
MRQPMS: Design of a Map Reduce Bioinspired Model for Solving
Quorum Planted Motif Search for High-Speed Deployments
Aditi R. Durge and Deepti D. Shrimankar
Visvesvaraya National Institute of Technology Nagpur, Maharashtra, India
Keywords: Motif, Quorum, GA, Bioinspired, Map, Reduce, Hadoop.
Abstract: Quorum Planted Motif Search (qPMS) is a specialized field of PMS which provides matching outputs only
if the search motif appears in q% of the results. Designing qPMS models is a multidomain task, that
involves collection of application-specific datasets, pre-processing of these datasets for identification of
frequent patters, matching of these patterns, and contextual post-processing operations. Due to large-length
sequences, the search process is highly complex, and requires dataset-specific optimizations. To perform
these optimizations, a wide variety of tools are developed by researchers and each of them vary in terms of
their qualitative & quantitative characteristics. Most of these models are non-reconfigurable, and can be
used only for specific datasets, while others present highly complex search mechanisms, which limits their
applicability. To overcome these limitations, this text proposes design of a Map Reduce Model for solving
Quorum Planted Motif Search for high-speed deployments. The proposed model initially stores input
genomic sequences via a Map Reduce framework, which assists in faster search via use of unique entity-
level keys for different sequence types. These keys are stored via the Apache Hadoop framework, which
assists in improving search performance under large dataset scenarios. Due to use of Map Reduce, the
model is capable of higher scalability, better flexibility, low delay, and security via parallel processing
operations. This was possible due to pre-processing of input DNA sequences and reducing them into index-
based searchable formats. The model also deploys a Genetic Algorithm (GA) for identification of optimum
Q values for enhanced accuracy under different use cases. It was tested for protein & DNA sequences, and
its performance was evaluated in terms of accuracy, retrieval delay, precision, & throughput parameters, and
compared with various state-of-the-art models under different use case scenarios. Based on this comparison,
it was observed that the proposed model was capable achieving 3.5% higher accuracy, 9.4% lower delay,
2.9% higher precision, and 8.5% higher throughput under different scenarios. Due to these advantages the
proposed model is capable of deployment for a wide variety of real-time use cases.
1
INTRODUCTION
Design of Quorum based Planted Motif Search
(QPMS) requires researchers to integrate multiple
data representation & search models for improving
performance under different use cases. These use
cases include, genomic searches, DNA
representational searches, protein sequence analysis,
etc. A typical PMS Model (Semwal et al.,2022) that
uses 𝑙,𝑑-mers for feature extraction is depicted in
figure 1, wherein Firefly Algorithm is used with
Freeze Techniques for optimization of motif search
process. The model initially converts DNA
sequences into static search sequences based on
common pattern analysis. These sequences marked
as ‘local freeze’ sequences, and are used for the
search process. To perform this search process, 𝑙
gram matching is used, where 𝑑 sequences are fused
together to form final ‘global freeze’ sequence sets.
These sets are presented at the output, and are used
for representation of search results under different
use cases.
To perform this search, a score is evaluated via
equation 1,
𝑆=
𝐽
𝑎𝑐𝑐𝑎𝑟𝑑(𝑄
(
𝑖,
𝑗
)
,𝑆
(
𝑖,
𝑗
)
)
(1)
Where, 𝑄 & 𝑆 represents input query, and the
sequence to be used for matching purposes. This
model uses Jaccard similarity index, which is
optimized via use of Firefly optimizations, thereby
increasing overall complexity of such deployments.
Durge, A. and Shrimankar, D.
MRQPMS: Design of a Map Reduce Bioinspired Model for Solving Quorum Planted Motif Search for High-Speed Deployments.
