
Benchmarking Disease Modeling Techniques on the Philippines’
COVID-19 Dataset
Christian Pulmano
a
and Proceso Fernandez
b
Department of Information Systems and Computer Science, Ateneo de Manila University, Quezon City, Philippines
Keywords:
Mathematical Modeling, Parameter Estimation, ARIMA, COVID-19.
Abstract:
The COVID-19 pandemic has emphasized the importance of timely and accurate prediction of disease out-
breaks. Mathematical disease models can help simulate the trajectory of diseases and guide policymakers in
identifying priorities and gaps in current policies. This study evaluates the performance, on various metrics,
of three different parameter estimation algorithms in compartmental models, i.e., Nelder-Mead, Simulated
Annealing, and L-BFGS-B, together with the ARIMA time-series modeling, in modeling COVID-19 cases.
Using the daily number of confirmed cases of COVID-19 in the Philippines as the dataset, the models were
trained on 90 different periods, with each period having 30 days of case data. After training, the models were
used to predict the cases up to 30 days later. The Negative Log Likelihood (NLL), time spent, iterations per
second, and memory allocation were all measured. The results show that ARIMA performed better in terms
of accuracy, time, and space efficiency than each of the other algorithms. This suggests that ARIMA should
be preferred for predicting the number of cases. However, policymaking sometimes requires scenario-based
modeling, which ARIMA is unable to provide. For such requirements, any of the three compartmental models
may be preferred, as each performed generally very well, too.
1 INTRODUCTION
Disease modeling has always been vital for health
planning and policymaking during disease outbreaks
and epidemics. In some areas, disease modeling
has been a standard practice for decision-making
(Kretzschmar, 2020) at the local and national levels
(de Lara-Tuprio et al., 2022). The COVID-19 pan-
demic recently emphasized further the need to pro-
duce accurate disease models as the world raced to
mitigate the spread of the disease.
Some standard methodologies used for disease
modeling are compartmental models using ordi-
nary differential equations and time-series modeling.
Mathematical models are often implemented when
studying the spread of diseases (Panovska-Griffiths,
2020). The increase in computing power has already
advanced the development of disease models. How-
ever, further optimizations can still be applied to gen-
erate more sophisticated models more efficiently.
One crucial step in compartmental disease mod-
eling is parameter estimation. When model parame-
ters are unknown and cannot be derived from existing
datasets and models, the values can be estimated us-
a
https://orcid.org/0000-0001-7870-8197
b
https://orcid.org/0000-0001-5370-4544
ing parameter estimation methodologies. This kind of
optimization problems is also relevant to other fields
aside from epidemiology, including finance, physics,
biology, and engineering (Rica and Ruz, 2020). Mod-
elers need to perform the parameter estimation pro-
cess on a regular basis (de Lara-Tuprio et al., 2022)
as new data are generated. In some cases, the param-
eter estimation problem can get computationally ex-
pensive, especially when the model designs become
complex (Akman et al., 2018).
Alternatively, time-series approaches such
as Auto-Regressive Integrated Moving Average
(ARIMA) can also be used to model the spread of
diseases (Tandon et al., 2020). During the COVID-19
pandemic, ARIMA models have been used to do
short-term predictions (Anne and Jeeva, 2020) on
the number of cases (Tandon et al., 2020). ARIMA
has been shown to provide valuable insights for
COVID-19 epidemiological surveillance efforts (Roy
et al., 2021).
This study evaluates the performance of differ-
ent parameter estimation algorithms, and the ARIMA
time-series modeling, in modeling the trend of
COVID-19 cases in the Philippines. The study aims to
create a benchmark that may provide insights toward
developing more efficient disease models that can be
embedded into health decision support systems.
264
Pulmano, C. and Fernandez, P.
Benchmarking Disease Modeling Techniques on the Philippines’ COVID-19 Dataset.
DOI: 10.5220/0011626400003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 264-270
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
The paper is organized as follows: Section 2 pro-
vides a review of past researches and related works,
Section 3 outlines the methodology for the study, Sec-
tion 4 presents the results of the experiments, and Sec-
tion 5 summarizes the conclusions of the study.
