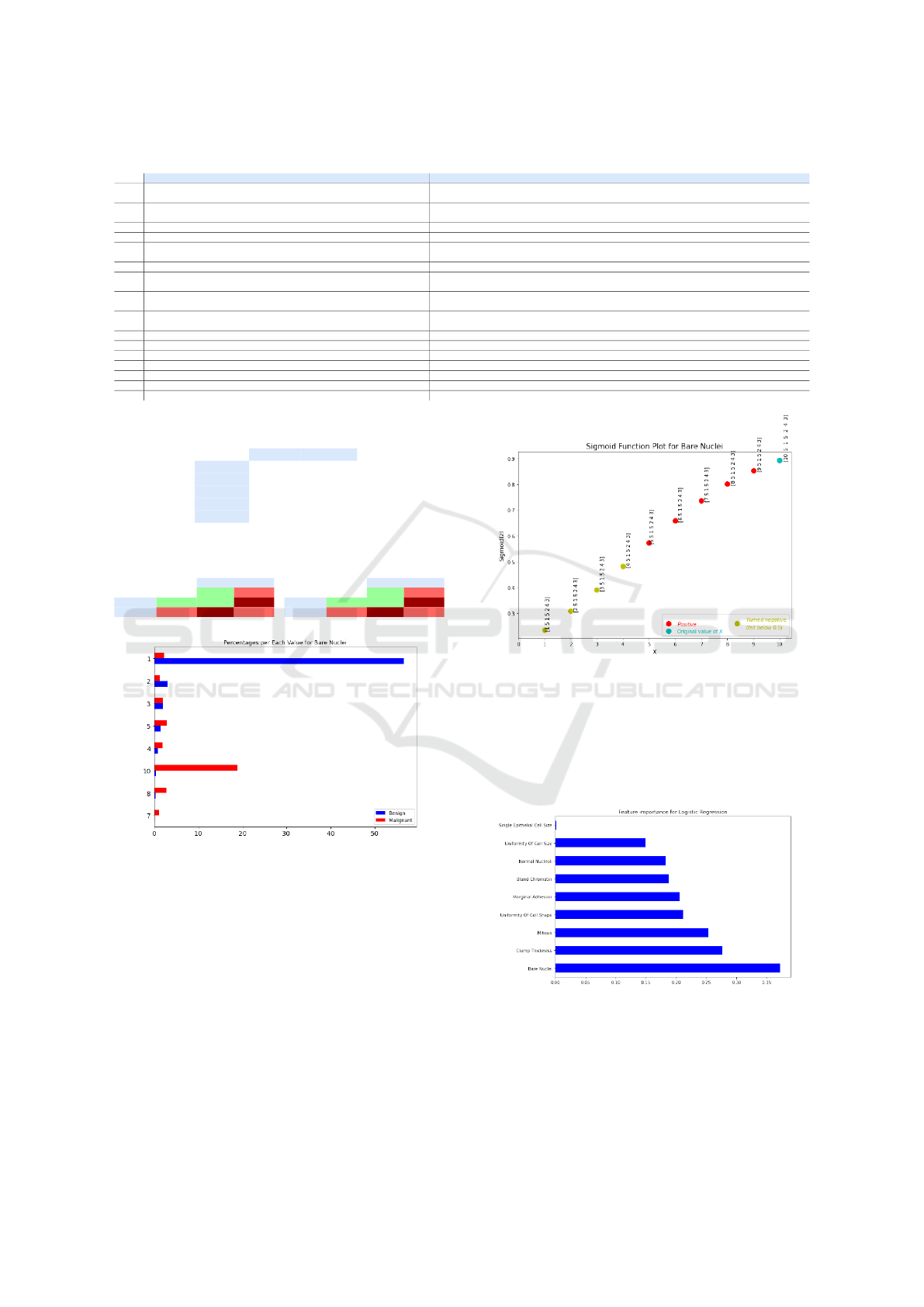
Table 1: Questions database.