DOI: 10.5220/0011616500003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 123-130
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
123
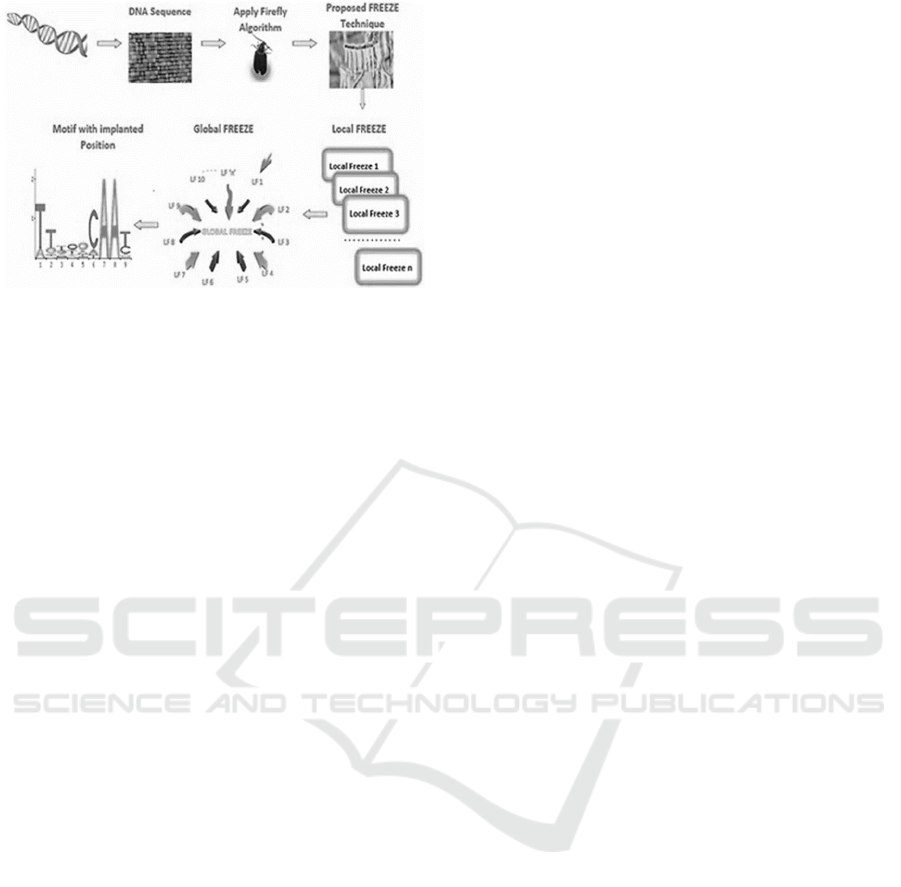
Figure 1: Design of a Firefly based Model for QPMS
applications.
To reduce this complexity, next section discusses
design of various PMS Models (Xiao et al, 2021; Yu
et al, 2019; Li et al, 2021) and evaluates them in term
of their application-specific nuances, contextual
advantages, functional limitations, and deployment
based future research scopes. Based on this
discussion, it was observed that most of these models
are non-reconfigurable, and can be used only for
specific datasets, while others present highly complex
search mechanisms, which limits their applicability.
To overcome these limitations, section 3 proposes
design of a Map Reduce Model for solving Quorum
Planted Motif Search for high-speed deployments.
The proposed model was evaluated in terms of
accuracy, precision, delay & throughput parameters,
and compared with various state-of-the-art models,
which assists in validating its performance under real-
time use cases. Finally, this text concludes with some
context-specific & performance-specific observations
about the proposed model, and recommends methods
to further improve its performance levels for different
application scenarios.
2
LITERATURE REVIEW
A wide variety of models are available for Motif
Search, and each of them vary in terms of their
internal performance & use case types. For instance,
work in (Zhang et al, 2018) (Zhao et al,2021)
discusses Discriminative Motif Learning Algorithm
(DMLA), and Motif Based PageRank (MBP), which
assists in integrating multiple datasets for searching
different Motif types. But the model is highly context
specific, this cannot be scaled for real time scenarios.
To overcome this limitation, work in (Sun et al, 2019)
proposes use of Time First Search (TFS), which
reduces number of Motifs to be searched per query,
thereby improving search speed, and scalability under
multiple use cases. Similar models are discussed in
(Xing et al,2020) (Yu et al,2021) (Chaudhry et
al,2018) which propose use of Graph Neural
Network, Artificial Generation of Searching
Conditions, and Monte Carlo Tree Search which
assists in improving its search performance via high
density feature representations. Extensions to these
models are discussed in (Nicolae et al,2015) (Yu et
al,2019) (Shrimankar, 2019), which propose use of
DNA (ℓ, d), Approximate qPMS (AqPMS), and DNA
Regulatory Networks, which assists in enhancing
search speed under multiple scenarios. These models
are highly superior when applied to large-scale
datasets, and thus can be used under different
scenarios.
Models that use Edit-distance based Motif Search
(EMS) (Xiao P. et al,2021), Intrinsically Disordered
Proteins (IDPs) (Schultz et al, 2022), Stochastic
Search Models (Merlin et al, 2013), Motif Stem
Search (MSS) (Yu et al, 2015), and Simple Motif
Search (SMS) (Pathak et al, 2013), aim at reducing
complexity of search under different use cases. These
models are used when large search sequences are to
be parsed, and their performance is to be evaluated
under multiple use cases. Extensions to these models
are discussed in (Reddy et al, 2010) (Kashiwabara et
al,2018), which propose use of Particle Swarm
Optimization (PSO), and Memetic Algorithm (MA),
that introduce bioinspired computing models for
continuous parametric tuning under different use
cases. But these models vary widely in terms of their
qualitative & quantitative characteristics and most of
them are non-reconfigurable, thus, can be used only
for specific datasets, while others present highly
complex search mechanisms, which limits their appli-
cability. To overcome these limitations, next section
proposes design of a Map Reduce Model for solving
Quorum Planted Motif Search for high-speed deploy-
ments. The proposed model was evaluated in terms of
accuracy, precision, recall, and delay metrics under
multiple datasets, which will assist in validating its
real-time performance levels for different use cases.
