
Machine Learning Algorithms for Mouse LFP Data Classification in
Epilepsy
Antonis Golfidis
1
, Michael Vinos
2,3 a
, Nikos Vassilopoulos
2,3
, Eirini Papadaki
2,3
,
Irini Skaliora
1,2,3 b
and Vassilis Cutsuridis
1,4 c
1
Athens International Master’s Programme in Neurosciences, Department of Biology,
National and Kapodistrian University of Athens, Athens, Greece
2
Department of History and Philosophy of Science, National and Kapodistrian University of Athens, Athens, Greece
3
Center for Basic Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece
4
School of Computer Science, University of Lincoln, Lincoln, U.K.
Keywords: Machine Learning, Classification, HCTSA, Epilepsy, Animal, LFP, Endogenous Activity, Interictal Activity,
Seizure like Activity.
Abstract: Successful preictal, interictal and ictal activity discrimination is extremely important for accurate seizure
detection and prediction in epileptology. Here, we introduce an algorithmic pipeline applied to local field
potentials (LFPs) recorded from layers II/III of the primary somatosensory cortex of young mice for the
classification of endogenous (preictal), interictal, and seizure-like (ictal) activity events using time series
analysis and machine learning (ML) models. Using the HCTSA time series analysis toolbox, over 4000
features were extracted from the LFPs after applying over 7700 operations. Iterative application of correlation
analysis and random-forest-recursive-feature-elimination with cross validation method reduced the
dimensionality of the feature space to 22 features and 27 features, in endogenous-to-interictal events
discrimination, and interictal-to-ictal events discrimination, respectively. Application of nine ML algorithms
on these reduced feature sets showed preictal activity can be discriminated from interictal activity by a radial
basis function SVM with a 0.9914 Cohen kappa score with just 22 features, whereas interictal and seizure-
like (ictal) activities can be discriminated by the same classifier with a 0.9565 Cohen kappa score with just
27 features. Our preliminary results show that ML application in cortical LFP recordings may be a promising
research avenue for accurate seizure detection and prediction in focal epilepsy.
1 INTRODUCTION
Epilepsy, the sacred disease, is one of the oldest
recognizable neurological conditions with written
records dating back to 2000 BCE (Chang and
Lowenstein, 2003; Magiorkinis et al., 2010). As of
2020 around 50 million people worldwide were
affected by epilepsy (Ghosh et al., 2021). The causes
of epilepsy are mostly unknown (idiopathic), but
often epilepsy is caused from brain damage, stroke,
and trauma (Goldberg and Coulter, 2013). The
disease is characterized by recurrent violent episodes
of involuntary movements called seizures, which may
be partial (involve only one part of the body) or
a
https://orcid.org/0000-0001-9961-1079
b
https://orcid.org/0000-0002-7528-7208
c
https://orcid.org/0000-0001-9005-0260
generalized (involve the whole body) followed at
times by loss of consciousness and/or control of
bowel or bladder function (Duncan et al., 2006).
Seizures are the result of excessive electrical
discharges in neuronal populations (Colmers and
Maguire, 2020). Seizures measured by
electroencephalography (EEG) or LFP recordings
have been shown to vary in frequency, from one per
year to several episodes per day. Because they occur
so sporadically and at unknown times, the availability
of seizure-like (ictal) activity is scarce and thus
interictal activity is often used in diagnosis.
The best way for detecting interictal activity is a
visual inspection of the EEG/LFP signal by an expert
36
Golfidis, A., Vinos, M., Vassilopoulos, N., Papadaki, E., Skaliora, I. and Cutsuridis, V.
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy.
DOI: 10.5220/0011625600003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 4: BIOSIGNALS, pages 36-47
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
(Lodder et al., 2014). This approach, however, has
several limitations including a very long learning
curve and extensive analysis time, especially for long
recordings. Human error, subjectivity, intra and
interobserver variability often result in misdiagnosis
leading to lack of treatment or prescription of
medication with potentially harmful side effects
(Lodder et al., 2014).
Overcoming these drawbacks requires the
development of an artificial intelligence system for an
automatic pre-ictal, interictal, and ictal detection that
can match or even outperform experts, hence
reducing the time and resources spent on visual
analysis, as well as misdiagnosis rates.
Herein, we introduce an algorithmic pipeline
applied to LFP signals recorded from mouse
somatosensory cortical slices to extract features used
for the classification of endogenous (preictal),
interictal, and seizure-like (ictal) events using time
series analysis and ML models.
2 MATERIALS AND METHODS
2.1 LFP Data
2.1.1 Animals
Thirty-one C57Bl/6J mice were bred in the animal
facility of the Center for Experimental Surgery of the
Biomedical Research Foundation of the Academy of
Athens, registered as a breeding and experimental
facility according to the Presidential Decree of the
Greek Democracy 160/91, which harmonizes the
Greek national legislation with the European Council
Directive 86/609/EEC on the protection of animals
used for experimental and other scientific purposes.
The present study was approved by the Regional
Veterinary Service, in accordance with the National
legal framework for the protection of animals used for
scientific purposes (reference number 2834/08-05-
2013). Mice were weaned at 21 days old, housed in
groups of 5 – 7, in 267 × 483 × 203 mm cages
supplied with bedding material and kept at a 12/12 h
dark-light schedule. Food was provided ad libitum.
2.1.2 Slice Preparation
Coronal brain slices (400 μm) were prepared from the
primary somatosensory cortex of young mice (P18-
20) as described before (Rigas et al 2015; 2018;
Sigalas et al 2015; 2017). Briefly slices were placed
in a holding chamber with artificial cerebrospinal
fluid (ACSF) containing (in mM): NaCl 126; KCl
3.53; NaH2PO4.H2O 1.25; NaHCO3 26; MgSO4 1;
D-Glucose 10 and CaCl2.2H2O 2 [osmolarity (mean
± SD): 317 ± 4 mOsm, pH: 7.4±0.2], where they were
left to recover at room temperature (RT: 24–26 °C).
2.1.3 ex Vivo Electrophysiology
Twenty minutes LFP recordings of endogenous
cortical activity in the form of recurring Up and Down
states were obtained. Subsequently, epileptiform
activity was induced by replacing the ACSF with low
Mg
2+
ACSF (Avoli & Jefferys, 2016; Dreier &
Heinemann, 1991) for up to 80 minutes to ensure that
the pattern of epileptiform activity had stabilized.
Network activity was assessed by LFP recordings
which were obtained from cortical layers II/III of
S1BF using low impedance (∼0.5 MΩ) glass pipettes
filled with ACSF. Recordings were obtained in
current-clamp mode with a Multiclamp 700B
amplifier (Molecular Devices, San Jose, CA, USA).
LFP signals were low-pass filtered at 6 kHz (by an
analog anti-aliasing filter) and subsequently digitized
at 15 kHz by means of a 16-bit multi-channel
interface (InstruTECH ITC-18; HEKA Elektronic,
Lambrecht, Germany). Data acquisition was
accomplished using AxoGraph X (version 1.3.5;
https://axograph.com; RRID: SCR_014284).