2 REVIEW OF RELATED
LITERATURE
2.1 Mathematical Modeling and
Parameter Estimation
Mathematical models are commonly used to study the
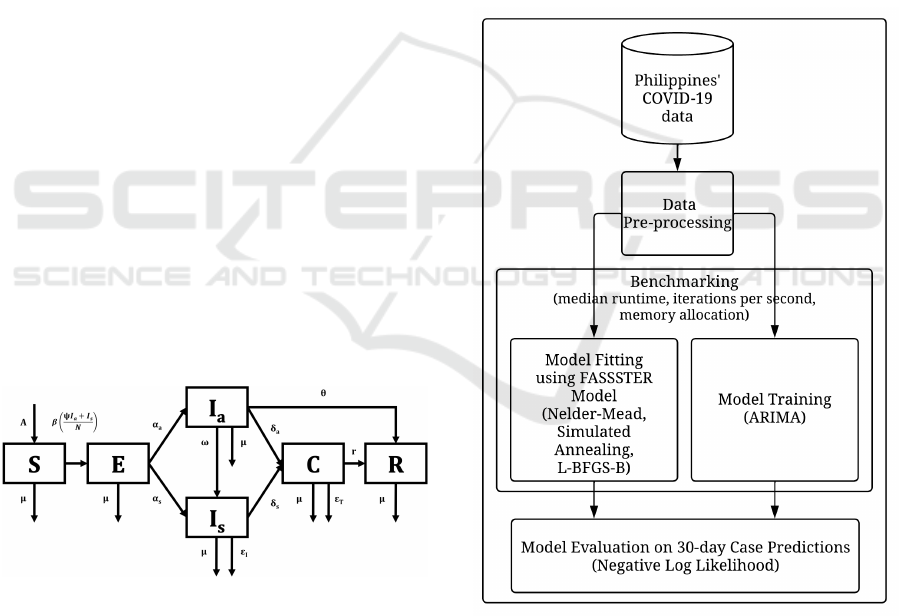
dynamics and spread of diseases (Mohamadou et al.,
2020). In the Philippines, the FASSSTER compart-
mental model, whose visual representation is shown
in Figure 1, is used as a toolkit for modeling the
spread of COVID-19. The model is composed of
six compartments: Susceptible (S), Exposed (E), In-
fectious but asymptomatic (I
a
), Infectious but symp-
tomatic (I
s
), Confirmed (C), and Recovered (R). The
transmissions between compartments are computed
using ordinary differential equations, where param-
eter values describe the rate of transmission from one
compartment to another. In the FASSSTER model,
some parameters were derived from existing data and
taken from literature, but some unknown parameters
and state values were estimated using parameter es-
timation by model fitting. The results of the model
have contributed significantly to the policymaking ef-
forts of the Philippines’ national government through-
out the pandemic (de Lara-Tuprio et al., 2022).
Figure 1: FASSSTER COVID-19 compartmental model.
2.2 Time-Series Modeling
Statistical methods such as time-series models have
also been applied in disease forecasting and predic-
tion. Studies in the past have used time-series mod-
els in estimating incidence and prevalence of dis-
eases like influenza, malaria, and COVID-19 (Cey-
lan, 2020). One study used ARIMA to predict the
number of deaths related to COVID-19. In said study,
ARIMA was able to forecast the possible number of
deaths where the average absolute percentage error
validated the model by 99.09% (Chaurasia and Pal,
2020) indicating a good performance of the model.
As shown in past studies, both methods pro-
vide reliable solutions for disease modeling prob-
lems. However, they also have their own strengths
and weaknesses. As such, the remaining sections of
this paper describe an evaluation of the performance
of these methods when used in a disease modeling
problem.
3 METHODOLOGY
The methodology of the study is illustrated in Figure
2.
Figure 2: Methodology of the study.
3.1 Dataset and Pre-Processing
The Philippines’ COVID-19 dataset was extracted as
a CSV file from the Department of Health - Philip-
pines (DOH Philippines) COVID-19 tracker (Depart-
ment of Health, 2020). The initial dataset consisted
of anonymized individual case data from March 2020
Benchmarking Disease Modeling Techniques on the Philippines’ COVID-19 Dataset
265

to September 2022. However, the study only focused
on data from April 2020 to July 2020 since this was
when the identified version of the FASSSTER model
was heavily adopted, and thus the model parameter
values are more appropriate. The individual case data
were aggregated to get the daily number of new cases
in the Philippines. Although the FASSSTER model
was also used for modeling COVID-19 for smaller
administrative areas, this study only focused on the
national level. The national daily case data was used
for the model fitting/training and evaluation.
3.2 Model Development and Simulation
The objective of the study is to evaluate various pa-
rameter estimation algorithms in a compartmental
disease model, and the ARIMA time-series model.
Both the compartmental and ARIMA time-series
models were developed using the R programming lan-
guage.