3
DESIGN OF A MAP REDUCE
BIOINSPIRED MODEL FOR
SOLVING QUORUM PLANTED
MOTIF SEARCH FOR
HIGH-SPEED DEPLOYMENTS
Based on the literature review, it can be observed
that existing models vary in terms of their qualitative
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
124
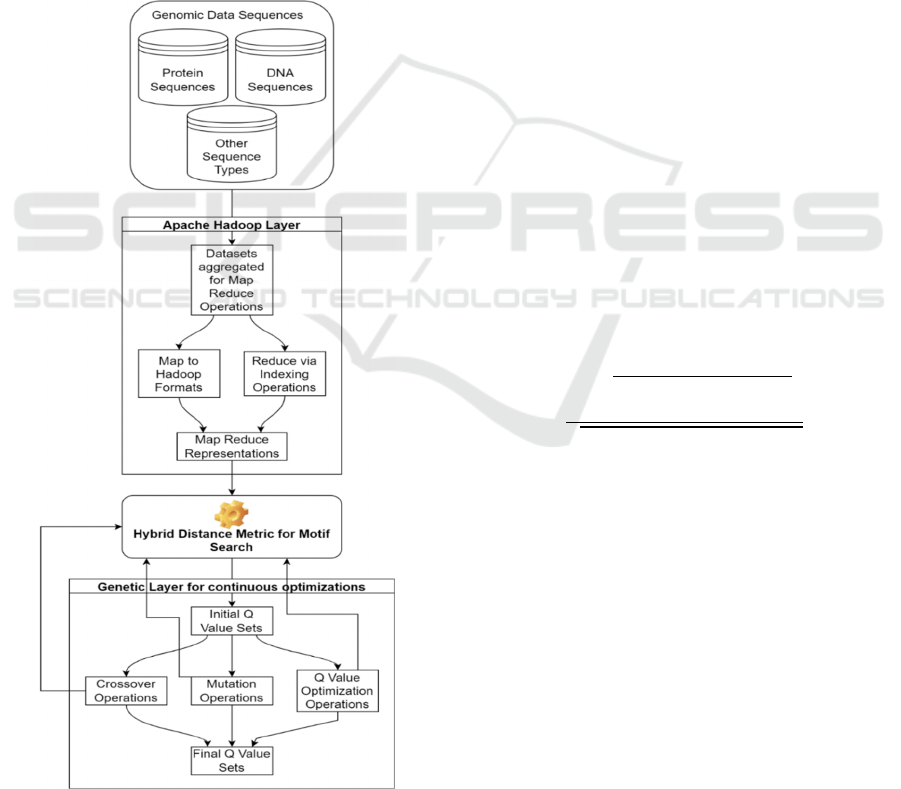
& quantitative characteristics. There are a number of
models that can only be used for specific datasets,
while others have complex search mechanisms that
limit their applicability. A Map Reduce Model for
high-speed deployments of Quorum Planted Motif
Search (QPMS) is proposed in this section to
overcome these limitations. Figure 2 shows the
model's flow, which uses a Map Reduce framework
to store input genomic sequences and then uses
unique entity-level keys for different sequence types
to speed up search. The Apache Hadoop framework
then stores these keys, which helps to improve
search performance when dealing with large
datasets. The model's parallel processing operations
enable it to achieve greater scalability, better
flexibility, lower latency, and increased security
thanks to the use of Map Reduce. Input DNA
Figure 2: Overall flow of the proposed model for
optimized QPMS.
sequences were pre-processed before being reduced
to index-based searchable formats, allowing for this
to be accomplished Additionally, the model uses a
Genetic Algorithm (GA) to find the best Q values
for different use cases.
Collecting massive datasets that contain a wide
variety of protein sequences, DNA sequences, and
other genomic sets is the first step in the process of
building the search corpus for the model. This step is
part of the process of building the search engine.
This corpus is stored via the Apache Hadoop in
a Map Reduce format, which works via the
following process,
• Assign each symbol a unique numeric value via
equation 2,
𝑁
=𝑖,𝑤ℎ𝑒𝑟𝑒𝑖∈1,𝑁
(2)
W
here, 𝑁
represents numerical value for each of
the unique 𝑁
• Repeat this process for bigram, trigram, and N
gram sequences
• Store all sequence sets into the Map Reduce
databases in the format depicted in reduce
representation index of Map Reduce.