Questions Answers
1 Why is the tumor classified as malignant?
The parameters of this tumor produced a similar function value to the tumors
which have previously been marked malignant by a human doctor. Your parameters are:
2 Why is the tumor classified as benign?
The parameters of this tumor produced a similar function value to the tumors
which have previously been marked benign by a human doctor. Your parameters are:
3 What does ’malignant’ mean? Malignant tumors are cancerous (i.e., they invade other sites).
4 What does ’benign’ mean? Benign tumors are those that stay in their primary location without invading other sites of the body.
5 How similar is this tumor to tumors which have been classified as False Positive?
Your tumor falls in the range of tumors that are known to have been correctly classified
as negative, i.e. were True Positive and had no similar False Positive points nearby.
6 How similar is this tumor to tumors which have been classified as False Negative? Your tumor is not classified as benign, therefore it is not similar to other such data points.
7 Should I get additional tests done?
Additional tests are not considered necessary in cases similar to yours.
However, consult your specialist about the best course of action.
8 What do ’benign’ and ’malignant’ mean?
Benign tumors are those that stay in their primary location without invading other sites of the body.
Malignant tumors are cancerous (i.e., they invade other sites).
9 What is the difference between ’benign’ and ’malignant’?
Benign tumors are those that stay in their primary location without invading other sites of the body.
Malignant tumors are cancerous (i.e., they invade other sites).
10 What is the next course of action? The next course of action for your tumor should be consulted with your specialist.
11 What are some similar samples and their likelihoods? Point X was found to be in the 5 most similar points to your sample. It has a Y probability of being malignant.
12 What is the treatment plan for my tumor? The treatment plan should be consulted with your specialist.
13 Can I see my tumor’s data point visualized? Here is your tumor data visualized:
14 Which feature values prevail for malignant tumors? Here you can see approximate percentages of either classification per feature per value:
15 Which feature values prevail for benign tumors? Here you can see approximate percentages of either classification per feature per value:
16-22 How does the classification shift with changing the value of feature X? This is how your tumor classification changes with changing the value of feature X:
Table 2: Evaluation of LR model.
Test set Whole set
CV score / 100.00%
Accuracy 98.55% 96.93%
Precision 95.83% 95.42%
Recall 100.00% 95.82%
F1-score 97.87% 95.62%
Table 3: Confusion matrix for the test set (left) and the
whole dataset (right) for LR model trained on 7 features.
Actual Actual
Positive Negative Positive Negative
Positive 45 1 Positive 433 11
Predicted
Negative 0 23
Predicted
Negative 10 229
Figure 4: Class percentages plot for bare nuclei.
is shown along with the answer to questions no. 16-22
(one question per feature), for the purposes of visually
clarifying the impact of the individual features on the
classification shift of the currently observed cancer
sample. In this way, we clearly show how changing
the value of just one feature impacts the classification.
5.3 Explainability Results
Figure 6 shows the feature importance scores for our
LR model, which are also the model weights. These
scores explain which features had the most impact on
the prediction. It can be noticed from the plot that
Figure 5: Changing the value of bare nuclei for the observed
tumor sample.
the feature bare nuclei contributes most to the clas-
sification outcome, which means that it will have the
greatest ”pull” in deciding how the tumor is classified.
This plot clearly shows how important each feature is
for the prediction outcome.
Figure 6: Feature importance scores for the LR model.
Figure 7 shows the heat maps for the malignant
and benign portions of the data. Upon visual inspec-
tion, it becomes obvious that these two portions differ.