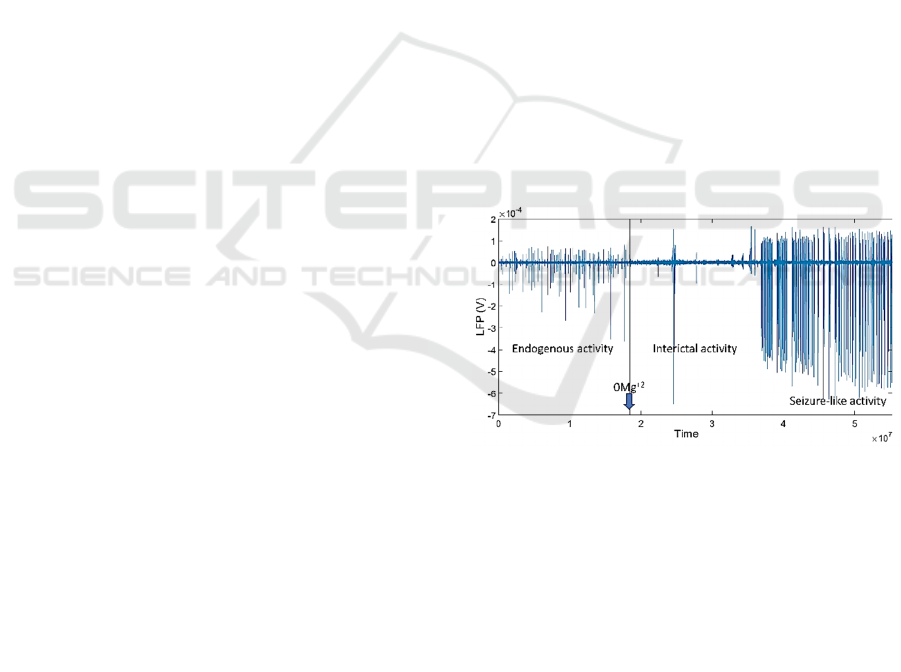
Figure 1: Exemplary 20-min LFP recording trace from a
coronal slice of the primary somatosensory cortex of a
young mouse. Blue downward pointing arrow indicates the
time the ACSF was replaced with zero Mg
2+
ACSF.
2.1.4 Data Analysis
The detection of spontaneous network events was
performed semi-automatically from the LFP
recordings. Traces were exported to MATLAB
format and analyzed with LFPAnalyzer, an in-house-
developed software (Tsakanikas et al., 2017;
Kaplanian et al., 2022). Briefly: (i) input signals were
pre-processed by DC offset subtraction and low-pass
filtering at 200 Hz; (ii) two feature sequences were
extracted for each segment, based on two
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
37

complementary mathematical transformations
(Hilbert and Short energy); (iii) a dynamic, data-
driven threshold based on Gaussian mixture models
was estimated for each feature sequence and used to
create a mask; and (iv) the two masks were combined
using a logical OR operator and used for the detection
of the onset and offset of the LFP events. After
identification of their onset and offset, events were
manually classified as endogenous activity (EA) (up-
states), interictal activity (IA), or seizure-like activity
(SLA) (see Figure 1 for traces of these three types of
events) based on their shape (waveform), and on the
basis of previous simultaneous whole-cell patch
clamp recordings (Sigalas et al 2015; Kaplanian et al
2022).
2.2 Algorithmic pipeline
Our high-level algorithmic pipeline is depicted in
Figure 2. Every step in the pipeline is described in
detail in the following sections.
Figure 2: General algorithmic pipeline.
2.2.1 Data Preparation
The steps followed for preparing the data for further
analysis are depicted in Figure 3.
Figure 3: Data preparation pipeline.
All digitized recordings were downsampled (f
s
= 962
Hz). A segmentation window with a 5 sec duration
and a 50% overlap was slid to all signals to segment
them into 18542 samples. Out of these samples 8357
were identified as EA, 1318 as IA and 8872 as SLA
by expert users (see Figure 4 for segmented data
distributions). The HCTSA suite of time series
methods (Fulcher et al., 2013) was then used to
extract features. HCTSA consists of thousands of
time-series analysis methods allowing users to
convert a time series into a vector of thousands of
informative features, corresponding to different
outputs of time-series analysis operations (Fulcher et
al., 2013; Fulcher and Jones, 2017). HCTSA has been
successfully used to a wide range of problems
including the diagnosis of Parkinson’s disease from
speech signals, monitoring sleep-stage progression,
predicting schizophrenia from brain imaging data,
Figure 4: Segmented signal events distribution. EA:
endogenous activity; IA: interictal activity; SLA: seizure-
like activity. Time in parenthesis is the total cumulative
duration of each event class in minutes.
and forecasting catastrophes in financial and
ecological systems. The features we extracted with
HCTSA were from the time, frequency, time-
frequency, and chaotic domains of the segmented
LFP signals by performing over 7700 operations to
them. For all segmented signals from our LFP
recordings a total of 4476 meaningful (non-zero, non-
constant, etc) features were extracted. All features
were then normalized to a common scale (0-1),
without distorting differences in the ranges of values.
These features constituted the Full Feature Set.
2.2.2 Dimensionality Reduction
The steps followed for reducing the dimensionality of
the extracted features of the LFP data are depicted in
Figure 5. To further reduce the high-dimensional
space of the extracted features we calculated the
correlation scores of all features in the Full Feature
Set. Any features whose score was higher than ρ were
removed. The remaining features constituted the
Uncorrelated Feature Set.
Figure 5: Dimensionality reduction pipeline.
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
38
On the uncorrelated feature set we used Random
Forrest (RF) (Breiman, 2001), a machine learning
method that operates by constructing a multitude of
decision trees at training time (Ho, 1995, 1998). For
classification tasks, RF performs implicit feature
selection, using a small subset of "strong variables"
for classification only, resulting in superior
performance on high-dimensional data (Menze et al,
2009). The mean decrease of Entropy (or increase of
Information Gain) over all the decision trees is an
indicator of feature relevance derived from this
implicit feature selection of the random forest. A
feature importance score indicates the relative
importance of features, which is a by-product of
random forest classifier training. Several studies
(Menze et al., 2007; Díaz-Uriarte & Alvarez de
Andres, 2006) have shown that this feature selection
step can significantly reduce the number of features
while increasing the model’s accuracy. The output of
RF is the class selected by most trees according to
some predefined criterion (Ho, 1998). The criterion in
our case was the importance score (we kept those
features with score greater than 6*mean importance
score). Each of these feature sets constituted the
Selected Features Set. A recursive feature elimination
with cross-validation (RFECV) method was then
used to remove the weakest features and find from
each Selected Features Set the optimum number of
features that gave the best accuracy results. Because
it was not known in advance how many features
would be valid, cross validation was used with RFE
to score different feature subsets and find the average
optimum number of features. Each of these optimum
number of features constituted the RFE Feature Set.
2.2.3 Classification
We employed 9 machine learning classifiers: a linear
SVM (SVMlin), a polynomial of the 2
nd
degree SVM
(SVMpol), a polynomial of the 3
rd
degree SVM
(SVMpol), a polynomial of the 4
th
degree SVM
(SVMpol), a polynomial of the 5
th
degree SVM
(SVMpol), a radial basis function with a Gaussian
kernel SVM (SVMrbf), an RF, a decision tree (DT)
and a k-nearest neighbours (kNN). The data (N
samples x M features) were split into a training set
(80%) and a validation test set (20%). A stratified 5-
fold cross validation was used to preserves the
percentages of samples of each fold. A
GridSearchCV function was used for hyperparameter
tuning of every classifier (see Table 1).