This structure is internally used by the Hadoop
Framework to find matching between query input
sequences and stored sequence sets. This matching
is performed via a combination of Jaccard & Cosine
Distance Metrics (DM), which is evaluated via
equation 3, which combines these metrics in order to
find top Q search sequences,
𝐷𝑀 =
𝐶
(
𝑄𝑢𝑒𝑟𝑦
)
⋂
𝐶
(
𝐼𝑛𝑝𝑢𝑡
)
𝐶
(
𝑄𝑢𝑒𝑟𝑦
)
⋃
𝐶
(
𝐼𝑛𝑝𝑢𝑡
)
∑
𝐶
(
𝑄𝑢𝑒𝑟𝑦
)
∗𝐶
(𝐼𝑛𝑝𝑢𝑡)
∑
𝐶
(
𝑄𝑢𝑒𝑟𝑦
)
∗
∑
𝐶
(
𝐼𝑛𝑝𝑢𝑡
)
(3)
Where, 𝐶
(
𝑄𝑢𝑒𝑟𝑦
)
𝐶
(𝐼𝑛𝑝𝑢𝑡) represents Map
Reduced values for query & input sequences, which
are evaluated via equation 1, and assist in improving
overall search performance under different sequence
types. The model selects a Q value via Genetic
Algorithm (GA), that works as follows,
• Initialize following optimization parameters of
the model,
o Total selected iterations for optimization (𝑁
)
o Total selected solutions for optimization (𝑁
)
o Rate at which the model learns via cognitive
process (𝐿
)
o Maximum value of 𝑄 required for the
optimization process (𝑀𝑎𝑥𝑄)
• To start the optimization process, setup all
solutions to be ‘mutate’
MRQPMS: Design of a Map Reduce Bioinspired Model for Solving Quorum Planted Motif Search for High-Speed Deployments
125
• Scan all solutions for each of the iteration via
the following process,
o If current solution is setup as ‘to be
crossover’, then skip it and go to next solution
in sequence
o For ‘mutated’ sequences, generate Q Value
via equation 4,
𝑄 = 𝑆𝑇𝑂𝐶𝐻
(
𝐿
∗𝑀𝑎𝑥𝑄,𝑀𝑎𝑥𝑄
)
(4)
Where, 𝑆𝑇𝑂𝐶𝐻 represents a stochastic Markovian
process used for generation of numbers between
range sets.
o Based on this Q Value, identify top Q
matching sequences, and evaluate solution
fitness via equation 5,
𝑓
=
1
𝑄
∗
𝑁𝐶
𝑁𝑇
(5)
Where, 𝑁𝐶 & 𝑁𝑇 represents Number of Correctly
identified sequences, & Total Number of sequences
extracted during the identification process.
o Evaluate the fitness value for all solutions, and
then calculate iteration fitness via equation 6,
𝑓
=
1
𝑁
𝑓
∗𝐿
(6)
o Scan each solution, and mark it as ‘mutate’ if
𝑓
≤𝑓
, else mark it as ‘crossover’, and go
the next iterations
• At the end of final iteration, identify Q Value
with maximum fitness levels, and use it for the
process
Based on this process, values of Q are evaluated,
