
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic
Segmentation
Arthur B. A. Pinto
1 a
, Jefersson A. dos Santos
1,5 b
, Hugo Oliveira
2 c
and Alexei Machado
3,4 d
1
Department of Computer Science, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
2
Institute of Mathematics and Statistics, University of S
˜
ao Paulo, Brazil
3
Department of Anatomy and Imaging, Universidade Federal de Minas Gerais, Brazil
4
Department of Computer Science, Pontif
´
ıcia Universidade Catolica de Minas Gerais, Brazil
5
Computing Science and Mathematics, University of Stirling, Scotland, U.K.
Keywords:
Few-Shot, Domain Adaptation, Image Translation, Semantic Segmentation, Generative Adversarial Networks.
Abstract:
Due to ethical and legal concerns related to privacy, medical image datasets are often kept private, prevent-
ing invaluable annotations from being publicly available. However, data-driven models as machine learning
algorithms require large amounts of curated labeled data. This tension between ethical concerns regarding
privacy and performance is one of the core limitations to the development of artificial intelligence solutions in
medical imaging analysis. Aiming to mitigate this problem, we introduce a methodology based on few-shot
domain adaptation capable of leveraging organ segmentation annotations from private datasets to segment pre-
viously unseen data. This strategy uses unsupervised image-to-image translation to transfer annotations from
a confidential source dataset to a set of unseen public datasets. Experiments show that the proposed method
achieves equivalent or better performance when compared with approaches that have access to the target data.
The method’s effectiveness is evaluated in segmentation studies of the heart and lungs in X-ray datasets, often
reaching Jaccard values larger than 90% for novel unseen image sets.
1 INTRODUCTION
The Internet provides a virtually unlimited amount of
unlabeled, weakly-labeled, or even fully labeled im-
ages in the visible spectrum. Specific imaging do-
mains as medical data, however, deal with privacy
and ethical concerns during the creation of public
datasets, while also being harder and highly more ex-
pensive to annotate. As the literature of medical im-
age analysis migrates from shallow feature extraction
to deep feature learning, the main limitation to the
performance of machine learning models becomes the
lack of labeled data.
Deep Neural Networks (DNNs) for visual recog-
nition (Krizhevsky et al., 2012) require extensive and
representative datasets for training, that may be un-
available for most clinical scenarios. While the lack
of annotated data is an issue that can be alleviated
with techniques such as transfer learning and semi-
a
https://orcid.org/0000-0003-2057-9489
b
https://orcid.org/0000-0002-8889-1586
c
https://orcid.org/0000-0001-8760-9801
d
https://orcid.org/0000-0001-8077-3377
supervised learning, one aspect that makes this task
difficult is that most labeled datasets are private or
not fully publicly accessible. In order to protect the
patients’ privacy, hospitals decline to share medical
records to train machine learning models, even when
these are expected to help diagnosis counseling.
Domain Adaptation (DA) is traditionally handled
with the aid of supervised, semi-supervised, weakly-
supervised or even unsupervised methods (Zhang
et al., 2017) by leveraging source data/labels and tar-
get data. Unsupervised Domain Adaptation (UDA)
can be used to transfer representations between do-
mains or tasks without requiring any target labels,
while Semi-Supervised Domain Adaptation (SSDA)
considers the case of a few labeled samples on the
target set. However, as such DA methods demand si-
multaneous access to both source and target data, they
do not fit Few-Shot Domain Adaptation (Few-Shot
DA) cases, where the target-domain data for the task
of interest are unavailable. An example of Few-Shot
DA is the case of medical image datasets, where the
source or the target sets are often not publicly avail-
able due to privacy and ethical concerns. This limita-
Pinto, A., Santos, J., Oliveira, H. and Machado, A.
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation.
DOI: 10.5220/0011616800003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 4: VISAPP, pages
715-726
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
715

tion represents reproducibility hurdles, as annotations
from specialized physicians end up being used only
for local research, remaining inaccessible to other in-
stitution.
In this paper, we introduce a novel DA architec-
ture applicable in Few-Shot DA cases where the tar-
get domain data for the tasks of interest is unavailable.
The approach is based on the Conditional Domain
Adaptation Generative Adversarial Networks (CoDA-
GANs)(Oliveira et al., 2020) and the Few-Shot Un-
supervised Image-to-Image Translation (FUNIT)(Liu
et al., 2019) framework, specifically applied to the
context of biomedical image segmentation tasks.
For the current study, we claim the following con-
tributions:
1. We propose an innovative method that combines
Few-shot Image-to-Image translation with a seg-
mentation model to perform successful Few-shot
DA in biomedical image segmentation task;
2. A strategy with a more consistent training phase,
i.e., less instability from the Generative Adversar-
ial Networks (GANs);
3. A thorough test of our technique on a large collec-
tion of Chest x-ray (CXR) datasets utilizing vari-
ous source dataset combinations.
The method’s improved stability in the training
phase and its performance with unseen images are
demonstrated by extensive evaluation of a large col-
lection of Chest X-Ray (CXR) datasets using different
combinations of source datasets for two segmentation
tasks: lungs and heart.
2 BACKGROUND AND RELATED
WORK
2.1 Image-to-Image Translation
Image-to-Image (I2I) translation aims to learn the
mapping from a source image domain to a target im-
age domain. I2I often employs Generative Adversar-
ial Networks (GANs) (Goodfellow et al., 2014) that
are capable of transforming samples from one image
domain into images from another. These networks
use paired images to simplify the learning process
and loss functions, comparing the original and trans-
lated images at pixel or patch levels. Pix2Pix (Isola
et al., 2017) uses a GAN to create the mapping func-
tion according to a source image that serves as con-
ditioning to the model. On the other hand, BiCycle-
GANs (Zhu et al., 2017b) generate diverse outputs in
I2I problems, promoting the one-to-one relationship
between the network results and the latent vector by
modeling continuous and multi-modal distributions.
Although high-quality results have been shown both
in Pix2Pix and BiCycleGANs experiments(Zhu et al.,
2017b), the training procedure of these architectures
requires paired training data that reduces the appli-
cability of I2I translation to a small and limited sub-
set of image domains where there is the possibility of
generating paired datasets. This limitation motivated
the conception of Unpaired Image Translation meth-
ods such as CycleGAN (Zhu et al., 2017a), Unsuper-
vised Image-To-Image Translation (UNIT) (Liu et al.,
2017), and the Multimodal Unsupervised Image-To-
Image Translation (MUNIT) (Huang et al., 2018)
method that aim to learn a conditional image gen-
eration mapping function able to translate input im-
ages of a source domain to analog images of a target
domain without pairing supervision. These methods
leverage Cycle-Consistency to regularize the training
and to model the translation process between two im-
age domains as an invertible process.
2.2 Few-Shot Unsupervised Image
Translation
The FUNIT framework (Liu et al., 2019) proposes to
map an image of a source domain to a similar image
of an unseen target domain by leveraging only the few
target samples available at test time. During train-
ing, FUNIT uses images from a set of source datasets
(e.g. images of several animal species, or, closer to
our context, public medical imaging datasets) to train
a multi-source I2I translation model.
In the deploy phase, few images from a novel do-
main are presented to the model. The model leverages
the few target samples to translate any source sample
to analogous images of the target class. Then, when
the model is fed the few target images from a different
unseen class, it morphs source images to their analo-
gous target translation.