Table 1: Machine learning classifiers, their
hyperparameters and their hyperpameter values. C:
Controls the amount of misclassified data points allowed by
introducing a penalty. Low C values lead to decision
boundaries with large margin. High C values add greater
penalty thus minimizing the number of misclassified
examples; Gamma: The distance of influence of a training
point. Low values of gamma indicate greater distance
resulting in more points taken into account for the
calculation of the separation line; N_estimators (RF): The
number of decision trees being built in the forest;
Max_depth (RF): The number of splits that each decision
tree is allowed to make; N_estimators (kNN): The number
of nearest neighbors; Criterion (DT): How the impurity of
a split will be measured; Max_depth (DT): The number of
splits that the decision tree is allowed to make.
Classifier Hyperparameter Values
SVMlin C [0.01, 0.1, 1, 10]
SVMpol (2
nd
degree)
C [0.01, 0.1, 1, 10]
SVMpol (3
rd
degree)
C [0.01, 0.1, 1, 10]
SVMpol (4
th
degree)
C [0.01, 0.1, 1, 10]
SVMpol (5
th
degree)
C [0.01, 0.1, 1, 10]
SVMrbf C [0.01, 0.1, 1, 10]
Gamma [0.1, 1, 10]
RF N_estimators [100, 200, 300,
400, 500, 600,
700, 800, 900]
Max_depth [10, 20, 30]
DT Criterion [gini, entropy]
Max_depth
[1, 2, 3, 4, 5, 10,
20, 30]
kNN N_estimators [1, 2, 3, 4, 5, 6,
7, 8, 9, 10]
Performance Metrics. We used the following
metrics for evaluating the performances of our
classifiers:
𝐶𝑜ℎ𝑒𝑛 𝜅 =
(∗∗)
(
)
∗
(
)
(
)
∗()
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 =
𝑅𝑒𝑐𝑎𝑙𝑙 =
𝐹1 𝑠𝑐𝑜𝑟𝑒 = 2 ∗
∗
where TP are the true positives, TN are the true
negatives, FP are the false positives and FN are the
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
39
false negatives. The Cohen kappa score was used
because our classes (EA, IA and SLA) were
unbalanced. The Cohen kappa score values ranged
from -1 (worst) to +1 (best). The Precision, Recall,
and F1-score values ranged from 0 (worst) and 1
(best).
3 RESULTS
3.1 EA vs IA Classification
We started our analysis from the downsampled and
segmented 9674 samples (8357 EA + 1318 IA) and
applied the HCTSA toolbox on them to extract 4476
meaningful features (Full Feature Set). Then, starting
with the full feature set we followed the
“Dimensionality Reduction” pipeline depicted in
figure 5 and described in section 2.2.2. In every step
of this pipeline, we evaluated the performances of all
nine classifiers to determine how much of the
performance will be lost as the feature space is
reduced. The performances of all nine classifiers
tested against the Full Feature Set are summarized in
Table 2. We then calculated the correlation score of
each feature in the full feature set, compared it to the
ρ criterion (see section 2.2.2 for details) and kept only
those features whose correlation score was lower than
ρ. We tried different values for ρ (ρ = 0.8 or ρ = 0.9).
We kept ρ = 0.9 because it gave the best Cohen kappa
scores when only an RF was tested against the derived
number of features (1933 features). These 1933
features constituted the Uncorrelated Feature Set.
We tested the performances of our classifiers
including the RF one on the Uncorrelated Feature Set
and found that SVMpol of the 2
nd
degree had the best
Cohen kappa score (see Table 3). In the next step and
to further reduce the feature space we employed the
RF with a criterion method (we kept those features
with score greater than 6*mean importance score =
0.0031) on the uncorrelated feature set to find the 62
most important features (Selected Features Set). We
tested once again the performances of our classifiers
on the Selected Features Set. The SVMrbf displayed
the best performance (Cohen kappa score = 0.98,
precision = 0.9893, recall = 0.9893, F1-score =
0.9893) (see Table 4). Finally, the RFECV method
was employed to find the optimum feature set (RFE
Feature Set). RFECV resulted in 31 optimum
features. Once more we tested performances of our
classifiers on this feature set and found that the best
performance was SVMrbf (Cohen kappa score =
0.9871, precision = 0.9951, recall = 0.9920, F1-score
= 0.9936) (see Table 5).
Table 2: Classifiers’ performances on the full feature set
(4476 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.959 0.9833 0.9758 0.9795
SVMpol
2
nd
degree
0.9678 0.9861 0.9817 0.9839
SVMpol
3
rd
degree
0.9657 0.9841 0.9814 0.9828
SVMpol
4
th
degree
0.9657 0.9843 0.9814 0.9828
SVMpol
5
th
degree
0.9613 0.9836 0.9777 0.9806
SVMrbf - - - -
RF 0.9316 0.9658 0.9658 0.9658
kNN 0.9027 0.9564 0.9465 0.9513
DT 0.9056 0.9542 0.9514 0.9528
Table 3: Classifiers’ performances on the uncorrelated
feature set (1933 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.9461 0.9768 0.9694 0.9731
SVMpol
2
nd
degree