which assists in improving classification accuracy
for different dataset types. The accuracy levels along
with precision, recall & delay needed for search is
compared with various state-of-the-art models under
different sequence types, and is evaluated in the next
section of this text.
4
RESULT ANALYSIS &
COMPARISON WITH
STANDARD METHODS
Due to integration of GA with QMS and Map
Reduce operations, the proposed model was
observed to perform faster and showcase higher
accuracy when compared with other models under
different scenarios. This performance estimation was
done for the following datasets,
• David Reich Lab Dataset, which is available at
https://reich.hms.harvard.edu/datasets
• Structural Protein Sequences, available at
https://www.kaggle.com/datasets/shahir/protein-
data-set
• Plant Genomic Dataset, which is available at
https://www.plantgdb.org/
When combined together, these datasets have n
aggregated 200k DNA sequences, with unequal
lengths, which makes them a perfect candidate for
QPMS operations. The combination of datasets was
done as follows,
• Sequences were integrated to form a combined
sequence set via aggregation operations
• Classes of these sequences were arranged
sequentially to obtain final search sets
The full dataset was used for searching different
Motifs, that varied in lengths from 20 characters to
50 characters. These sequences were searched for
different Test Set Sizes (TSS), and their accuracy
performance was compared with DMLA (Zhang et
al, 2018), AQ PMS (Yu et al, 2019) and PSO
(Reddy et al,2010) which can be observed from table
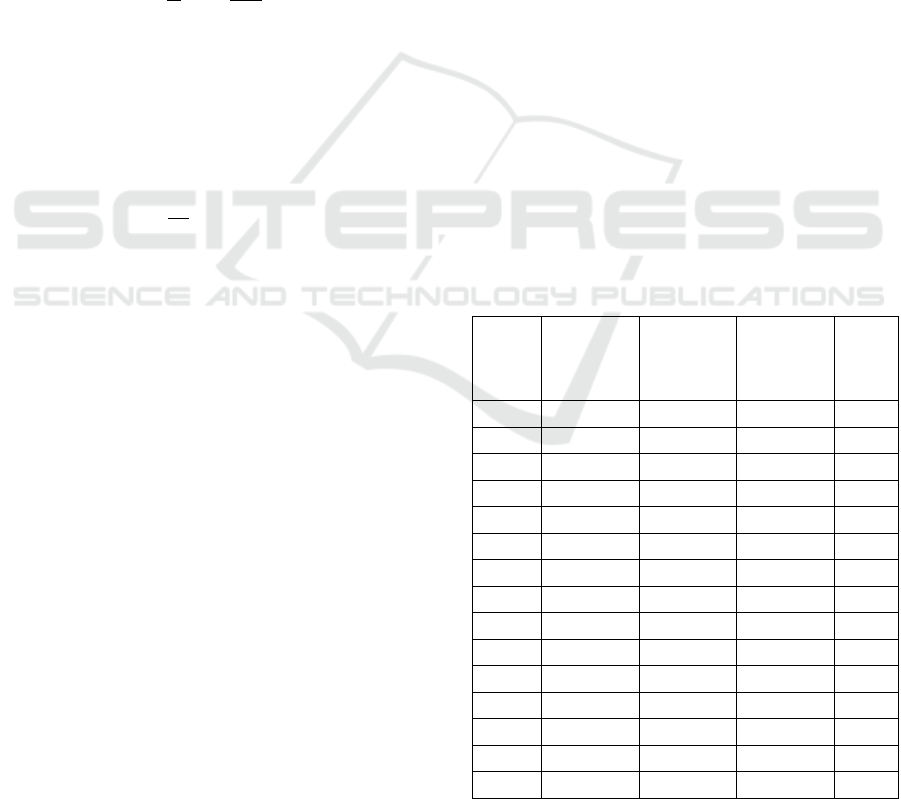
2 as follows:
Table 2: Average search accuracy for different Test Set
Sequences.
TSS A (%)
DMLA
(Zhang et
al, 2018)
A (%)
AQ PMS
(Yu et al,
2019)
A (%)
PSO
(Reddy et
al, 2010)
A (%)
MRQ
PMS
25
k
79.58 62.65 64.68 98.37
37.5
k
79.82 62.85 64.88 98.67
5k0 79.95 62.94 64.98 98.82
62.5
k
80.05 63.02 65.06 98.95
75
k
80.09 63.06 65.10 99.00
87.5
k
80.11 63.07 65.11 99.02
100
k
80.11 63.08 65.12 99.03
112.5
k
80.12 63.08 65.12 99.03
125
k
80.12 63.08 65.12 99.04
137.5
k
80.12 63.08 65.12 99.04
150
k
80.18 63.13 65.17 99.11
162.5
k
80.25 63.18 65.23 99.20
175
k
80.33 63.25 65.29 99.30
187.5
k
80.42 63.32 65.36 99.41