2.3 Domain Adaptation
A method often used in tasks such as classification,
detection, and segmentation is transfer learning via
fine tuning. This method adapts DNNs pre-trained
on larger source datasets to perform similar tasks
on smaller labeled target datasets. Although use-
ful, Fully Supervised Domain Adaptation (FSDA) ap-
proaches have the limitation of requiring at least small
quantities of labeled target datasets, while their unsu-
pervised counterpart (i.e. UDA) allows for zero su-
pervision on target domains.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
716

In recent years, modern alternatives to perform
UDA in neural networks have emerged such as the
ones based on Maximum Mean Discrepancy (MMD)
(Yan et al., 2017; Sun and Saenko, 2016; Tzeng et al.,
2017). Aiming to improve MMD by exploiting the
prior probability on the source and target domains,
Yan et al. (Yan et al., 2017) propose a weighted MMD
that includes domain-specific auxiliary weights into
MMD. Sun and Saenko (Sun and Saenko, 2016) dis-
cuss the case when the target domain is unlabeled and
extend the Correlation Alignment method to layer ac-
tivations in DNNs. Tzeng et al. (Tzeng et al., 2017)
combine discriminative modeling, untied weight shar-
ing, and an adversarial loss in a method called Adver-
sarial Discriminative Domain Adaptation (ADDA).
A vast number of works have used I2I Translation
for Domain Adaptation in order to perform segmen-
tation. Among these works, the Cycle-Consistent Ad-
versarial Domain Adaptation (CyCADA) (Hoffman
et al., 2018) accomplishes UDA by adding an FCN
to the end of a CycleGAN (Zhu et al., 2017a). Other
important works to be mentioned are the I2IAdapt
(Murez et al., 2018), that uses a CycleGAN (Zhu
et al., 2017a) coupled with segmentation architec-
tures to perform UDA; and the Dual Channel-wise
Alignment Network (DCAN) (Wu et al., 2018) that
attaches a segmentation architecture to the target end
of a translation architecture.
DA using Cycle-Consistency GANs have also
been applied to medical imaging, aiming to im-
prove cross-dataset generalization (Zhang et al., 2018;
Tang et al., 2019b; Tang et al., 2019a), transferring
knowledge between imaging modalities (Yang et al.,
2019) and even domain generalization (Oliveira et al.,
2020). However, all of these methods, except CoDA-
GANs (Oliveira et al., 2020), have the limitation of
not being multi-source/multi-target. In addition to
that, all of the previously mentioned GANs for med-
ical imaging DA need the source and target datasets
to be available during the training phase, which limits
their use to private target data.
2.4 CoDAGANs
CoDAGAN (Oliveira et al., 2020) is a framework
that combines I2I translation architectures (Liu et al.,
2017; Huang et al., 2018) with Encoder-Decoder seg-
mentation models (Ronneberger et al., 2015) to per-
form UDA, SSDA, or FSDA between various im-
age sets from the same imaging modality. The base
translation models of CoDAGANs rely on Autoen-
coders as generators, containing down-sampling and
up-sampling residual blocks. The intermediate rep-
resentations from the generator’s encoders are used
as basis for the isomorphic representation that serves
as input for the supervised segmentation module. By
employing supervision on an isomorphic space shared
across all datasets, CoDAGANs use the supervision
of the source datasets to perform inference across tar-
get data. Due to the nature of adversarial training, one
main disadvantage of CoDAGANs is the lack of sta-
bility in its DA performance. This limitation can be
mitigated by using historical averages, as discussed in
Section 3.
3 METHODOLOGY
We propose a new approach for Few-Shot DA in
cross-dataset semantic segmentation tasks applied to
medical imaging, henceforth referred to as CoDA-
Few. CoDA-Few is based on previous developments
in the UDA/SSDA translation (Oliveira et al., 2020)
and Few-Shot I2I (Liu et al., 2019), and is there-
fore an incremental improvement for CoDAGANS
(Oliveira et al., 2020). It uses the same proposition
of generating a mid-level isomorphic representation I
as CoDAGANs (Oliveira et al., 2020), with the dis-
tinction that a Few-Shot I2I translation network (Liu
et al., 2019) is used to compute I instead of the orig-
inal MUNIT/UNIT architectures (Huang et al., 2018;
Liu et al., 2017). During training, CoDA-Few uses the
Few-Shot I2I translation network to learn to generate
I from unseen datasets. Then, I is fed to a super-
vised model M based on I capable of inferring over
several datasets. At test time, we can use CoDA-Few
to infer over a dataset that was never seen in training.
The unsupervised translation process, followed by a
supervised learning model, can be seen in Figure 1.
This change effectively allows our Few-Shot DA net-
work to perform predictions on fully-unseen datasets,
while CoDAGANs can only infer over target distribu-
tions seen during training.
A few-shot segmentation task F is defined as a
task where the dataset has a small number of labeled
samples. In particular, we define F as a zero-shot task
when we have a source dataset S used in training, and
an unseen target dataset F used for testing. The chal-
lenge is to segment images from F using information
from S .
The proposed method allows the multi-
source/multi-target configuration on the Few-Shot
DA scenario involving two meta-datasets, i.e., the
source meta-dataset S = {S
1
,S
2
,... ,S
N
} with an
arbitrary number of labeled datasets N ≥ 2, and the
target dataset F = {F
1
,F
2
,. .. ,F
M
} with an arbitrary
number M of unlabeled unseen datasets. This allows
the proposed method to be trained with multiple
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation
717

source datasets and be applied in many target sets,
as CoDA-Few does not need the presence of a target
dataset in the training phase. For simplicity we will
refer to each target dataset F
i
individually as F .
For this work, FUNIT (Liu et al., 2019) was used
as a base to generate I . Similarly to CoDAGANs
(Oliveira et al., 2020), a supervised model M based
on a U-Net (Ronneberger et al., 2015) was attached
on top of that, with some considerable changes to
the translation approaches, regarding the architecture
and conditional distribution modeling of the origi-
nal GANs. Specifically, the first two layers of the
segmentation network were removed, resulting in an
asymmetrical U-Net to compensate for the loss of
spatial resolution introduced by the Encoder. Also,
the number of input channels in M was changed in
order to match the number of channels of I . As in
the case of MUNIT (Huang et al., 2018), FUNIT (Liu
et al., 2019) also separates the content of an image
from its style. The U-Net is only fed the content infor-
mation, as the style vector can be ignored since it has
no spatial resolution. In contrast to MUNIT/UNIT,
FUNIT (Liu et al., 2019) uses a progressive growth by
historical average with a weighted update, resulting in
a final generator G
µ
= {E
µ
,D
µ
} that is an epochal ver-
sion of the intermediate generators. With that, the sta-
bility in the training phase is considerably improved
for both translation and DA.
A training iteration on a CoDA-Few follows the
sequence shown in Figure 1. The generator network
G
µ
is an Encoder-Decoder translation architecture.
The encoding half (E
µ
) receives images from the dif-
ferent source domains S and generates an isomorphic
representation I within the image domains in a high
dimensional space. Decoders (D
µ
) are fed with I
and produce synthetic images from the same or dif-
ferent domains used in the learning process. Then,
a Discriminator D evaluates whether the fake im-
ages generated by G
µ
according to the style of the
target dataset are convincing samples to have been
drawn from the target distribution. At last, E
µ
is used
to generate the isomorphic representation I that are
forwarded to a supervised model M that learns how
to segment images. The aforementioned isomorphic
representation is an essential part of CoDA-Few, as
the whole supervised learning process is performed
using I . At each training iteration of CoDA-Few,
there are three routines for training the networks: (a)
Dis Update, when the generator is frozen and the dis-
criminator is updated; (b) Gen Update, when the dis-
criminator is frozen and the generator is updated; and
(c) Sup Update, when the supervised model is up-
dated. These routines will be further detailed in the
following paragraphs.