For example, features marginal adhesion and bland
chromatin have a correlation of 0.33 for the posi-
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
166
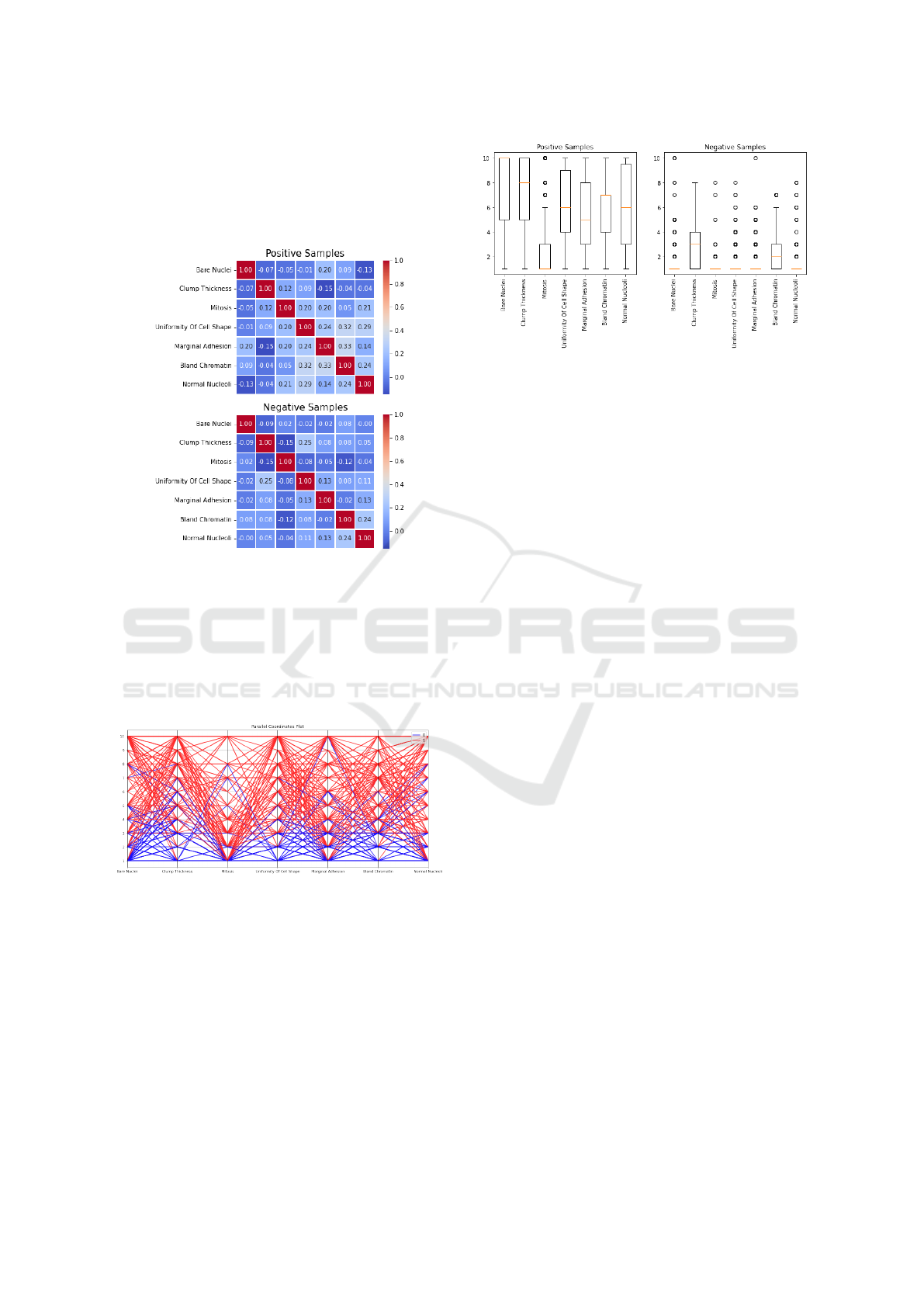
tive portion, and -0.02 for the negative portion of the
dataset. Many more similar examples can be found in
the heat maps. This plot shows how features manifest
different relationships between malignant and benign
tumors.