200
k
80.52 63.40 65.45 99.53
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
126
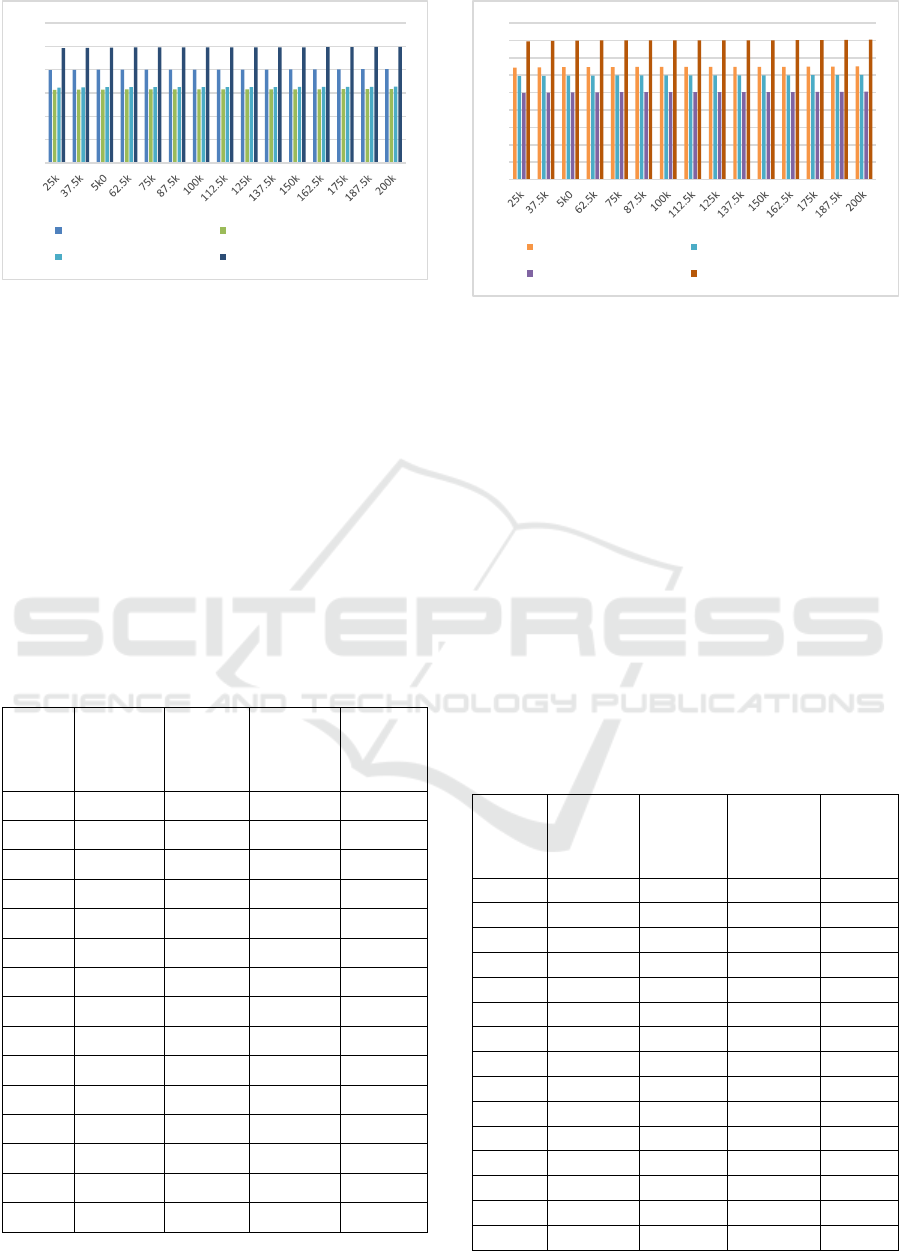
Figure 3: Average search accuracy for different Test Set
Sequences.
Based on this evaluation, and figure 3, it can be
observed that the proposed model is capable of
achieving 18.4% better accuracy than DMLA
(Zhang et al, 2018), 25.5% higher accuracy than AQ
PMS (Yu et al, 2019), and 23.9% better accuracy
than PSO (Reddy et al,2010) under different test
sequence sizes. This is due to combination of GA
with multiple distance metrics, and use of Map
Reduce, which assists in improving search
performance under different scenarios. Similar
evaluations for search precision can be observed
from table 3 as follows:
Table 3: Average search precision for different Test Set
Sequences.
TSS
P (%)
DMLA
(Zhang et
al, 2018)
P (%)
AQ PMS
(Yu et al,
2019)
P (%)
PSO
(Reddy et
al,2010)
P (%)
MRQ
PMS
25k 64.27 59.42 49.83 79.44
37.5k 64.47 59.60 49.99 79.68
5k0 64.57 59.70 50.06 79.81
62.5k 64.65 59.77 50.12 79.91
75k 64.68 59.80 50.15 79.95
87.5k 64.70 59.82 50.16 79.97
100k 64.70 59.82 50.16 79.97
112.5k 64.70 59.83 50.17 79.98
125k 64.71 59.83 50.17 79.98
137.5k 64.71 59.83 50.17 79.98
150k 64.75 59.87 50.21 80.04
162.5k 64.81 59.92 50.25 80.11
175k 64.87 59.98 50.30 80.19
187.5k 64.95 60.05 50.35 80.28
200k 65.03 60.13 50.42 80.38
Figure 4: Average search precision for different Test Set
Sequences
Based on this evaluation, and figure 4, it can be
observed that the proposed model is capable of
achieving 15.9% better precision than DMLA
(Zhang et al, 2018), 20.5% higher precision than AQ
PMS (Yu et al, 2019) and 19.4% better precision
than PSO (Reddy et al,2010) under different test
sequence sizes. This is due to combination of GA
with multiple distance metrics, and use of Map
Reduce, which assists in improving search
performance under different scenarios. Precision and
recall values are observed to be lower than accuracy,
which indicates that there are higher true negative
instances, and lower false positive instances in the
results. Similar evaluations for search recall can be
observed from table 4 as follows:
Table 4: Average search recall for different Test Set
Sequences.