Generative Update. This routine is responsible for
the generator updates. First, a pair of source domains
a ∼ p
S
and b ∼ p
S
are randomly selected from the
N domains used in training. A batch X
a
of images
from S
a
is then appended to a code h
a
generated by a
one-hot encoding scheme, intending to inform the en-
coder E
µ
of the samples’ domain. The encoded batch
of samples X
a
is passed to the encoder E
µ
, producing
an intermediate isomorphic representation I for the
input X
a
according to the marginal distributions com-
puted by E
µ
for domain S
a
. Next, I is passed through
the decoder D
µ
and produces X
a→b
, a translation of
images in batch X
a
with the style of domain S
b
.
Discriminative Update. This routine is responsible
for the discriminator updates. At the end of the De-
coder D
µ
, the synthetic image X
a→b
is presented. The
original samples X
a
and the translated images X
a→b
are merged into a single batch and passed to the dis-
criminator D, which uses the adversarial loss compo-
nent to classify between real and fake samples. In
routines where the generators are being updated, the
adversarial loss is computed instead.
Supervised Update. This routine is responsible for
updating the supervised model M. For each sam-
ple X
(i)
∈ S
a
that has a corresponding label Y
(i)
a
, the
isomorphisms I
(i)
a
, I
(i)
a→b→a
are both fed to the same
supervised model M. Then the model M performs
the desired supervised task, generating the predictions
ˆ
Y
(i)
a
and
ˆ
Y
(i)
a→b→a
. These predictions can be compared
in a supervised way to Y
(i)
a
by employing L
S
if there
are labels for the image i in this batch. Since there are
always some labeled samples in this case, M is trained
to infer over isomorphic representations of both origi-
nal labeled data and translated data by the CoDA-Few
for the style of other datasets.
If domain shift is calculated and correctly ad-
justed during the training procedure, the properties
X
a
≈ X
a→b→a
and I
a
≈ I
a→b→a
are both achieved, sat-
isfying the Cycle-Consistency and Isomorphism, re-
spectively. Then, after training, we achieve a state
where I
a
≈ I
a→b→a
≈ I
T
. Now, it does not matter
which domain S or F is fed to E
µ
to generate the iso-
morphism I since samples from all datasets must be-
long to the same joint distribution in I -space. There-
fore, any learning performed in I
S
and I
a→b→a
is uni-
versal for all domains used in the training procedure
and for any future unseen domains.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
718

real
fake
......
(a) Training (b) Deployment
Figure 1: CoDA-Few architecture for visual DA. Training: A single Generative network G
µ
, divided into Encoder (E
µ
)
and Decoder (D
µ
) blocks, performs translations between the datasets. A Discriminator D evaluates whether the fake images
generated by G
µ
according to the style of the target dataset are convincing samples to have been drawn from the target
distribution. A single supervised model M is trained on the isomorphic representation I . Deployment: Images from the
target dataset (F ) are presented to the trained model, although the model has never seen a single sample from F during
training. E
µ
generates the isomorphic representation I , which is used by the supervised model M to segment the images.
3.1 CoDA-Few Loss
FUNIT jointly optimizes adversarial L
adv
, image re-
construction L
rec
, and feature matching L
f ea
loss
components. The content reconstruction loss (L
rec
)
helps G
µ
to learn a translation model in an unsuper-
vised fashion through cycle-consistency, mostly con-
tributing to the low-frequency components and se-
mantic consistency of the translation (Isola et al.,
2017). The adversarial component (L
adv
) encourages
the network to produce images with higher fidelity
and more accurate high-frequency components. The
feature matching loss (L
f ea
) helps regularizing the
training, handles the instability of GANs by specify-
ing a new objective for the generator that prevents it
from overfitting the current discriminator (Liu et al.,
2019). Instead of directly maximizing the output of
the discriminator, this new objective instructs the gen-
erator to yield data that matches the statistics of the
authentic samples. In this case, the discriminator
is used only to specify the statistics that are worth
matching (Salimans et al., 2016). A feature extractor
f
D
is created by removing the prediction layer from
the discriminator. Then, the features from the trans-
lation output and the target image are extracted using
f
D
and used to calculate the complete loss function of
FUNIT, L
F
:
L
F
= λ
adv
[L
adv
(X
b
,X
a→b
) +
L
adv
(X
a
,X
a→b→a
)] +
λ
f ea
[L
f ea
( f
D
(X
b
), f
D
(X
a→b
)) +
L
f ea
( f
D
(X
a
), f
D
(X
a→b→a
))] +
λ
rec
[L
rec
(X
a
,X
a→b→a
)]. (1)
More details about the FUNIT loss components can
be found in the original paper (Liu et al., 2019).
Aiming to tackle the unbalance from semantic
segmentation datasets, as a supervised loss compo-
nent L
sup
, CoDA-Few uses a combination of the
Cross-Entropy loss (L
CE
(Y, ˆy) = −Y log( ˆy) − (1 −
Y )log(1− ˆy)), and the Dice loss (L
DSC
(Y, ˆy) = (2Y ˆy+
1)/(Y + ˆy+ 1)), where Y represents the pixel-wise se-
mantic map and ˆy the probabilities for each class for a
given sample. Therefore, the supervised loss is given
as L
sup
= L
CE
(Y, ˆy)+ L
DSC
(Y, ˆy). The final loss L for
CoDA-Few is consequently defined as:
L = λ
adv
[L
adv
(X
b
,X
a→b
) +
L
adv
(X
a
,X
a→b→a
)] +
λ
f ea
[L
f ea
( f
D
(X
b
), f
D
(X
a→b
)) +
L
f ea
( f
D
(X
a
), f
D
(X
a→b→a
))] +
λ
rec
[L
rec
(X
a
,X
a→b→a
)] +
λ
sup
[L
sup
(Y
a
,M(I
a
)) +
L
sup
(Y
b
,M(I
b
)) +
L
sup
(Y
a
,M(I
a→b
)) +
L
sup
(Y
b
,M(I
b→a
))]. (2)
4 EXPERIMENTAL SETUP
The method was implemented using the PyTorch
framework and FUNIT repository (Liu et al., 2019).
All experiments were executed on an NVIDIA Titan
X Pascal GPU with 12GB of memory
1
.
CoDA-Few was trained for 10,000 iterations in the
experiments. This number of iterations was empiri-
cally found to be a good stopping point for conver-
gence (Oliveira et al., 2020). The learning rate was set
1
https://github.com/Arthur1511/CoDA-Few
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation
719

to 10
−4
with L2 normalization by weight decay with a
value of 10
−4
and the RMSProp solver. The values for
λ
adv
= 1, λ
rec
= 0.1, λ
f ea
= 1, and λ
sup
= 1 were also
empirically chosen based on exploratory experiments
and previous knowledge from CoDAGANs. Due to
GPU memory constraints, a batch size of 3 was used.