Figure 7: Heat maps for the LR model for positive (malig-
nant) and negative (benign) portions of the data.
Figure 8 shows the parallel coordinates plot for the
malignant and benign portions of the data. One can
infer simply by looking at this graph that the features
tend to take lower values for the benign portion of
the dataset, while the malignant portion tends to take
higher values. This can also be inferred from figure 4.
Figure 8: Parallel coordinates plot for positive (1=malig-
nant) and negative (0=benign) portions of the dataset.
Figure 9 shows the box plots for positively and
negatively classified samples. Again, it is clear that
the positive and negative samples behave differently.
For instance, the median values and ranges are dif-
ferent for all features. Feature values for 5 out of 7
features in the negative portion are scattered across
the plot. These plots confirm that the malignant and
benign portions of the data have different statistical
properties.
To better explain the effect that the values of the
features have on the classification of the tumor, we
Figure 9: Box plots for the whole dataset.
find the space of possible solutions for Sigmoid(Z) −
0.5 < ε, ε = 1 × 10
−6
. This way, we show how mov-
ing the values along their respective ranges affects the
shift in classification at the classification border.
Figure 10 shows the classification shift at the clas-
sification border while moving the values of the fea-
ture bare nuclei along its range. Analogous figures
are generated for all other features. These figures help
explain how changing even a single feature’s value af-
fects the classification.
If we try to imagine the vector of 7 features as a
vector in 7-dimensional Euclidean space, where each
feature represents a single dimension, then we can
also imagine that the illustrated changes are step-wise
changes per dimension. This way, especially illus-
trative are the figures where the classification had
changed, but the step-wise shift per dimension is only
one step away from the original vector. This can,
for example, be observed for feature bare nuclei,
where changing the value of bare nuclei from 3 to
2, while the values of the other features remain con-