0.9590 0.9833 0.9758 0.9795
SVMpol
3
rd
degree
0.9611 0.9851 0.9761 0.9806
SVMpol
4
th
degree
0.9568 0.9829 0.974 0.9784
SVMpol
5
th
degree
0.9524 0.9823 0.9703 0.9762
SVMrbf - - - -
RF 0.9275 0.9769 0.9516 0.9638
kNN 0.9105 0.9649 0.9462 0.9553
DT 0.8460 0.9230 0.9230 0.9230
Table 4: Classifiers’ performances on the selected feature
set (62 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.9202 0.9637 0.9566 0.9601
SVMpol
2
nd
degree
0.9367 0.9783 0.959 0.9684
SVMpol
3
rd
degree
0.9674 0.9772 0.9801 0.9787
SVMpol
4
th
degree
0.9617 0.9780 0.9838 0.9809
SVMpol
5
th
degree
0.9660 0.9801 0.986 0.983
SVMrbf 0.9786 0.9893 0.9893 0.9893
RF 0.9654 0.9888 0.9767 0.9827
kNN 0.9491 0.9703 0.9789 0.9746
DT 0.8736 0.9375 0.9361 0.9368
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
40
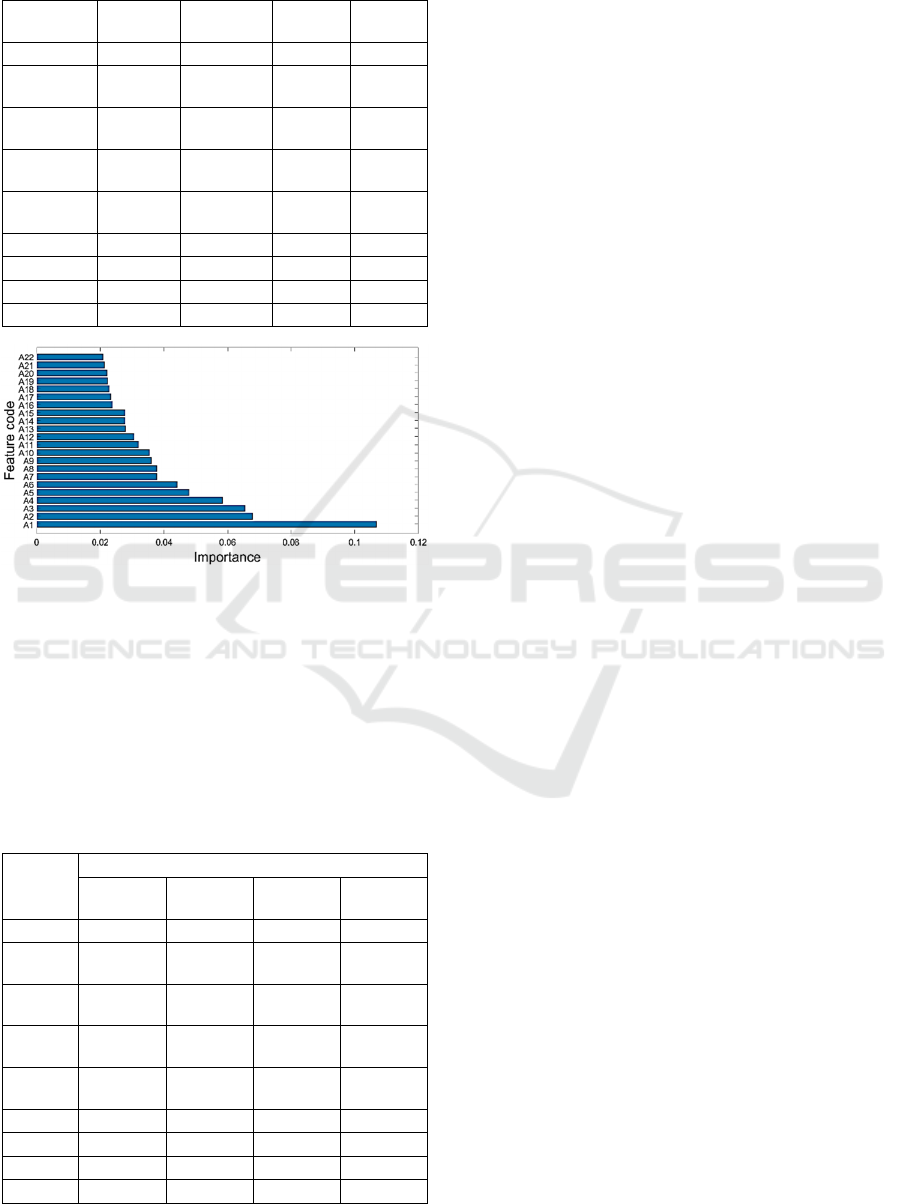
Table 5: Classifiers’ performances on the RFE feature set
(31 features).
Classifier Cohen
kappa
Precision Recall F1-
score
SVMlin 0.9295 0.9641 0.9655 0.9648
SVMpol
2
nd
degree
0.8890 0.9628 0.9281 0.9445
SVMpol
3
rd
degree
0.9638 0.9797 0.9841 0.9819
SVMpol
4
th
degree
0.9700 0.9865 0.9835 0.985
SVMpol
5
th
degree
0.9677 0.9876 0.9801 0.9838
SVMrbf 0.9871 0.9951 0.9920 0.9936
RF 0.9740 0.9932 0.9810 0.9870
kNN 0.9680 0.9832 0.9847 0.9840
DT 0.8954 0.9470 0.9484 0.9477
Figure 6: Importance scores of all 22 features in the RFE
feature set. See appendix for description of each feature
code.
Figure 6 depicts the importance scores of the 22 out
of the 31 features from the RFE feature set. See
Appendix for detailed description of each coded
feature in Fig. 6. The bottom feature has the highest
importance score value (IS > 0.1). We then
Table 6: Classifiers’ Cohen kappa scores on the RFE
feature set for different IS values. IS: importance score.
Classifier Cohen kappa score
IS > 0.02
(22 features)
IS > 0.03
(12 features)
IS > 0.04
(6 features)
IS > 0.05
(4 features)
SVMlin 0.9146 0.8597 0.7361 0.6113
SVMpol
2
nd
degree
0.9328 0.9128 0.8348 0.6015
SVMpol
3
rd
degree
0.9722 0.9442 0.8780 0.6078
SVMpol
4
th
degree
0.9722 0.9464 0.8941 0.6031
SVMpol
5
th
degree
0.9659 0.9440 0.9064 0.5908
SVMrbf 0.9914 0.9525 0.9073 0.6300
RF 0.9761 0.9566 0.9241 0.7518
kNN 0.9724 0.9534 0.9059 0.7202
DT 0.9033 0.9028 0.9059 0.6932
investigated combinations of these features to see if
we can improve the performances of our classifiers
and to also assess when their performances worsen as
feature space is further reduced. These results (Table
6) reveal that almost all classifiers’ performances
improved (compare values in Tables 5 and 6).
SVMrbf had an almost perfect Cohen kappa score
(0.9914) for 22 features. For smaller number of
features all classifiers’ Cohen kappa scores
progressively became worse (see Table 6).
3.2 IA vs SLA Classification
For the binary classification of IA vs SLA, we started
our analysis from the downsampled and segmented
10190 samples (8872 SLA + 1318 IA) and applied the
HCTSA toolbox on them to extract 4476 meaningful
features (Full Feature Set). We followed the
“Dimensionality Reduction” pipeline depicted in
figure 5 and described in section 2.2.2. In every step
of this pipeline, we evaluated the performances of our
nine classifiers to determine how they were affected
as the feature space was reduced. The classifiers’
performances on the Full Feature Set are summarized
in Table 7. As before we then calculated the
correlation score of each feature in the Full Feature
Set, compared it to the ρ criterion (see section 2.2.2
for details) and kept only those features whose
correlation score was lower than ρ. We tried different
values for ρ (0.8 and 0.9) and kept ρ = 0.9 because it
gave the best Cohen kappa scores when only an RF
was tested against the derived number of features
(1944 features). These 1944 features constituted the
Uncorrelated Feature Set. We tested the
performances of our classifiers including the RF one
on this reduced set and found that SVMpol of the 5
th
degree had the best Cohen kappa score (see Table 8).
Next, we employed the RF with a criterion method
(kept as before those features with score greater than
6*mean importance score = 0.0031) on the
Uncorrelated Feature Set to find the 40 most
important features (Selected Features Set). We tested
once again the performances of our classifiers on the
Selected Features Set. The SVMrbf had the best
performance (Cohen kappa score = 0.9217, precision
= 0.9849, recall = 0.9399, F1-score = 0.9608) (see
Table 9). Finally, the RFECV method was employed
to find the optimum feature set (RFE Feature Set).
RFECV resulted in 27 optimum features. We tested
again the performances of the nine classifiers on this
feature set and found that the best performance was
still SVMrbf (see Table 10).