As in FUNIT, the final generator is a historical aver-
age version of the intermediate generators where the
update weight is 10
−3
(Karras et al., 2017).
The proposed method was applied to a total of 11
Chest X-Ray (CXR) datasets, including the Chest X-
Ray 8 (Wang et al., 2017), the Japanese Society of Ra-
diological Technology (JSRT) (Shiraishi et al., 2000),
the Montgomery and Shenzhen sets (Jaeger et al.,
2014), PadChest (Bustos et al., 2020), NLMCXR
(Demner-Fushman et al., 2016) and the OpenIST
2
datasets. A specialist manually labeled lungs and
heart for a random subset of 10 samples from the
Chest X-Ray 8, PadChest, Montgomery, and Shenzen
datasets, which were used for evaluation purposes.
Two sets of baselines were defined:
a) CoDA-Unfair: In this case, unlabeled tar-
get images were included in the training procedure.
We used the original CoDAGANs training procedure
where the unlabeled images of the target datasets were
used in the training procedure to perform unsuper-
vised domain adaptation between two or more image
datasets. This baseline was called CoDA-Unfair.
b) CoDA-Fair: In this setting, images of the
target datasets were not available during training.
As the original CoDAGAN method is not designed
for this setting, a baseline was created by extend-
ing the CoDAGAN framework based on MUNIT.
The CoDAGAN model was trained purely using the
source datasets. Through testing, we evaluate the
performance of the predictions in the target unseen
datasets. This baseline was called CoDA-Fair.
To properly compare CoDA-Few, CoDA-Fair,
and CoDA-Unfair, all datasets were randomly split
into the same training and test sets according to an
80%/20% division. Aiming to simulate real-world
scenarios wherein the absence of labels is a significant
problem, no samples were kept for validation pur-
poses. Results were evaluated from the last iteration
for computing the mean and standard deviation val-
ues to consider the statistical variability of the meth-
ods during the final iterations. Quantitative evaluation
was conducted according to the well-known Jaccard
score metric.
2
github.com/pi-null-mezon/OpenIST
5 RESULTS AND DISCUSSION
Two segmentation tasks were evaluated: CXR lungs
and heart segmentation. Source datasets included the
JSRT, OpenIST, Shenzhen, and Montgomery repos-
itories due to the presence of labels for these tasks
in these sets. Different combinations with three and
two datasets being used as source were tested. Since
Chest X-Ray 8, PadChest, and NLMCXR do not have
training labels, they were only used as target datasets.
Among the source datasets in the heart segmentation
task, only JSRT has training labels, so the remain-
ing source datasets were used to improve the gen-
eralization of the isomorphic representations. The
cross-sample average Jaccard and confidence inter-
vals with p ≤ 0.05 values for the lungs and heart seg-
mentation are shown in Figures 2 and 3. Tables 1, 2,
3, and 4 present jaccard results and standard devia-
tion, bold values represent the best overall results in a
given source dataset configuration for a specific target
dataset.
The proposed CoDA-Few framework outperforms
the baselines in most of the target datasets for both
lung and heart segmentation tasks. In the lung seg-
mentation task on CXRs, (a-d) in Figure 2 and (a-f)
in Figure 3, CoDA-Few presents better results for tar-
get datasets than CoDA-Unfair, even when only two
source datasets are employed to train CoDA-Few. In
the rare cases where the baselines outperform the pro-
posed method, CoDA-Few narrowly misses and, in
some circumstances, has a slightly smaller variation.
Heart segmentation proved to be a more difficult
task, with J values below 85%, as shown in (e-g) of
Figure 2 and (g-i) of Figure 3. One of the reasons that
caused the heart segmentation task to deliver worse
results when compared to the lung is the low contrast
that the heart has with the surrounding tissues, unlike
lungs that have well-defined boundaries. Once more
the proposed CoDA-Few framework outperforms the
baselines in most of the targets datasets, mainly when
three source datasets are used in the training phase,
implying that the method is able to learn from multi-
ple dataset source distributions. When the baselines
surpass the proposed method, they do it by a small
gap.
Figure 2f and 3h clearly shows that CoDA-
Few outperforms all baselines for heart segmenta-
tion when well-behaved datasets, such as JSRT and
OpenIST are used as source datasets and not well-
behaved datasets are used as targets datasets, such as
Padchest. One should notice that the target datasets,
in this case, are considerably harder than the source
ones due to poor image contrast, the presence of un-
foreseen artifacts such as pacemakers, rotation, and
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
720

M C P N
60
65
70
75
80
85
90
95
100
Sources: J, O, S
(a)
S C P N
60
65
70
75
80
85
90
95
100
Sources: J, O, M
(b)
O C P N
60
65
70
75
80
85
90
95
100
Sources: J, S, M
(c)
J C P N
60
65
70
75
80
85
90
95
100
Sources: O, S, M
(d)
M C P
60
65
70
75
80
85
90
95
100
Sources: J, O, S
(e)
S C P
60
65
70
75
80
85
90
95
100
Sources: J, O, M
(f)
O C P
60
65
70
75
80
85
90
95
100
Sources: J, S, M
(g)
Legend
CoDA-Few
CoDA-Unfair
CoDA-Fair
Figure 2: Jaccard results (in %) achieved for JSRT (J), OpenIST (O), Shenzhen (S), Montgomery (M), Chest X-Ray 8 (C),
PadChest (P), and NLMCXR (N) using 3 sources for the segmentation of lungs (a-d) and heart (e-g). CoDA-Few, Unfair and
Fair baselines are represented by blue, orange, and green bars, respectively.
Table 1: Jaccard results (in %) and standard deviation for lungs segmentation using 3 source datasets. Bold cells indicate the
best Jaccard values for each target dataset.
Source Target Coda-Few Coda-Unfair Coda-Fair
JSRT
OpenIST
Shenzhen
Montgomery
CXR8
Padchest
NLMCXR
93.47 ± 6.13
86.87 ± 1.98
86.48 ± 4.05
87.24 ± 2.06
89.56 ± 10.94
83.86 ± 3.43
83.74 ± 4.37
84.55 ± 3.47
89.83 ± 16.85
86.59 ± 1.64
86.72 ± 3.65
87.32 ± 1.90
JSRT
OpenIST
Montgomery
Shenzhen
CXR8
Padchest
NLMCXR
91.62 ± 5.37
85.58 ± 2.42
87.64 ± 2.24
86.67 ± 2.43
91.02± 5.43
85.31 ± 2.53
86.11 ± 3.25
82.82 ± 6.91
91.34 ± 6.13
85.46 ± 1.81
85.61 ± 4.41
86.24 ± 3.22
JSRT
Shenzhen
Montgomery
OpenIST
CXR8
Padchest
NLMCXR
92.54 ± 1.35
83.65 ± 4.27
85.58 ± 3.88
85.83 ± 2.22
91.04 ± 1.57
82.02 ± 5.27
82.94 ± 4.48
84.23 ± 2.80
92.35 ± 1.30
83.66 ± 2.49
83.87 ± 5.64
85.08 ± 2.36
OpenIST
Shenzhen
Montgomery
JSRT
CXR8
Padchest
NLMCXR
88.32 ± 2.48
84.27 ± 1.24
84.06 ± 4.22
84.61 ± 2.62
88.47 ± 1.68
84.00 ± 1.82
83.47 ± 5.40
85.54 ± 2.64
87.14 ± 3.27
84.46 ± 1.31
85.95 ± 2.93
85.14 ± 3.24
scale differences, and health conditions. Those fac-
tors, paired with the fact that the samples from the
JSRT dataset are the only source of labels for this
task evidencing CoDA-Few’s capability of generating
a better isomorphic representation of unseen datasets.