The FASSSTER model was adopted for the
COVID-19 compartmental model. The initial con-
ditions, known parameters, and parameters to be es-
timated were also based from the FASSSTER study
(de Lara-Tuprio et al., 2022). Three parameter esti-
mation algorithms, i.e., Nelder-Mead, Simulated An-
nealing, and L-BFGS-B, were evaluated for the com-
partmental model. The models were fitted to the ac-
tual case data based on the minimum negative log
likelihood (NLL) using Poisson distribution.
Both the compartmental disease models and
ARIMA time-series models were fitted/trained using
training data consisting of 30 days of daily COVID-
19 case counts starting from April 1 2020 to June 30
2020, totalling to 91 time periods. The resulting mod-
els were then used to predict the cases up to 30 days
forward.
3.3 Model Evaluation
The bench package of R was used to measure the me-
dian run time in seconds, total memory allocation in
megabytes (MB), and iterations per second on 5 it-
erations during the model fitting/training. The 30-
day model predictions were then compared with ac-
tual case data as reported by DOH Philippines. The
NLL was computed to measure the accuracy of the
case predictions.
One-way Analysis of Variance (ANOVA) was
used to test if there are statistical differences among
the model outputs. Further, the Tukey Honest Signifi-
cance Difference (Tukey HSD) test was implemented
to determine which specific pairs of modeling tech-
niques have significant differences. Tukey HSD in-
corporates some corrections, with such corrections
becoming necessary when multiple pairs are tested
for differences. An alternative correction is used in
the Bonferroni test, which is probably the simplest
among post hoc tests, and is conservative on Type I
errors but is however more prone to Type II errors.
4 RESULTS AND DISCUSSIONS
4.1 Negative Log Likelihood
Table 1 summarizes the NLL values based on the 30-
day predictions of new cases using the different dis-
ease modeling methods. The ARIMA model had the
lowest mean NLL value at 13074.85 and the lowest
median value at 9178.40. Although the overall mini-
mum NLL value was produced using Simulated An-
nealing at 1070.86, the minimum NLL for ARIMA,
1104.75, is only at a slight difference. ARIMA
also had the lowest standard deviation with a value
of 13442.28, signifying that the NLL values com-
puted from the ARIMA model predictions are closer
to the mean value. Among the group, L-BFGS-B
had the highest mean NLL value of 438883.18, fol-
lowed by Nelder-Mead, with a mean NLL value of
39248.45. Nelder-Mead produced the overall maxi-
mum NLL value of 1041587.03 and the highest NLL
standard deviation at 112435.38. L-BFGS-B and
Nelder-Mead also provided the highest median NLL
values at 13434.54 and 13432.61, respectively.
Table 1: Summary of negative log likelihood values for each
method.
method mean median min max stdev
NM 39248.45 13432.61 3477.48 1041587.03 112435.38
SANN 27801.88 11956.06 1070.86 454141.05 57790.56
L-BFGS-B 43883.18 13434.54 2976.53 391733.97 77462.12
ARIMA 13074.85 9178.40 1104.75 81025.72 13442.28
A box plot representation of the NLL values in
logarithmic scale for each method is shown in Figure
3. The resulting NLL values suggest that the case pre-
dictions using Nelder-Mead and L-BFGS-B have the
least likelihood. ARIMA mostly provided the best
NLL values, which suggests that ARIMA should be
preferred to predict new cases with the most likeli-
hood. However, it might still be worth investigat-
ing if the parameters, initial conditions, and hyper-
parameters that were used for the parameter estima-
tion algorithms are approximately optimal.
A one-way ANOVA was performed to compare
the NLL values of the four disease-modeling meth-
ods. The one-way ANOVA revealed that there was
a significant difference in the mean NLL scores be-
HEALTHINF 2023 - 16th International Conference on Health Informatics
266

Figure 3: Box plot representation of NLL values for each
method in logarithmic scale.
tween at least two groups (p = 0.0298). The sum-
mary is shown in Table 2. Tukey HSD Test for
pairwise comparisons found the mean NLL value
was significantly different between L-BFGS-B and
ARIMA (p=0.034), and no significant differences in
the mean NLL scores of Nelder-Mead and ARIMA
(p=0.084), SANN-ARIMA (p=0.541), Nelder-Mead
and L-BFGS-B (p=0.977), SANN and L-BFGS-B
(p=0.485), and SANN and Nelder-Mead (p=0.727).
The summary of the results of Tukey HSD is shown
in Table 3.