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
41
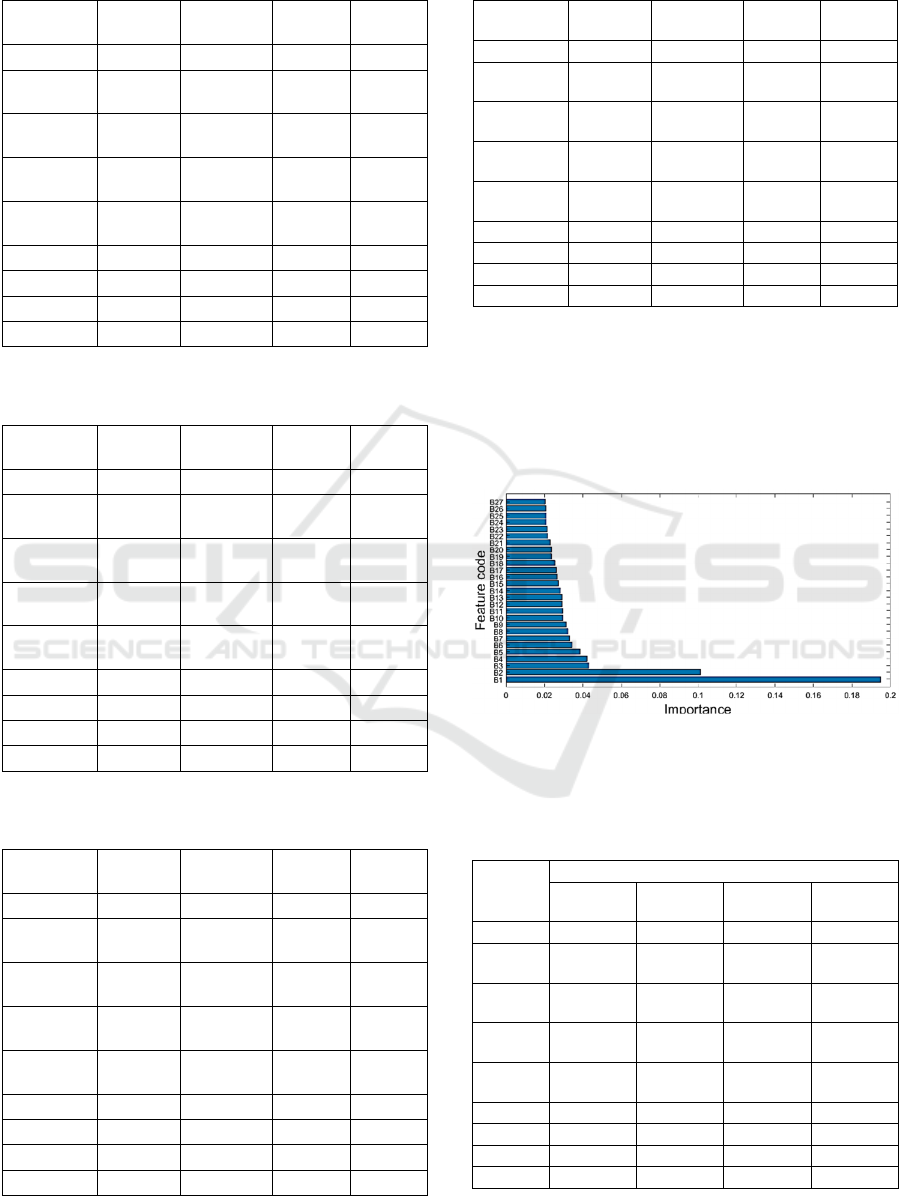
Table 7: Classifiers’ performances on the full feature set
(4476 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.7675 0.8991 0.8699 0.8837
SVMpol
2
nd
degree
0.8003 0.9227 0.8807 0.9001
SVMpol
3
rd
degree
0.8112 0.9293 0.8852 0.9056
SVMpol
4
th
degree
0.8298 0.9373 0.8953 0.9148
SVMpol
5
th
degree
0.8253 0.935 0.8932 0.9226
SVMrbf - - - -
RF 0.8157 0.9702 0.86 0.9076
kNN 0.7649 0.8985 0.8680 0.8824
DT 0.7379 0.8713 0.8666 0.8690
Table 8: Classifiers’ performances on the uncorrelated
feature set (1944 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.7222 0.8770 0.8470 0.8611
SVMpol
2
nd
degree
0.7573 0.9007 0.8596 0.8796
SVMpol
3
rd
degree
0.7524 0.9082 0.8502 0.876
SVMpol
4
th
degree
0.7760 0.9193 0.8625 0.8879
SVMpol
5
th
degree
0.7837 0.9271 0.8636 0.8917
SVMrbf - - - -
RF 0.7663 0.9697 0.8291 0.8826
kNN 0.7751 0.8852 0.89 0.8876
DT 0.7369 0.8852 0.8736 0.8684
Table 9: Classifiers’ performances on the selected feature
set (40 features).
Classifier
Cohen
kappa
Precision Recall
F1-
score
SVMlin 0.7503 0.9019 0.8528 0.8750
SVMpol
2
nd
degree
0.9145 0.9704 0.945 0.9572
SVMpol
3
rd
degree
0.9089 0.9619 0.9473 0.9545
SVMpol
4
th
degree
0.9133 0.9641 0.9495 0.9566
SVMpol
5
th
degree
0.9147 0.9689 0.9466 0.9574
SVMrbf 0.9217 0.9849 0.9399 0.9608
RF 0.9154 0.9788 0.9390 0.9577
kNN 0.8905 0.9573 0.9341 0.9453
DT 0.7942 0.9078 0.8871 0.8971
Table 10: Classifiers’ performances on the RFE feature set
(27 features).
Classifier Cohen
kappa
Precision Recall F1-
score
SVMlin 0.6773 0.8853 0.8046 0.8383
SVMpol
2
nd
degree
0.9219 0.9685 0.9537 0.961
SVMpol
3
rd
degree
0.9158 0.9631 0.9529 0.9579
SVMpol
4
th
degree
0.9206 0.9625 0.9582 0.9603
SVMpol
5
th
degree
0.9203 0.9639 0.9566 0.9602
SVMrbf 0.9565 0.9878 0.9692 0.9782
RF 0.9227 0.9781 0.9462 0.9614
kNN 0.9209 0.9746 0.9475 0.9605
DT 0.7981 0.9016 0.8965 0.899
Figure 8 depicts the importance scores of all 27
features from the RFE Feature Set. See Appendix for
detailed description of each coded feature in Fig. 8.
The bottom feature has the highest importance score.
We investigated combinations of these features to see
if we can further improve the classification
performance of our classifiers and also when their
Figure 8: Importance scores of all 27 features in the RFE
feature set. See appendix for description of each feature
code.
Table 11: Classifiers’ Cohen kappa scores on the RFE
feature set for different IS values. IS: importance score.
Classifier Cohen kappa score
IS > 0.02
(27 features)
IS > 0.03
(9 features)
IS > 0.04
(4 features)
IS > 0.05
(2 features)
SVMlin 0.6773 0.2969 - -
SVMpol
2
nd
degree
0.9219 0.8457 0.3829 -
SVMpol
3
rd
degree
0.9158 0.8730 0.3565 0.1438
SVMpol
4
th
degree
0.9206 0.8932 0.4522 0.2079
SVMpol
5
th
degree
0.9204 0.8856 0.4860 0.2963
SVMrbf 0.9565 0.9080 0.7410 0.6523
RF 0.9227 0.9054 0.7749 0.6036
kNN 0.9209 0.8942 0.7258 0.6572
DT 0.7981 0.8227 0.7491 0.5969
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
42

performance worsen as feature space is further
reduced. These results are depicted in Table 11. As the
number of features decreased all classifiers’ Cohen
kappa score progressively become worse. kNN had the
best score (0.6572) with only two features.