TSS
R (%)
DMLA
(Zhang et
al, 2018)
R (%)
AQ PMS
(Yu et al,
2019)
R (%)
PSO
(Reddy et
al,2010)
R (%)
MRQ
PMS
25k 63.46 58.68 49.21 78.45
37.5k 63.66 58.86 49.36 78.69
5k0 63.76 58.96 49.44 78.81
62.5k 63.84 59.03 49.50 78.91
75k 63.87 59.06 49.53 78.96
87.5k 63.89 59.07 49.54 78.97
100k 63.89 59.08 49.54 78.98
112.5k 63.89 59.08 49.54 78.98
125k 63.90 59.08 49.54 78.99
137.5k 63.90 59.09 49.55 78.99
150k 63.95 59.13 49.58 79.04
162.5k 64.00 59.18 49.62 79.11
175k 64.07 59.24 49.67 79.19
187.5k 64.14 59.30 49.73 79.28
200k 64.22 59.38 49.79 79.38
0
20
40
60
80
100
120
A (%) DMLA (Zhang et al, 2018) A (%) AQ PMS (Yu et al, 2019)
A (%) PSO (Reddy et al,2010) A (%) MRQ PMS
0
10
20
30
40
50
60
70
80
90
P (%) DMLA (Zhang et al, 2018) P (%) AQ PMS (Yu et al, 2019)
P (%) PSO (Reddy et al,2010) P (%) MRQ PMS
MRQPMS: Design of a Map Reduce Bioinspired Model for Solving Quorum Planted Motif Search for High-Speed Deployments
127
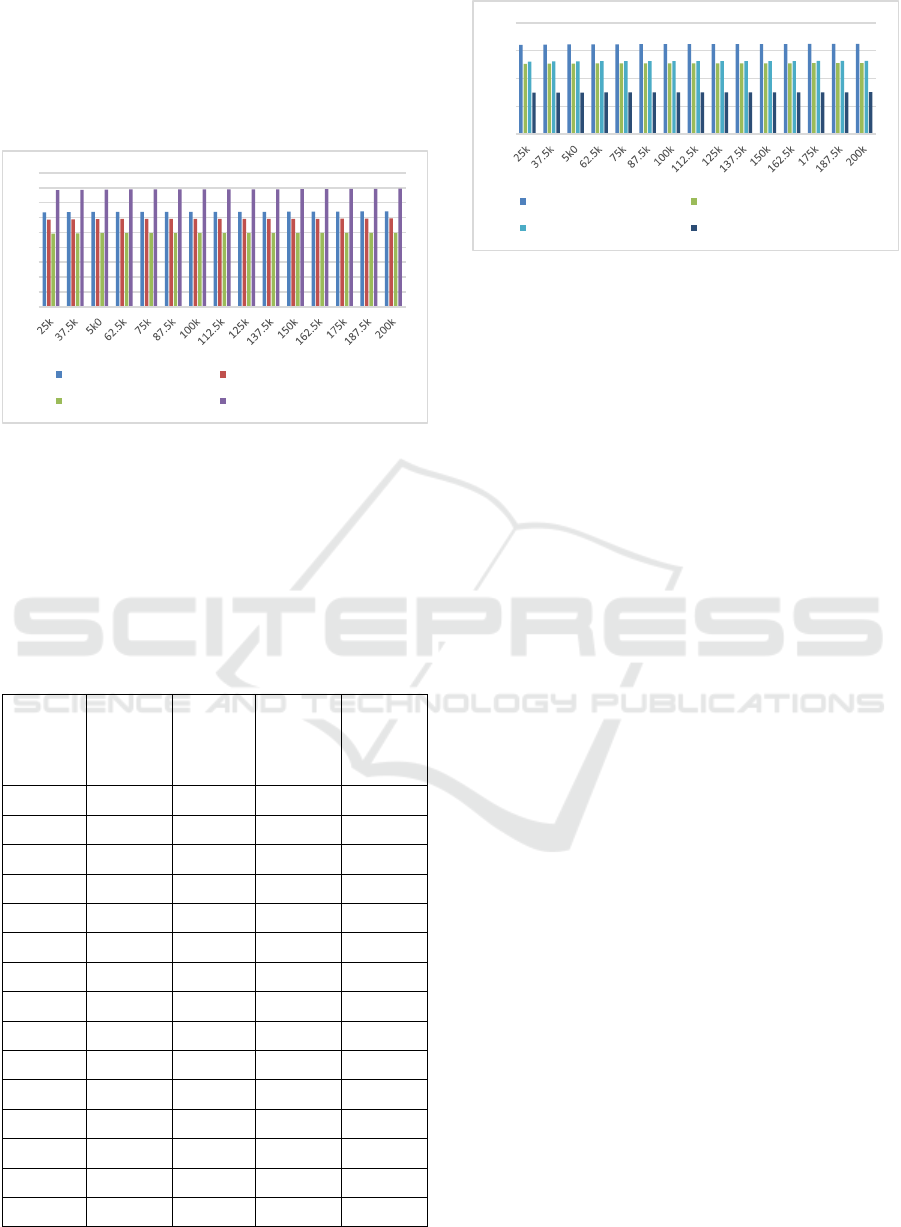
Based on this evaluation, and figure 5, it can be
observed that the proposed model is capable of
achieving 10.5% better recall than DMLA (Zhang et
al, 2018), 12.5% higher recall than AQ PMS (Yu et
al, 2019) and 23.4% better recall than PSO (Reddy
et al,2010) under different test sequence sizes.
Figure 5: Average search recall for different Test Set
Sequences
This is due to combination of GA with multiple
distance metrics, and use of Map Reduce, which
assists in improving search performance under
different scenarios. Similar evaluations for search
delay can be observed from table 5 as follows:
Table 5: Average search delays for different Test Set
Sequences.