5.1 Qualitative Results
Figures 5 and 7 show qualitative results for lungs seg-
mentation in CXR. Examples of predictions wherein
CoDA-Few outperformed the baselines are depicted
in Figure 5 while Figure 7 shows erroneous pre-
dictions achieved by the baselines and the proposed
method. Columns in both figures present the original
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation
721
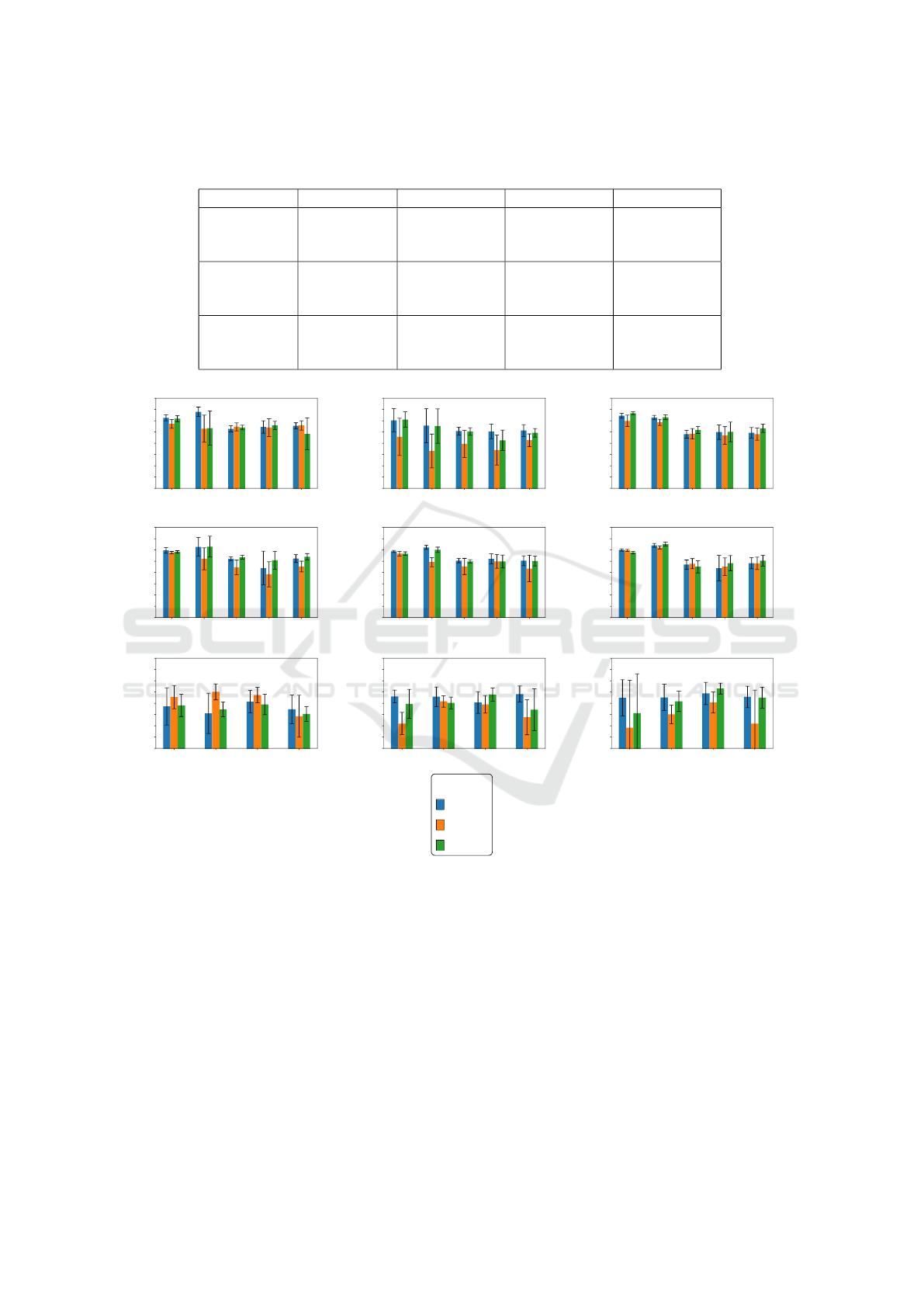
Table 2: Jaccard results (in %) and standard deviation for heart segmentation using 3 source datasets. Bold cells indicate the
best Jaccard values for each dataset.
Source Target CoDA-Few CoDA-Unfair CoDA-Fair
JSRT
OpenIST
Shenzhen
Montgomery
CXR8
Padchest
84.56 ± 4.34
85.646 ± 3.79
83.09 ± 5.33
83.82 ± 3.12
82.56 ± 6.62
76.57 ± 11.29
81.56 ± 3.11
81.59 ± 5.05
78.64 ± 7.44
JSRT
OpenIST
Montgomery
Shenzhen
CXR8
Padchest
82.53 ± 6.55
84.89 ± 5.93
85.86 ± 5.72
76.80 ± 3.73
75.23 ± 5.00
68.56 ± 10.07
78.50 ± 3.72
83.83 ± 2.73
72.48± 16.86
JSRT
Shenzhen
Montgomery
OpenIST
CXR8
Padchest
81.10 ± 12.93
85.65 ± 3.57
84.16 ± 5.23
72.37 ± 18.73
79.89 ± 4.67
75.06 ± 7.16
79.26 ± 12.08
86.20 ± 3.95
77.35 ± 7.70
S M C P N
60
65
70
75
80
85
90
95
100
Sources: J, O
(a)
O M C P N
60
65
70
75
80
85
90
95
100
Sources: J, S
(b)
O S C P N
60
65
70
75
80
85
90
95
100
Sources: J, M
(c)
J M C P N
60
65
70
75
80
85
90
95
100
Sources: O, S
(d)
J S C P N
60
65
70
75
80
85
90
95
100
Sources: O, M
(e)
J O C P N
60
65
70
75
80
85
90
95
100
Sources: S, M
(f)
O M C P
60
65
70
75
80
85
90
95
100
Sources: J, S
(g)
S M C P
60
65
70
75
80
85
90
95
100
Sources: J, O
(h)
O S C P
60
65
70
75
80
85
90
95
100
Sources: J, M
(i)
Legend
CoDA-Few
CoDA-Unfair
CoDA-Fair
Figure 3: Jaccard results (in %) achieved for JSRT (J), OpenIST (O), Shenzhen (S), Montgomery (M), Chest X-Ray 8 (C),
PadChest (P), and NLMCXR (N) using 2 sources for the segmentation of lungs (a-f) and heart (g-i). CoDA-Few, Unfair and
Fair baselines are represented by blue, orange, and green bars, respectively.
sample, the segmentation ground truth, and predic-
tions from CoDA-Few, CoDA-Unfair, and CoDA-Fair
for visual comparison. Each row presents an image
from each one of the target datasets.