4 DISCUSSION
Our study has produced several interesting results
concerning the usefulness of time series analysis and
ML in LFP based epileptology. Most importantly it
showed that discriminating endogenous (pre-ictal)
activity from interictal activity is more successful
(and easier) than discriminating interictal from
seizure-like (ictal) activity. This result confirms past
research findings (Fischer, 2014). By using feature
extraction methods from the time, frequency, time-
frequency and chaotic domains and standard (single
and ensemble) ML methods such as kNN, RF, SVM,
and DT we achieved an over 0.9 Cohen kappa score
and an over 0.95 precision and recall scores when the
Full Feature Set (4476 features) was used in the EA
vs IA discrimination task. As the feature space was
reduced (4476 to 22) the discriminability of the ML
classifiers changed. The classifier with the best
performance was SVMrbf (Cohen kappa score =
0.9914), whereas the classifier with the worst
performance was DT (Cohen kappa score = 0.9033).
The average Cohen kappa score was 0.96. Out of the
22 most important features, the feature with the
highest importance score (IS ~ 0.12) was the ratio of
autocorrelation (using lag = 2) of the transformed
time series over the original time series when 5% of
time points closest to the mean were removed. When
only the first 4 features with the highest importance
scores (A1-A4 in Fig. 6) were used, then the
discriminability of the classifiers ranged from 0.59-
0.75 (Average Cohen kappa score = 0.6455).
Addition of just two more features (4 to 6) increased
the performances of the classifiers by 23% on average
(Average Cohen kappa score = 0.8769). Addition of
6 more features (6 to 12) increased the performances
of the classifiers by only 5% (Average Cohen kappa
score = 0.9302).
In the interictal vs seizure-like (ictal) activity
discrimination task the landscape was different. As
before using feature extraction methods from the
time, frequency, time-frequency and chaotic domains
and the same ML methods we achieved an over 0.73
Cohen kappa score, an over 0.87 precision score, and
an over 0.86 recall score when the Full Feature Set
(4476 features) was used. As the feature space was
reduced (4476 to 27) the discriminability of the ML
classifiers changed. The classifier with the best
performance was once again the SVMrbf (Cohen
kappa score = 0.9565), whereas the classifier with the
worst performance was SVMlin (Cohen kappa score =
0.6773). The average Cohen kappa score was 0.88. Out
of the 27 most important features, the feature with the
highest importance score (IS > 0.19) was the mean
power spectrum density. When only the first 2 features
with the highest importance scores (A1-A2 in Fig. 8)
were used, then the discriminability of the classifiers
ranged from 0.14-0.65 (Average Cohen kappa score =
0.45). Addition of just two more features (2 to 4)
increased the performances of the classifiers by 13%
on average (Average Cohen kappa score = 0.58).
Addition of 5 more features (4 to 9) the inverse effect
to EA vs IA was seen: the average performance of the
classifiers increased by an additional 23% (Average
Cohen kappa score = 0.8103).
From these results it is evident that even though in
both discrimination tasks the first feature had a much
higher importance score than other features in the set
(see Figs 6 and 8), on each own it was not enough to
discriminate the pre-ictal (endogenous) from the
interictal, and the interictal from the ictal (seizure-
like) events. The performances of the classifiers on
average were poor (not shown here). Thus, the
discrimination ability of the classifiers depends on the
cumulative effect of the features, and not on the
individual effect of each feature. It is yet to be
determined whether this cumulative effect is additive
or multiplicative.
5 CONCLUSIONS
A novel algorithmic pipeline was successfully
applied to LFP recordings from layers II/III of the
primary somatosensory cortex of young mice to
discriminate with high accuracy the endogenous
(preictal), interictal and seizure-like (ictal) activity
events using time series analysis and ML modelling.
Over 4000 features were successfully extracted using
over 7700 operations applied to the LFPs. The high
dimensionality of the feature space was then reduced
via an iterative process of correlation analysis and
RF-RFECV to only 22 features for the EA vs IA
discrimination case and to 27 features for the IA vs
SLA one. ML algorithms were then applied to these
reduced feature sets and a radial basis function SVM
with a Gaussian kernel has been discovered to
discriminate with a 0.99 Cohen kappa score the EA
from IA and with a 0.9565 Cohen kappa the IA and
SLA. Our preliminary results show that ML
application in intracortical LFPs may be a promising
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
43

research avenue for accurate seizure detection and
prediction in focal epilepsy.
ACKNOWLEDGEMENTS
This work was supported by the European Union’s
Horizon 2020 Research and Innovation programme
under the Marie-Sklodowska Curie grant no778062
ULTRACEPT (VC) and the Human Resources and
Development, Education and Lifelong learning
programme no MIS-5049391 (IS).
REFERENCES
Avoli M., Jefferys J.G.R. (2016). Models of drug-induced
epileptiform synchronization in vitro. Journal of
Neuroscience Methods, 260: 26-32
Breiman L. (2001). Random forests. Mach Learn., 45:5–32.
Chang B.S., Lowenstein D.H. (2003). Epilepsy. The New
England Journal of Medicine, 349 (13): 1257–66
Chicco D., Warrens M.J., Jurman G. (2021). The Matthews
Correlation Coefficient (MCC) is More Informative
Than Cohen’s Kappa and Brier Score in Binary
Classification Assessment. IEEE Access, 9:78368-
78381. doi: 10.1109/ACCESS.2021.3084050.
Colmers P.L.W., Maguire J. (2020). Network Dysfunction in
Comorbid Psychiatric Illnesses and Epilepsy. Epilepsy
Curr., 20(4):205-210
Díaz-Uriarte R, Alvarez de Andrés S. (2006). Gene selection
and classification of microarray data using random forest.
BMC Bioinformatics, 7(1): 1-13.
Dreier J.P., Heinemann U. (1991). Regional and time
dependent variations of low Mg
2+
induced epileptiform
activity in rat temporal cortex slices. Experimental Brain
Research, 87(3): 581-596
Duncan J.S., Sander J.W., Sisodiya S.M., Walker M.C.
(2006). Adult epilepsy. Lancet, 367(9516): 1087–1100.
Fischer R. (2014). How can we identify ictal and interictal
abnormal activity? Adv Exp Med Biol., 813: 3–23
Fulcher B.D., Little M.A., Jones N.S. (2013). Highly
comparative time-series analysis: the empirical structure
of time series and their methods. J. Roy. Soc. Interf., 10:
20130048
Fulcher B.D., Jones N.S. (2017). hctsa: A Computational
Framework for Automated % Time-Series Phenotyping
Using Massive Feature Extraction. Cell Systems, 5: 527.
Ghosh S., Sinha J.K., Khan T., Devaraju K.S., Singh P.,
Vaibhav K., Gaur P. (2021). Pharmacological and
Therapeutic Approaches in the Treatment of Epilepsy.
Biomedicines, 9 (5): 470.