Table 2: Results of one-way ANOVA on NLL values.
df Sum of
Squares
Mean
Square
F p-value
Between
Groups
3 5.011E+10 1.670E+10 3.02 0.0298
Within
Groups
352 1.947E+12 5.530E+09
Total 355 1.997E+12
Table 3: Pairwise tests using Tukey HSD on NLL values.
Mean Difference 95% Confidence Interval p-value
Lower Bound Upper Bound
L-BFGS-B
-ARIMA
30808.332 1672.273 59944.390 0.034
Nelder-Mead
-ARIMA
26173.600 -2284.784 54631.980 0.084
SANN
-ARIMA
14727.030 -13731.354 43185.410 0.541
Nelder-Mead
-L-BFGS-B
-4634.733 -33770.792 24501.330 0.977
SANN
-L-BFGS-B
-16081.303 -45217.362 13054.760 0.485
SANN
-Nelder-
Mead
-11446.570 -39904.954 17011.810 0.727
4.2 Median Run Time and Iterations
per Second
The median run time (measured in seconds) and it-
erations per second measure the latency and through-
put of model training, respectively. The results were
aligned with the expectation that ARIMA will per-
form better than the other techniques due to its sim-
pler model design as compared to the other disease
modeling techniques. The summary values are shown
in Tables 4 and 7.
In terms of median run time, ARIMA was the
fastest having a mean value of 0.02 seconds, median
value of 0.02 seconds, and overall minimum value of
0.01 seconds. The maximum value of ARIMA at 0.07
seconds was also faster than the minimum values of
the other methods. Between Nelder-Mead and Simu-
lated Annealing, Nelder-Mead was consistently faster
having a mean value of 2.38 seconds, median value
of 2.27 seconds, and minimum value of 1.31 seconds.
Simulated Annealing provided a mean value of 2.93
seconds, median value of 2.81 seconds and minimum
value of 2.07 seconds. L-BFGS-B was the slowest
among the group having a mean value of 16.83 sec-
onds, median value of 9.20 seconds, minimum value
of 2.94 seconds, and reaching a maximum value of
66.84 seconds. Figure 4 shows a box plot representa-
tion of the median runtime values for each method.
Table 4: Summary of median run time values for each
method.
method mean median min max stdev
NM 2.38 2.27 1.31 3.99 0.63
SANN 2.93 2.81 2.07 4.87 0.62
L-BFGS-B 16.83 9.20 2.94 66.84 16.76
ARIMA 0.02 0.02 0.01 0.07 0.01
The one-way ANOVA result, as shown in Table 5,
reveals that there is a significant difference between at
least two groups (p<2E-16). Tukey HSD test further
shows that there are significant differences between
L-BFGS-B and ARIMA (p=0), Nelder-Mead and L-
BFGS-B (p=0), and SAAN and L-BFGS-B (p=0). It
is revealed however that there are no significant differ-
ence between Nelder-Mead and ARIMA (p=0.203),
SANN and ARIMA (p=0.075), and between SANN
and Nelder-Mead (p=0.969). The summary of Tukey
HSD is displayed in Table 6.
In terms of iterations per second, only ARIMA
performed multiple iterations of the model training in
a second. ARIMA provided a mean value of 49 iter-
ations, a median of 46 iterations, minimum of 14 it-
erations, and a maximum of 114 iterations. The other
methods all resulted in having less than one iteration
Benchmarking Disease Modeling Techniques on the Philippines’ COVID-19 Dataset
267

Figure 4: Box plot representation of median run time (sec-
onds) values for each method in logarithmic scale.
Table 5: Results of one-way ANOVA on median runtime
values.
df Sum of
Squares
Mean
Square
F p-value
Between
Groups
3 14848 4949 75.44 <2E-16
Within
Groups
352 23094 66
Total 355 37942
per second. The values are represented in Figure 5.
The results of one-way ANOVA for iterations
per second values reveal that there is a significant
difference between at least two groups (p<2E-16).
The Tukey HSD test shows that there are signifi-
cant differences in the mean iterations per second val-
ues between L-BFGS-B and ARIMA (p=0), Nelder-
Mead and ARIMA (p=0), SANN and ARIMA (p=0),
but no significant difference between Nelder-Mead
and L-BFGS-B (p=0.998), SANN and L-BFGS-B
(p=0.999), and between SANN and Nelder-Mead
(p=1.000).
Table 6: Pairwise tests using Tukey HSD on median runtime
values.