Figure 5 shows DA results for lung field segmen-
tation using the JSRT, OpenIST, Shenzhen, and Mont-
gomery datasets both as source and target, and using
the Chest X-Ray 8, PadChest, and NLMCXR datasets
only as targets. The latter cases are considerably more
challenging than the others due to poor image con-
trast, the presence of unforeseen artifacts as pacemak-
ers, rotation and scale differences, as well as a much
wider variety of lung sizes, shapes, and health condi-
tions. However, the DA approach using CoDA-Few
for lung field segmentation was satisfactory for most
images, only showing errors on very challenging sam-
ples.
Figures 4 and 6 show qualitative results for
heart segmentation in CXR. Examples of predictions
wherein CoDA-Few outperformed the baselines are
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
722
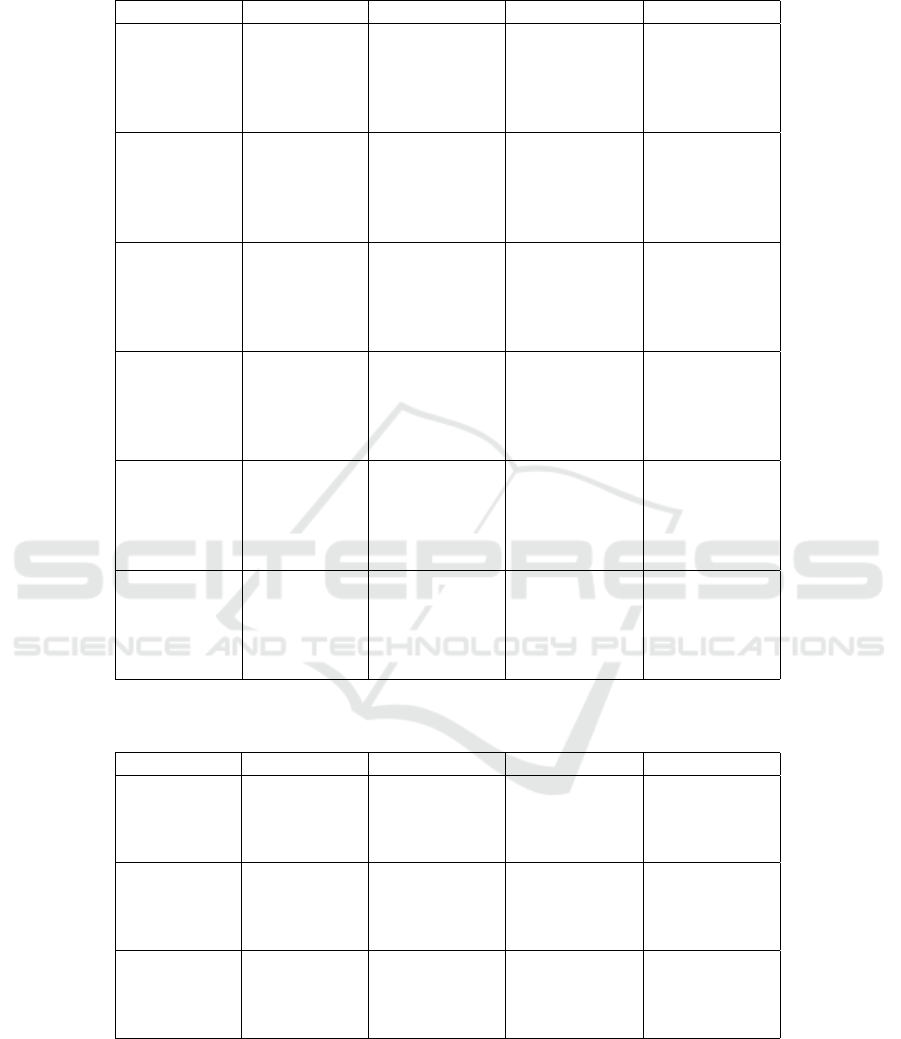
Table 3: Jaccard results (in %) and standard deviation for lungs segmentation using 2 source datasets. Bold cells indicate the
best Jaccard values for each dataset.
Source Target Coda-Few Coda-Unfair Coda-Fair
JSRT
OpenIST
Shenzhen
Montgomery
CXR8
Padchest
NLMCXR
91.27 ± 6.90
93.98 ± 5.27
86.39 ± 1.79
87.21 ± 3.65
87.71 ± 1.82
88.64 ± 10.60
86.43 ± 15.26
87.28 ± 2.30
86.98 ± 5.22
87.94 ± 2.75
90.91 ± 7.41
86.68 ± 19.19
86.98 ± 1.38
87.92 ± 2.36
84.15 ± 9.31
JSRT
Shenzhen
OpenIST
Montgomery
CXR8
Padchest
NLMCXR
90.20 ± 9.09
87.81 ± 19.06
85.38 ± 2.35
85.25 ± 4.26
85.65 ± 3.47
82.82 ± 14.41
76.58 ± 19.00
79.69 ± 7.95
76.95 ± 8.78
81.33 ± 3.76
90.52 ± 5.98
87.65 ± 19.33
85.22 ± 2.21
81.30 ± 5.98
84.64 ± 2.54
JSRT
Montgomery
OpenIST
Shenzhen
CXR8
Padchest
NLMCXR
92.24 ± 1.98
91.41 ± 5.38
84.02 ± 2.31
84.95 ± 4.32
84.65 ± 3.08
89.94 ± 4.40
89.31 ± 7.50
84.21 ± 3.12
83.42 ± 5.23
84.04 ± 3.53
93.34 ± 1.03
91.59 ± 5.98
85.87 ± 2.02
85.06 ± 5.77
86.61 ± 2.40
OpenIST
Shenzhen
JSRT
Montgomery
CXR8
Padchest
NLMCXR
89.84 ± 4.47
91.42 ± 10.64
86.09 ± 1.24
82.02 ± 9.92
86.18 ± 2.31
88.86 ± 1.72
86.11 ± 12.37
82.25 ± 4.28
79.19 ± 7.35
82.66 ± 3.15
89.12 ± 2.04
91.50 ± 11.75
86.75 ± 1.14
85.43 ± 5.26
86.96 ± 1.93
OpenIST
Montgomery
JSRT
Shenzhen
CXR8
Padchest
NLMCXR
89.36 ± 1.66
91.14 ± 4.81
85.26 ± 1.25
86.04 ± 2.94
85.23 ± 2.89
88.36 ± 3.66
84.62 ± 10.74
82.68 ± 4.75
84.91 ± 4.03
81.76 ± 7.82
88.39 ± 2.76
90.06 ± 6.39
84.93 ± 1.05
84.88 ± 3.87
85.13 ± 2.93
Shenzhen
Montgomery
JSRT
OpenIST
CXR8
Padchest
NLMCXR
90.05 ± 1.77
91.08 ± 1.46
83.52 ± 2.72
81.92 ± 7.51
84.16 ± 3.34
89.79 ± 1.64
91.09 ± 1.25
83.90 ± 3.00
82.58 ± 5.15
84.06 ± 3.65
88.90 ± 1.88
92.63 ± 1.38
82.55 ± 3.62
84.15 ± 4.41
85.28 ± 3.21
Table 4: Jaccard results (in %) and standard deviation for heart segmentation using 2 source datasets. Bold cells indicate the
best Jaccard values for each dataset.