Goldberg E.M., Coulter D.A. (2013). Mechanisms of epile-
ptogenesis: a convergence on neural circuit dysfunction.
Nature Reviews Neuroscience 14 (5): 337–49.
Ho T.K. (1995). Random Decision Forests. In Proceedings
of the 3rd International Conference on Document
Analysis and Recognition, Montreal, QC, 14–16 August
1995. pp. 278–282.
Ho T.K. (1998). The Random Subspace Method for
Constructing Decision Forests. IEEE Transactions on
Pattern Analysis and Machine Intelligence, 20(8): 832–
844.
Kaplanian A, Vinos M, Skaliora I. (2022). GABAB - and
GABAA -receptor-mediated regulation of Up and Down
states across development. The Journal of Physiology,
600(10): 2401-2427.
Lodder S.S., Askamp J., van Putten M.J. (2014). Computer-
assisted interpretation of the EEG background pattern: a
clinical evaluation. PLoS ONE, 9(1): e85966.
Magiorkinis E., Sidiropoulou K., Diamantis A. (2010).
Hallmarks in the history of epilepsy: epilepsy in
antiquity. Epilepsy Behav., 17(1):103-8
Menze BH, Kelm BM, Masuch R, Himmelreich U, Bachert
P, Petrich W, Hamprecht FA. (2009). A comparison of
random forest and its Gini importance with standard
chemometric methods for the feature selection and
classification of spectral data. BMC bioinformatics,
10(1): 1-16.
Menze BH, Petrich W, Hamprecht FA. (2007). Multivariate
feature selection and hierarchical classification for
infrared spectroscopy: serum-based detection of bovine
spongiform encephalopathy. Anal Bioanalytical Chem,
387(5): 1801-1807.
Rigas P., Adamos D.A., Sigalas C., Tsakanikas P., Laskaris
N.A., Skaliora I. (2015). Spontaneous Up states in vitro:
A single-metric index of the functional maturation and
regional differentiation of the cerebral cortex. Frontiers
in Neural Circuits, 9: 59.
Rigas P., Sigalas C., Nikita M., Kaplanian A., Armaos K.,
Leontiadis L.J., Zlatanos C., Kapogiannatou A., Peta C.,
Katri A., Skaliora I. (2018) Long-Term Effects of Early
Life Seizures on Endogenous Local Network Activity of
the Mouse Neocortex. Front. Synaptic Neurosci., 10:43.
Sigalas C., Rigas P., Tsakanikas P., Skaliora I. (2015). High-
affinity nicotinic receptors modulate spontaneous
cortical up states in vitro. Journal of Neuroscience,
35(32): 11196-208.
Sigalas C., Konsolaki E., Skaliora I. (2017). Sex differences
in endogenous cortical network activity: spontaneously
recurring Up/Down states. Biol Sex Differ, 8:21.
Talkner P., Weber R.O. (2000). Power spectrum and
detrended fluctuation analysis: Application to daily
temperatures. Phys. Rev. E, 62(1): 150
Tsakanikas P., Sigalas C., Rigas P., Skaliora I. (2017). High-
Throughput Analysis of in-vitro LFP Electrophy-
siological Signals: A validated workflow/software
package. Scientific Reports, 7(1):3055
APPENDIX
All Matlab functions used to extract the features
described below are from the HCTSA time series
toolbox (Fulcher et al., 2013, 2017).
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
44

EA vs IA Classification
A1: How time-series properties change as 5% of time
points are removed. The time points being removed
are those that are the closest to the mean. The ratio of
autocorrelation (using lag = 2) of the transformed
time series over the original time series is the
extracted feature. The DN_RemovePoints.m function
was used to extract this feature.
A2: Fitting an AutoRegressive (AR) model to the
input time series. The range of the order of the fitted
model is [1, 8] and the optimum model order is being
chosen using Schwartz's Bayesian Criterion (SBC).
Eigendecomposition of the AR model is being
performed in order to compute the maximum of the
real part of eigenmodes. To extract this feature the
MF.arfir.m function was used with ‘pmin’, ‘pmax’,
and ‘selector’ (criterion to select optimal time series
model order) input arguments set to ‘1’, ‘8’, and
‘SBC’, respectively.
A3: Same as A1, but at the proportion of points
closest to the mean removed was set to 8%.
A4: Fits an AR model to 25 segments of length equal
to 10% of the input time series. The standard
deviation (std) of the optimal AR model order is the
extracted feature. The MF_FitSubsegments.m
function was used to extract this feature.
A5: Same as A4, but the extracted feature is the mean
of the optimal AR model order.
A6: AutoMutual information between the original
time-series and their respective delayed version
(delayed by 10 samples). The Gaussian estimation
method was used for the computation while the
maximum time delay to investigate equals to 20
samples. The IN_AutoMutualInfoStats.m function
was used to extract this feature.
A7: Interquartile range is defined as the spread of the
middle half of the distribution of the time-series. The
iqr.m function was used to extract this feature.
A8: The power spectrum of the input time-series is
being computed using the Welch’s method with
rectangular windows. A robust linear regression is
then performed using the logarithmic versions of the
frequencies and the acquired power spectrum. The
extracted feature is the gradient of the linear fit using
the SP_Summaries.m function.
A9: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 5 samples) is the extracted
feature. The Kraskov estimation method was used for
the computation while the maximum time delay was
20 samples. The IN_AutoMutualInfoStats.m function
was used to extract this feature.
A10: Coarse-grains the time series, turning it into a
sequence of symbols of a given alphabet of size
equals to 3. Quantifies measures of
surprise/information gain of a process with local
memory of the past memory values of the symbolic
string. Uses a memory of 50 samples and repeats over
500 random samples. The mean amount of
information over these 500 iterations is the extracted
feature A10. The FC_Surprise.m function was used to
extract this feature.
A11: An exponential function, f(x) = A*exp(bx), is
fitted to the variation across the first 10 successive
derivatives of the signal. The extracted feature is
parameter A of the above fitted exponential function.
The SY_StdNthDerChange.m was used to extract this
feature.
A12: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 5 samples) is this extracted
feature. Gaussian estimation method was used for the
computation while the maximum time delay was set
to 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
A13: Same as A2 the Eigendecomposition of the AR
model is being performed in order to compute the
maximum of the imaginary part of the eigenmodes.
A14: Implements fluctuation analysis using a
detrended RMS method (Talkner and Weber, 2000).
It first segments the input time-series into parts of log-
spaced lengths, then removes a polynomial trend of
order 3 in each segment. The average RMS over
different segment lengths is being computed along
with a linear fit between log-scales and log-RMS. The
mean squares residual of the fit is the extracted
feature. The SC_FluctAnal.m function is used to
extract this feature from the input time series.
A15: Input time-series is divided into 5 segments
with 50% overlap. The distribution entropy of each
segment is being computed using a kernel-smoothed
distribution. The mean of these entropies is the
extracted feature. The SY_SlidingWindow.m function
was used to extract this feature.
A16: measures the standard deviation of the first
derivative of the input time-series multiplied by a
constant value. The MD_rawHRVmeas.m function
was used to extract this feature.
A17: The AutoMutual information between the
original time-series and their respective delayed
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
45

version (delayed by 1 samples) is the extracted
feature. Gaussian estimation method was used for the
computation while the maximum time delay was 20
samples. The IN_AutoMutualInfoStats.m function
was used to extract this feature.