stant, changes the output of Sigmoid(Z) from close to
0.5 but positive, to significantly less than 0.5.
Another interesting connection can be made be-
tween the weights of the features and the classifica-
tion shifts plot. Namely, it can be observed that the
Sigmoid(Z) values always increase as X
i
increases,
where i is the currently observed feature. This is be-
cause the weights of the features are all positive.
6 CONCLUSION
In this paper, we presented a framework for breast
cancer prediction and output interpretation based on
Logistic Regression and NLP methods. The inherent
interpretability of Logistic Regression was used as the
foundation of the quest toward interpretability, while
the pursuit of explainability focused on explaining the
preferred data of each class. While Logistic Regres-
sion is commonly used in clinical and health services
Interpretability and Explainability of Logistic Regression Model for Breast Cancer Detection
167
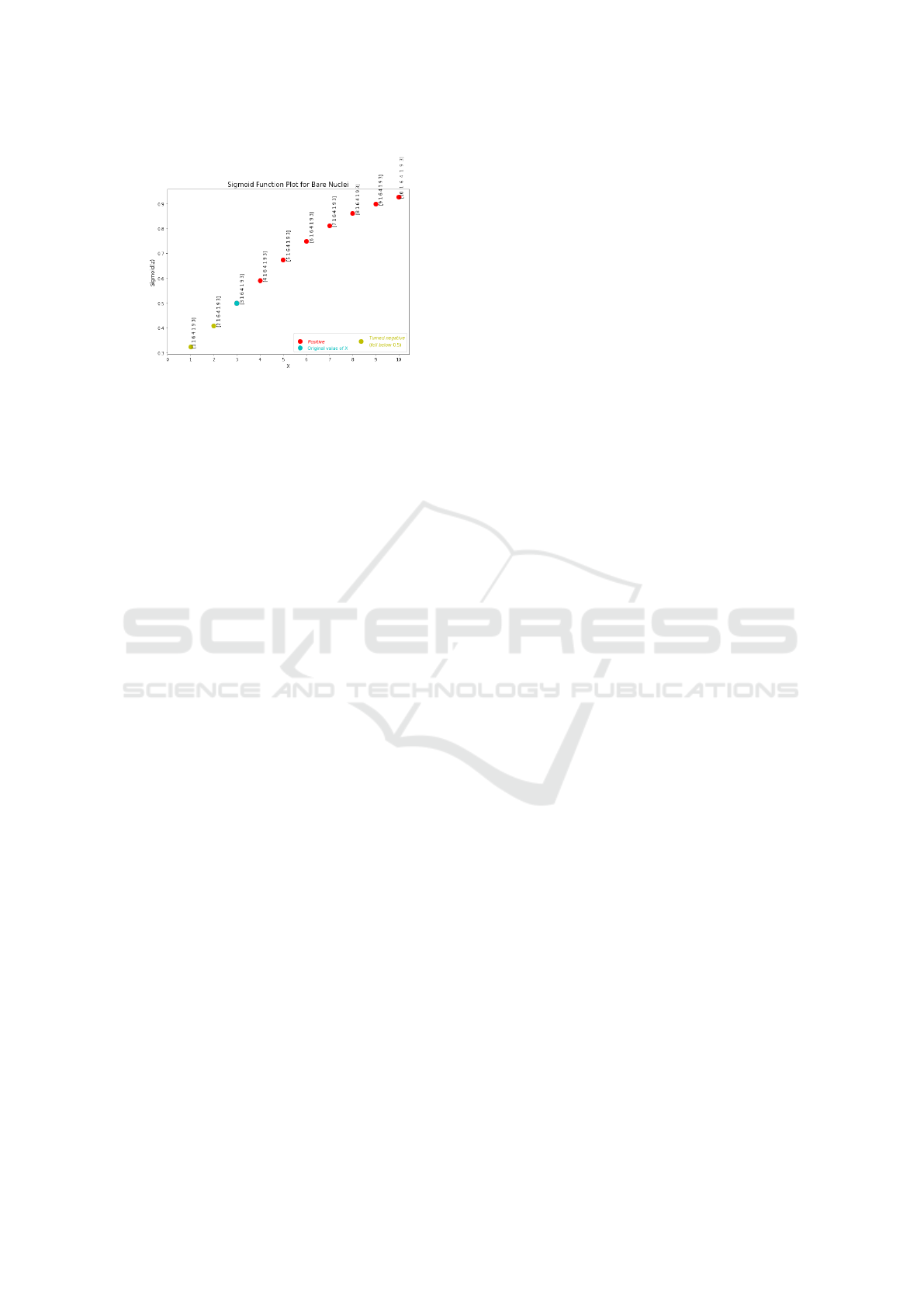
Figure 10: Classification shift at classification border for
bare nuclei.
as a powerful tool, it is important not to forget its lim-
itations, such as the assumption of the absence of high
intercorrelations among the predictors. Therefore, in
future work, we will examine the XAI setup for med-
ical applications with more powerful methods such as
Deep Learning.
Our contribution is a completely transparent, in-
terpretable, and explainable model, developed with
the purpose of aiding medical personnel in the
decision-making process. It contributes towards ex-
tending XAI to regression models, by adapting an
NLP method as a way to access desired explanations.
In future work, we plan to perform user experiments
in order to rate the helpfulness of our model.
REFERENCES
Anisha, P., Reddy C, K., Apoorva, K., and Mangipudi, C.
(2021). Early diagnosis of breast cancer prediction
using random forest classifier. IOP Conference Series:
Materials Science and Engineering, 1116:012187.
Aviv, R. I., d’Esterre, C. D., Murphy, B. D., Hopyan, J. J.,
Buck, B., Mallia, G., Li, V., Zhang, L., Symons, S. P.,
and Lee, T.-Y. (2009). Hemorrhagic transformation of
ischemic stroke: Prediction with ct perfusion. Radiol-
ogy, 250(3):867–877.