Source Target CoDA-Few CoDA-Unfair CoDA-Fair
JSRT
OpenIST
Shenzhen
Montgomery
CXR8
Padchest
83.02 ± 3.50
82.92 ± 5.77
80.36 ± 6.42
84.11 ± 4.77
71.00 ± 6.12
80.84 ± 3.38
79.52 ± 5.18
73.76 ± 10.30
79.74 ± 7.93
80.16 ± 3.47
83.89 ± 3.89
77.19 ± 12.36
JSRT
Shenzhen
OpenIST
Montgomery
CXR8
Padchest
78.63 ± 10.96
75.56 ± 11.90
80.71 ± 6.65
77.32 ± 8.53
82.78 ± 6.87
85.08 ± 4.71
83.66 ± 4.60
74.21 ± 12.33
79.10 ± 6.62
77.27 ± 4.34
79.50 ± 6.06
75.24± 4.38
JSRT
Montgomery
OpenIST
Shenzhen
CXR8
Padchest
82.43 ± 10.63
82.61 ± 7.20
84.38 ± 6.69
82.82 ± 6.37
69.16 ± 27.98
75.10 ± 5.03
80.35 ± 6.20
70.94 ± 19.79
75.58 ± 23.27
80.86 ± 5.53
86.53 ± 3.21
82.48 ± 6.20
depicted in Figure 4 while Figure 6 shows erro-
neous predictions achieved by the baselines and the
proposed method. Columns in both figures present
the original sample, the segmentation ground truth,
and predictions from CoDA-Few, CoDA-Unfair, and
CoDA-Fair for visual comparison. Each row presents
an image from each one of the target datasets.
Figure 4 shows DA results for heart field segmen-
tation using the JSRT, OpenIST, Shenzhen, and Mont-
gomery datasets both as source and target, and us-
ing the Chest X-Ray 8 and PadChest datasets only as
targets. One should notice that the latter cases are
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation
723
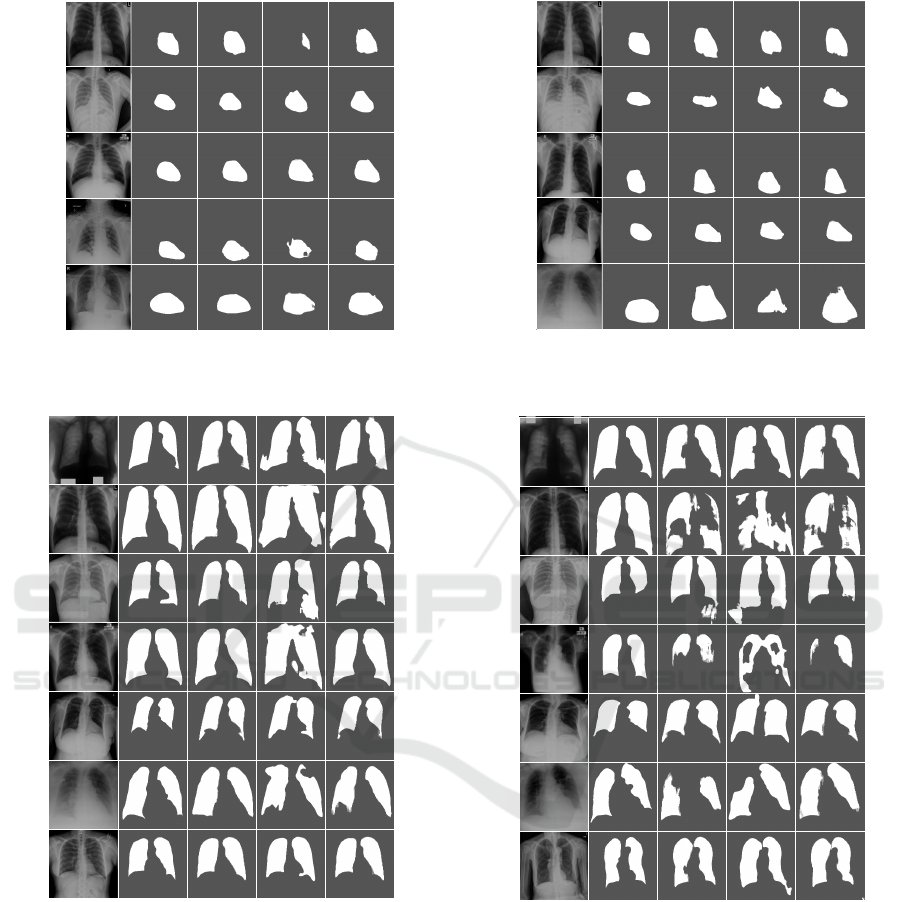
CoDA-FewImage Ground Truth
Chest-X-Ray 8
Padchest
CoDA-FairCoDA-Unfair
Montgomery Shenzhen
OpenIST
Figure 4: Qualitative heart segmentation results in CXR im-
ages for the unseen target datasets of heart segmentation.
CoDA-UnfairImage Ground Truth CoDA-Few CoDA-Fair
JSRTOpenISTShenzhenMontgomeryChest X-Ray 8PadChestNLMCXR
Figure 5: Qualitative lungs segmentation results in CXR
images for the unseen target datasets of lungs segmentation.
considerably more challenging than the others due to
poor image contrast, the presence of unforeseen arti-
facts as pacemakers, rotation and scale differences, as
well as a much wider variety of heart sizes, shapes,
and health conditions. However, the DA approach us-
ing CoDA-Few for heart field segmentation, yielded
consistent and satisfactory predictions maps across all
target datasets for most images, only showing errors
on very challenging samples from the dataset.
CoDA-Few
Chest-X-Ray 8
Padchest
CoDA-Fair CoDA-Unfair
Montgomery
Shenzhen
OpenIST
Image Ground Truth
Figure 6: Noticeable errors in CoDA-Few and baseline re-
sults for the unseen target datasets of heart segmentation.
.
JSRTOpenISTShenzhenMontgomeryChest X-Ray 8PadChestNLMCXR
CoDA-UnfairImage Ground Truth CoDA-Few CoDA-Fair
Figure 7: Noticeable errors in CoDA-Few and baseline re-
sults for the unseen target datasets of lungs segmentation.
6 CONCLUSION
This paper proposed and validated a method that per-
forms Few-Shot Domain Adaptation in dense label-
ing tasks for multiple sources and target biomedi-
cal datasets. Quantitative and qualitative experimen-
tal evaluation were performed on several distinct do-
mains, datasets, and segmentation tasks. We found
empirical evidence that CoDA-Few can segment im-
ages of an unseen target dataset made available at test
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
724

time based on the knowledge of seen source datasets.
CoDA-Few was shown to be a useful Domain
Adaptation method that could learn a single model
that performs satisfactory predictions for several dif-
ferent unseen target datasets in a domain, even when
the visual patterns of these data were different. The
proposed method was able to gather both labeled and
unlabeled data in the inference process, making it
highly adaptable to a wide variety of data scarcity sce-
narios.
CoDA-Few reached results in Few-Shot DA that
are comparable to DA methods that do have access to
the target data distribution. Furthermore, it presented
better Jaccard values in most experiments where la-
beled data was scarce, such as in heart segmentation
where only JSRT provided labeled training data. The
method also presented good performance in Few-Shot
DA tasks, even for highly imbalanced classes, such as
in the case of heart segmentation, wherein the region
of interest in images represented only a very small
slice of the number of pixels.
One should notice that CoDA-Few is conceptually
not limited to 2D dense labeling tasks or biomedical
images, despite being tested only for non-volumetric
segmentation tasks in this paper. Future works will
investigate Few-Shot DA in the segmentation of vol-
umetric images, such as Computed Tomography (CT)
scans, Positron Emission Tomography (PET scans),
and Magnetic Resonance Imaging (MRI). We also
plan to test CoDA-Few in other image domains, such
as traditional Computer Vision datasets and Remote
Sensing data.