A18: Simulates a hypothetical walker moving
through the time domain. The walker moves as if it
has a mass and inertia from the previous time step and
the time series acts as a force altering its motion in a
classical Newtonian dynamics framework. The sum
of the absolute distances between the original time-
series and the hypothetical walker is the extracted
feature. The PH_Walker.m function was used to
extract this feature.
A19: The mean AutoMutual information over the
span of 1 to 20 delay times between the original time-
series and their respective delayed version is the
extracted feature. The Kraskov estimation method
was used for this calculation. The
IN_AutoMutualInfoStats.m function was used to
extract this feature.
A20: The power spectrum of the input time-series is
being computed, using Periodogram method with
hamming windows. The extracted feature is the
frequency at which the cumulative sum of the Power
Spectrum Density reaches 25% of the maximum
value. The SP_Summaries.m function was used to
extract this feature.
A21: Same as A10 but with alphabet size equal to 2.
A22: Couples the values of the time series to a
dynamical system. The input time series forces a
simulated particle in a quartic double-well potential.
The time series contributes to a forcing term on the
simulated particle. The autocorrelation of the position
of the particle is calculated and the first zero-crossing
of the autocorrelation function is the extracted feature.
The PH_ForcePotential.m function is used to extract
this feature.
IA vs SLA Classification
B1: The power spectrum of the input time-series is
being computed using the Welch’s method with
rectangular windows. The extracted feature is the
mean Power Spectrum Density across windows. The
SP_Summaries.m function was used to extract this
feature from the time series.
B2: Measures the standard deviation of the first
derivative of the input time-series multiplied by a
constant value. The MD_rawHRVmeas.m function
was used to extract this feature.
B3: First fitting an AR model to the input time series.
The range of the order of the fitted model is [1, 8] and
the optimum model order is being chosen using
Schwartz's Bayesian Criterion. Aikake's final
prediction error is computed. The minimum value
divided by the mean of the adjacent points is the
extracted feature. To extract this feature the
MF.arfir.m function was used.
B4: A hypothetical walker was simulated moving
through the time domain. The walker moved as if it
had a mass equaled to 5 a.u. and inertia from the
previous time step and the time series acted as a force
altering its motion in a classical Newtonian dynamics
framework. The autocorrelation of the residuals
between the walker and the actual time-series was the
extracted feature. The PH_Walker.m function was
used to extract this feature.
B5: Same as A13.
B6: Same as A6.
B7: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 6 samples) is the extracted
feature. The Gaussian estimation method was used
for the calculation, while the maximum time delay
was 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
B8: Simple local linear predictors using the past two
values of the time series to predict its next value. The
autocorrelation of the residuals between the actual
time-series and the predictions is the extracted feature.
The FC_LocalSimple.m function was used to extract
this feature.
B9: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 16 samples) was the extracted
feature. The Gaussian estimation method was used
for the calculation, while the maximum time delay
was 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
B10: How time-series properties change as 1% of
time points are removed. The time points being
saturated are those that are the furthest from the mean.
The ratio of autocorrelation (using lag = 1) of the
transformed time series over the original time series
is the extracted feature. The DN_RemovePoints.m
function was used to extract this feature.
B11: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 19 samples) is the extracted
feature. The Gaussian estimation method was used
BIOSIGNALS 2023 - 16th International Conference on Bio-inspired Systems and Signal Processing
46

for the calculation, while the maximum time delay
was 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
B12: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 11 samples) is the extracted
feature. The Gaussian estimation method was used
for the calculation, while the maximum time delay
was 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
B13: The minimum value of the input time-series.
B14: Calculates a normalized nonlinear
autocorrelation function. Then the time lag at which
the first minimum of the automutual information
occurred was calculated. The CO_trev.m function was
used to extract this feature.
B15: Embeds the (z-scored) time series in a two-
dimensional time-delay embedding space with time-
delay equals to 3 and estimates the autocorrelation
function. The first zero-crossing of the
autocorrelation function is the extracted feature. The
CD_Embed2.m function was used to extract this
feature.
B16: An exponential function, f(x) = A*exp(bx), is
fitted to the variation across the first 10 successive
derivatives. The parameter b is the extracted feature.
The SY_StdNthDerChange.m was used to extract this
feature.
B17: Generates 100 surrogate time series and tests
them against the original time series according to
some test statistics: T_{rev}, using TSTOOL code
trev. The standard deviation of the times of the first
minimum of the mutual information is the extracted
feature. The SD_TSTL_surrogates.m function was
used to extract this feature.
B18: The root mean squared error of predictions
using different local window lengths ranging from 1
to 9 samples. The SD_LoopLocalSimple.m function
was used to extract this feature.
B19: Calculates the autocorrelation of the residuals
between the prediction and the actual time-series
using different local window lengths ranging from 1
to 9 samples. The mean autocorrelation score across
different window lengths is the extracted feature. The
SD_LoopLocalSimple.m function was used to extract
this feature.
B20: Finds maximums and minimums within 50-
sample segments of the time series and analyses the
results. The standard deviation of the local minimums
is the extracted feature. The function
ST_LocalExtrema.m was used to extract this feature
from the time series.
B21: Finds maximums and minimums within 50
segments of the time series. The proportion of zero-
crossings of the local extrema is the extracted feature.
The function ST_LocalExtrema.m was used to extract
this feature from the time series.
B22: The root mean squared value of the input time-
series is the extracted feature. The function rms.m was
used to extract this feature.
B23: The AutoMutual information between the
original time-series and their respective delayed
version (delayed by 7 samples) is the extracted
feature. The Gaussian estimation method was used
for the calculation, while the maximum time delay
was 20 samples. The IN_AutoMutualInfoStats.m
function was used to extract this feature.
B24: Simulates a hypothetical walker moving
through the time domain. The walker moves as if it
has a mass equal to 2 a.u. and inertia from the
previous time step and the time series acts as a force
altering its motion in a classical Newtonian dynamics
framework. The autocorrelation of the residuals
between the walker and the actual time-series is the
extracted feature. The PH_Walker.m function was
used to extract this feature.
B25: How time-series properties change as 1% of
time points are removed. The time points being
saturated are those that are the furthest from the mean.
The difference between the autocorrelation (using lag
= 3) of the transformed time series and the
autocorrelation of the original time series is the
extracted feature. To extract this feature the
DN_RemovePoints.m function was used.
B26: Simulates a hypothetical walker moving
through the time domain. The walker moves as if it
has a mass equal to 2 a.u. and inertia from the
previous time step and the time series acts as a force
altering its motion in a classical Newtonian dynamics
framework. The autocorrelation of the walker divided
by the autocorrelation of the actual time-series is the
extracted feature. The PH_Walker.m function was
used to extract this feature.
B27: Fitting an AR model to the input time series. The
range of the order of the fitted model is [1, 8] and the
optimum model order is being chosen using
Schwartz's Bayesian Criterion. Then it computes the
margins of error A
err
such that (A ± A
err
) are
approximate 95% confidence intervals. The
minimum error margin is the extracted feature. To
extract this feature the MF.arfir.m function was used.
Machine Learning Algorithms for Mouse LFP Data Classification in Epilepsy
47