Ayer, T., Chhatwal, J., Alagoz, O., Kahn, C. E., Woods,
R. W., and Burnside, E. S. (2010). Comparison of
logistic regression and artificial neural network mod-
els in breast cancer risk estimation. RadioGraphics,
30(1):13–22.
Cashmore, M., Collins, A., Krarup, B., Krivic, S., Maga-
zzeni, D., and Smith, D. E. (2019). Towards explain-
able AI planning as a service. CoRR, abs/1908.05059.
Chakrobartty, S. and El-Gayar, O. F. (2021). Explainable
artificial intelligence in the medical domain: A sys-
tematic review. In Chan, Y. E., Boudreau, M., Aubert,
B., Par
´
e, G., and Chin, W., editors, 27th Americas
Conference on Information Systems. Association for
Information Systems.
Gunning, D. and Aha, D. W. (2019). Darpa’s explain-
able artificial intelligence (XAI) program. AI Mag.,
40(2):44–58.
Hastie, T., Tibshirani, R., and Friedman, J. H. (2009). The
Elements of Statistical Learning: Data Mining, Infer-
ence, and Prediction, 2nd Edition. Springer Series in
Statistics. Springer.
Krarup, B., Krivic, S., Magazzeni, D., Long, D., Cashmore,
M., and Smith, D. E. (2021). Contrastive explanations
of plans through model restrictions. J. Artif. Intell.
Res., 72:533–612.
Krieger, H. (2016). Capturing graded knowledge and uncer-
tainty in a modalized fragment of OWL. In van den
Herik, H. J. and Filipe, J., editors, Proceedings of the
8th International Conference on Agents and Artificial
Intelligence, Vol.2, pages 19–30. SciTePress.
Letzgus, S., Wagner, P., Lederer, J., Samek, W., M
¨
uller, K.,
and Montavon, G. (2021). Toward explainable AI for
regression models. CoRR, abs/2112.11407.
Lipton, Z. C. (2018). The mythos of model interpretability.
Commun. ACM, 61(10):36–43.
Lou, Y., Caruana, R., and Gehrke, J. (2012). Intelligible
models for classification and regression. In The 18th
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining, KDD ’12, 2012,
pages 150–158. ACM.
Mangasarian, O. L., Street, W. N., and Wolberg, W. H.
(1995). Breast cancer diagnosis and prognosis via lin-
ear programming. Oper. Res., 43(4):570–577.
Miller, T. (2019). Explanation in artificial intelligence: In-
sights from the social sciences. Artif. Intell., 267:1–
38.
Naji, M. A., Filali, S. E., Aarika, K., Benlahmar, E. H., Ab-
delouhahid, R. A., and Debauche, O. (2021). Machine
learning algorithms for breast cancer prediction and
diagnosis. Procedia Computer Science, 191:487–492.
Sultana, J. and Jilani, A. K. (2018). Predicting breast cancer
using logistic regression and multi-class classifiers.
International Journal of Engineering & Technology,
7(4.20).
Wolberg, W. H. and Mangasarian, O. L. (1990). Multisur-
face method of pattern separation for medical diag-
nosis applied to breast cytology. Proceedings of the
National Academy of Sciences, 87(23):9193–9196.
Yang, G., Ye, Q., and Xia, J. (2022). Unbox the black-box
for the medical explainable ai via multi-modal and
multi-centre data fusion: A mini-review, two show-
cases and beyond. Information Fusion, 77:29–52.
Zhao, Y., Yang, F., Fang, Y., Liu, H., Zhou, N., Zhang,
J., Sun, J., Yang, S., Menze, B. H., Fan, X., and
Yao, J. (2020). Predicting lymph node metastasis
using histopathological images based on multiple in-
stance learning with deep graph convolution. In 2020
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition, CVPR 2020, Seattle, WA, USA,
June 13-19, 2020, pages 4836–4845. Computer Vision
Foundation / IEEE.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
168