ACKNOWLEDGEMENTS
The authors would like to thank CAPES, CNPq
(424700/2018-2 and 306955/2021-0), FAPEMIG
(APQ-00449-17 and APQ-00519-20), FAPESP (grant
#2020/06744-5), and Serrapilheira Institute (grant
#R-2011-37776) for their financial support to this re-
search project.
REFERENCES
Bustos, A., Pertusa, A., Salinas, J.-M., and de la Iglesia-
Vay
´
a, M. (2020). Padchest: A large chest x-ray image
dataset with multi-label annotated reports. Medical
Image Analysis, 66:101797.
Demner-Fushman, D., Kohli, M. D., Rosenman, M. B.,
Shooshan, S. E., Rodriguez, L., Antani, S., Thoma,
G. R., and McDonald, C. J. (2016). Preparing a col-
lection of radiology examinations for distribution and
retrieval. Journal of the American Medical Informat-
ics Association, 23(2):304–310.
Goodfellow, I. J., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A. C., and Ben-
gio, Y. (2014). Generative adversarial nets. In NIPS.
Hoffman, J., Tzeng, E., Park, T., Zhu, J.-Y., Isola, P.,
Saenko, K., Efros, A., and Darrell, T. (2018). Cy-
cada: Cycle-consistent adversarial domain adaptation.
In ICML, pages 1989–1998. PMLR.
Huang, X., Liu, M.-Y., Belongie, S., and Kautz, J. (2018).
Multimodal unsupervised image-to-image translation.
In Proceedings of the European conference on com-
puter vision (ECCV), pages 172–189.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017).
Image-to-image translation with conditional adversar-
ial networks. In Proceedings of the IEEE conference
on computer vision and pattern recognition, pages
1125–1134.
Jaeger, S., Candemir, S., Antani, S., W
´
ang, Y.-X. J., Lu,
P.-X., and Thoma, G. (2014). Two public chest
x-ray datasets for computer-aided screening of pul-
monary diseases. Quantitative Imaging in Medicine
and Surgery, 4(6):475.
Karras, T., Aila, T., Laine, S., and Lehtinen, J. (2017). Pro-
gressive growing of gans for improved quality, stabil-
ity, and variation. arXiv preprint arXiv:1710.10196.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
ageNet classification with deep convolutional neural
networks. NIPS, 25:1097–1105.
Liu, M.-Y., Breuel, T., and Kautz, J. (2017). Unsupervised
image-to-image translation networks. In Proceedings
of the 31st International Conference on Neural Infor-
mation Processing Systems, pages 700–708.
Liu, M.-Y., Huang, X., Mallya, A., Karras, T., Aila, T.,
Lehtinen, J., and Kautz, J. (2019). Few-shot unsu-
pervised image-to-image translation. In Proceedings
of the IEEE/CVF International Conference on Com-
puter Vision, pages 10551–10560.
Murez, Z., Kolouri, S., Kriegman, D., Ramamoorthi, R.,
and Kim, K. (2018). Image to image translation for
domain adaptation. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition,
pages 4500–4509.
Oliveira, H. N., Ferreira, E., and Dos Santos, J. A.
(2020). Truly generalizable radiograph segmentation
with conditional domain adaptation. IEEE Access,
8:84037–84062.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. In International Conference on Medical
image computing and computer-assisted intervention,
pages 234–241. Springer.
Salimans, T., Goodfellow, I., Zaremba, W., Cheung, V.,
Radford, A., and Chen, X. (2016). Improved tech-
niques for training gans. Advances in neural informa-
tion processing systems, 29:2234–2242.
Shiraishi, J., Katsuragawa, S., Ikezoe, J., Matsumoto, T.,
Kobayashi, T., Komatsu, K.-i., Matsui, M., Fujita,
H., Kodera, Y., and Doi, K. (2000). Development
of a digital image database for chest radiographs with
CoDA-Few: Few Shot Domain Adaptation for Medical Image Semantic Segmentation
725

and without a lung nodule: receiver operating char-
acteristic analysis of radiologists’ detection of pul-
monary nodules. American Journal of Roentgenology,
174(1):71–74.
Sun, B. and Saenko, K. (2016). Deep coral: Correla-
tion alignment for deep domain adaptation. In Euro-
pean conference on computer vision, pages 443–450.
Springer.
Tang, Y., Tang, Y., Sandfort, V., Xiao, J., and Summers,
R. M. (2019a). Tuna-net: Task-oriented unsupervised
adversarial network for disease recognition in cross-
domain chest x-rays. In International Conference on
Medical Image Computing and Computer-Assisted In-
tervention, pages 431–440. Springer.
Tang, Y.-B., Tang, Y.-X., Xiao, J., and Summers, R. M.
(2019b). Xlsor: A robust and accurate lung segmen-
tor on chest x-rays using criss-cross attention and cus-
tomized radiorealistic abnormalities generation. In
International Conference on Medical Imaging with
Deep Learning, pages 457–467. PMLR.
Tzeng, E., Hoffman, J., Saenko, K., and Darrell, T. (2017).
Adversarial discriminative domain adaptation. In Pro-
ceedings of the IEEE conference on computer vision
and pattern recognition, pages 7167–7176.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Sum-
mers, R. (2017). Hospital-scale chest x-ray database
and benchmarks on weakly-supervised classification
and localization of common thorax diseases. In CVPR,
pages 3462–3471.
Wu, Z., Han, X., Lin, Y.-L., Uzunbas, M. G., Goldstein,
T., Lim, S. N., and Davis, L. S. (2018). Dcan:
Dual channel-wise alignment networks for unsuper-
vised scene adaptation. In Proceedings of the Euro-
pean Conference on Computer Vision (ECCV), pages
518–534.
Yan, H., Ding, Y., Li, P., Wang, Q., Xu, Y., and Zuo,
W. (2017). Mind the class weight bias: Weighted
maximum mean discrepancy for unsupervised domain
adaptation. In CVPR, pages 2272–2281.
Yang, J., Dvornek, N. C., Zhang, F., Chapiro, J., Lin,
M., and Duncan, J. S. (2019). Unsupervised domain
adaptation via disentangled representations: Appli-
cation to cross-modality liver segmentation. In In-
ternational Conference on Medical Image Computing
and Computer-Assisted Intervention, pages 255–263.
Springer.
Zhang, J., Li, W., and Ogunbona, P. (2017). Transfer learn-
ing for cross-dataset recognition: a survey. arXiv
preprint arXiv:1705.04396.
Zhang, Y., Miao, S., Mansi, T., and Liao, R. (2018). Task
driven generative modeling for unsupervised domain
adaptation: Application to x-ray image segmenta-
tion. In International Conference on Medical Im-
age Computing and Computer-Assisted Intervention,
pages 599–607. Springer.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017a).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Proceedings of
the IEEE international conference on computer vi-
sion, pages 2223–2232.
Zhu, J.-Y., Zhang, R., Pathak, D., Darrell, T., Efros, A. A.,
Wang, O., and Shechtman, E. (2017b). Multimodal
image-to-image translation by enforcing bi-cycle con-
sistency. In Advances in neural information process-
ing systems, pages 465–476.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
726