Mean Difference 95% Confidence Interval p-value
Lower Bound Upper Bound
L-BFGS-B
-ARIMA
16.804 13.631 19.978 0.000
Nelder-Mead
-ARIMA
2.361 -0.739 5.460 0.203
SANN
-ARIMA
2.907 -0.193 6.006 0.075
Nelder-Mead
-L-BFGS-B
-14.444 -17.617 -11.270 0.000
SANN
-L-BFGS-B
-13.898 -17.071 -10.724 0.000
SANN
-Nelder-
Mead
0.546 -2.554 3.646 0.969
Table 7: Summary of iterations per second values for each
method.
method mean median min max stdev
NM 0.44 0.43 0.23 0.75 0.11
SANN 0.33 0.33 0.23 0.44 0.05
L-BFGS-B 0.14 0.11 0.02 0.33 0.10
ARIMA 49.53 46.14 14.00 114.50 21.53
Figure 5: Box plot representation of iteration per second
values for each method in logarithmic scale.
4.3 Memory Allocation
Table 10 summarizes the total memory allocations
during the model training. Similar to the median run
time and iterations per second, it was expected that
ARIMA will also perform better in terms of mem-
ory allocation. The results show that ARIMA consis-
tently provided the lowest memory allocations having
a mean value of 3.05MB, median value of 2.74MB,
and overall minimum value of 0.58MB. This signi-
fies that ARIMA was the most space efficient method
among the group. Nelder-Mead was considered the
second most space efficient method among the group
having a mean value 92.29MB, median value of
87.43MB, and minimum value of 51.18MB. Simu-
lated Annealing was the third most efficient among
the group having a mean value of 120.01MB, median
value of 100.27MB, and minimum value of 84.77. L-
Table 8: Results of one-way ANOVA on iterations per sec-
ond values.
df Sum of
Squares
Mean
Square
F p-value
Between
Groups
3 164094 54698 461.60 <2E-16
Within
Groups
352 41710 118
Total 355 205804
HEALTHINF 2023 - 16th International Conference on Health Informatics
268

Table 9: Pairwise tests using Tukey HSD on iterations per
second values.
Mean Difference 95% Confidence Interval p-value
Lower Bound Upper Bound
L-BFGS-B
-ARIMA
-49.386 -53.650 -45.121 0.000
Nelder-Mead
-ARIMA
-49.090 -53.256 -44.924 0.000
SANN
-ARIMA
-49.193 -53.359 -45.027 0.000
Nelder-Mead
-L-BFGS-B
0.296 -3.969 4.560 0.998
SANN
-L-BFGS-B
0.193 -4.072 4.458 0.999
SANN
-Nelder-
Mead
-0.103 -4.269 4.063 1.000
BFGS-B was least efficient in terms of memory allo-
cations, having a mean value of 436.08MB, a median
value of 292.22MB, and overall maximum value of
1434.58MB. A box plot representation of memory al-
location values is shown in Figure 6.
Table 10: Summary of memory allocations for each
method.
method mean median min max stdev
NM 92.29 87.43 51.18 158.42 25.70
SANN 120.01 100.27 84.77 306.81 41.64
L-BFGS-B 436.08 292.22 119.98 1434.58 342.90
ARIMA 3.05 2.74 0.58 12.08 1.90
Figure 6: Box plot representation of memory allocation val-
ues for each method in logarithmic scale.
Table 11 shows the summary of one-way ANOVA
on memory allocation values. The results revealed
that there was a significant difference in the mean
memory allocation values between at least two groups
(p<2E-16). Tukey HSD test further reveals the mean
memory allocation value was significantly different
between L-BFGS-B and ARIMA (p=0), Nelder-Mead
and ARIMA (p=0.002), SANN and ARIMA (p=0),
Nelder-Mead and L-BFGS-B (p=0), and SANN and
L-BFGS-B (p=0), but no significant difference be-
tween SANN and Nelder-Mead (p=0.679).
Table 11: Results of one-way ANOVA on memory alloca-
tion values.
df Sum of
Squares
Mean
Square
F p-value
Between
Groups
3 9.127E-06 3.042E-06 108.60 <2E-16
Within
Groups
352 9.858E-06 2.800E-08
Total 355 1.899E-05
Table 12: Pairwise tests using Tukey HSD on memory allo-
cation values.
Mean Difference 95% Confidence Interval p-value
Lower Bound Upper Bound
L-BFGS-B
-ARIMA
4.3303E-04 3.6746E-04 4.9859E-04 0.000
Nelder-Mead
-ARIMA
8.9237E-05 2.5196E-05 1.5328E-04 0.002
SANN
-ARIMA
1.1696E-04 5.2920E-05 1.8100E-04 0.000
Nelder-Mead
-L-BFGS-B
-3.4379E-04 -4.0936E-04 -2.7822E-04 0.000
SANN
-L-BFGS-B
-3.1607E-04 -3.8163E-04 -2.5050E-04 0.000
SANN
-Nelder-
Mead
2.7724E-05 -3.6317E-05 9.1765E-05 0.679
5 CONCLUSIONS AND FUTURE
WORK
This study evaluates the performance of parameter es-
timation algorithms, and the ARIMA time-series in
model training and evaluation for COVID-19 disease
modeling using the Philippines’ COVID-19 dataset.
After measuring the performance of the different
methods on various metrics, the results show the
ARIMA has the best performance in terms of max-
imizing the likelihood of predictions, latency and
throughput of model training, and memory efficiency.
This may suggest that ARIMA is efficient in the goal
of predicting the trends of the COVID-19 disease.
However, a limitation of ARIMA is scenario-based
modeling. Although the simulations show that the
parameter estimation algorithms weren’t as efficient
as ARIMA, they are capable of accepting varying pa-
rameter values and modeling different scenarios of the
disease.
This work provides a groundwork for the develop-
ment of more efficient algorithms for disease model-
ing. Future work will incorporate the results of this
study in order to develop more optimal algorithms
that can be used for epidemic planning and decision-
making. Ideally, efficient disease models will be in-
corporated into health information systems such as
Benchmarking Disease Modeling Techniques on the Philippines’ COVID-19 Dataset
269

clinic management systems, pharmacy information
systems, and laboratory information systems to assist
health decisions-makers in their day-to-day decision-
making. The evaluations can also be done on data sets
other than the Philippines. Since disease modeling is
also a global need, it is also worth identifying how
the methodology and the outputs of this study can be
applied to a global environment.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Depart-
ment of Information Systems and Computer Science,
Ateneo de Manila University for their support to the
study.
REFERENCES
Akman, D., Akman, O., and Schaefer, E. (2018). Parameter
estimation in ordinary differential equations modeling
via particle swarm optimization. Journal of Applied
Mathematics, 2018.
Anne, W. R. and Jeeva, S. C. (2020). Arima modelling of
predicting covid-19 infections. medRxiv.
Ceylan, Z. (2020). Estimation of covid-19 prevalence in
italy, spain, and france. Science of The Total Environ-
ment, 729:138817.
Chaurasia, V. and Pal, S. (2020). Covid-19 pandemic:
Arima and regression model-based worldwide death
cases predictions. SN Computer Science, 1(5):1–12.
de Lara-Tuprio, E., Estadilla, C. D. S., Macalalag, J. M. R.,
Teng, T. R., Uyheng, J., Espina, K. E., Pulmano,
C. E., Estuar, M. R. J. E., and Sarmiento, R. F. R.
(2022). Policy-driven mathematical modeling for
covid-19 pandemic response in the philippines. Epi-
demics, 40:100599.
Department of Health (2020). Covid-19 tracker.
https://doh.gov.ph/covid19tracker.
Kretzschmar, M. (2020). Disease modeling for public
health: added value, challenges, and institutional con-
straints. Journal of public health policy, 41(1):39–51.
Mohamadou, Y., Halidou, A., and Kapen, P. T. (2020). A re-
view of mathematical modeling, artificial intelligence
and datasets used in the study, prediction and manage-
ment of covid-19. Applied Intelligence, 50(11):3913–
3925.
Panovska-Griffiths, J. (2020). Can mathematical modelling
solve the current covid-19 crisis?
Rica, S. and Ruz, G. A. (2020). Estimating sir model param-
eters from data using differential evolution: an appli-
cation with covid-19 data. In 2020 IEEE Conference
on Computational Intelligence in Bioinformatics and
Computational Biology (CIBCB), pages 1–6.
Roy, S., Bhunia, G. S., and Shit, P. K. (2021). Spatial pre-
diction of covid-19 epidemic using arima techniques
in india. Modeling earth systems and environment,
7(2):1385–1391.
Tandon, H., Ranjan, P., Chakraborty, T., and Suhag, V.
(2020). Coronavirus (covid-19): Arima based time-
series analysis to forecast near future. arXiv preprint
arXiv:2004.07859.
HEALTHINF 2023 - 16th International Conference on Health Informatics
270