TSS D (ms)
DMLA
(Zhang et
al, 2018)
D (ms)
AQ PMS
(Yu et al,
2019)
D (ms)
PSO
(Reddy et
al,2010)
D (ms)
MRQ
PMS
25k 160.53 126.39 130.47 74.41
37.5k 161.02 126.78 130.87 74.64
5k0 161.27 126.98 131.08 74.75
62.5k 161.47 127.13 131.24 74.85
75k 161.56 127.20 131.31 74.89
87.5k 161.59 127.23 131.34 74.91
100k 161.60 127.23 131.35 74.91
112.5k 161.61 127.24 131.36 74.91
125k 161.62 127.24 131.36 74.92
137.5k 161.62 127.25 131.37 74.92
150k 161.74 127.34 131.46 74.97
162.5k 161.88 127.45 131.57 75.04
175k 162.04 127.57 131.70 75.11
187.5k 162.22 127.72 131.86 75.20
200k 162.43 127.88 132.02 75.29
Figure 6: Average search delays for different Test Set
Sequences.
Based on this evaluation, and figure 6, it can be
observed that the proposed model is capable of
achieving 23.5% faster search performance than
DMLA (Zhang et al, 2018), 16.5% faster search
performance than AQ PMS (Yu et al, 2019) and
18.2% faster search performance than PSO (Reddy
et al,2010) under different test sequence sizes. This
is due to combination of GA with multiple distance
metrics, and use of Map Reduce, which assists in
improving search performance under different
scenarios. Due to these enhancements the proposed
model is capable of deployment for a wide variety of
real-time QPMS application scenarios.
5
CONCLUSION AND FUTURE
SCOPE
The proposed model uses a combination of Map
Reduce along with GA & QPMS for optimization of
Motif searches. Due to use of Map Reduce the
model was able to reduce the search delay, which
was further optimized via GA, which assisted in
estimation of Q values for search operations. These
when combined with a hybrid similarity metric,
assisted in improving overall search performance
under different use cases. This performance was
compared with various state-of-the-art methods,
where the proposed model was found to be capable
of 18.4% better accuracy than DMLA (Zhang et al,
2018), 25.54% higher accuracy than AQ PMS (Yu et
al, 2019) and 23.94% better precision than DMLA
(Zhang et al, 2018), 15.9% better precision than AQ
PMS (Yu et al, 2019) and 19.44% better precision
than PSO (Reddy et al,2010) as well as 10.54%
better recall than DMLA (Zhang et al, 2018),
12.54% higher recall than AQ PMS (Yu et al, 2019)
and 23.44% better recall than PSO (Reddy et
al,2010) under certain conditions. The proposed
0
10
20
30
40
50
60
70
80
90
R (%) DMLA (Zhang et al, 2018) R (%) AQ PMS (Yu et al, 2019)
R (%) PSO (Reddy et al,2010) R (%) MRQ PMS
0
50
100
150
200
D (ms) DMLA (Zhang et al, 2018) D (ms) AQ PMS (Yu et al, 2019)
D (ms) PSO (Reddy et al,2010) D (ms) MRQ PMS
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
128

model was found to be capable of achieving 23.5
percent faster search performance than DMLA
(Zhang et al, 2018), 16.5 percent faster search
performance than AQ PMS (Yu et al, 2019) and 18.2
percent faster search performance than PSO (Reddy
et al,2010) under various test sequence sizes. This
performance was also evaluated in terms of search
delay. This is a result of the combination of Map
Reduce and GA with various distance metrics,
which helps to enhance search performance in
various scenarios. These improvements enable the
proposed model to be deployed for numerous real-
time QPMS application scenarios. In future, the
proposed model must be validated on larger datasets,
and can be improved via use of Deep Learning
Models like Q-Learning, Autoencoders, and other
Convolutional Neural Networks (CNNs), which will
assist in further improving its scalability. This
performance can be further improved via use of
Gated Recurrent Units (GRUs), Generative
Adversarial Networks (GANs), along with
bioinspired computing models which will allow the
model to be continuously optimized for different
Motif Search based use cases.
ACKNOWLEDGEMENTS
The authors are thankful to the Director, Department
of Computer Science and Engineering, Visvesvaraya
National Institute of Technology (VNIT), Nagpur
for providing necessary facilities for this work.
REFERENCES
R. Semwal, I. Aier, U. Raj and P. K. Varadwaj, "Pr[m]:
An Algorithm for Protein Motif Discovery," in
IEEE/ACM Transactions on Computational Biology
and Bioinformatics, vol. 19, no. 1, pp. 585-592, 1 Jan.-
Feb. 2022, doi: 10.1109/TCBB.2020.2999262.
P. Xiao, X. Cai and S. Rajasekaran, "EMS3: An Improved
Algorithm for Finding Edit-Distance Based Motifs," in
IEEE/ACM Transactions on Computational Biology
and Bioinformatics, vol. 18, no. 1, pp. 27-37, 1 Jan.-
Feb. 2021, doi: 10.1109/TCBB.2020.3024222.
Q. Yu and X. Zhang, "A New Efficient Algorithm for
Quorum Planted Motif Search on Large DNA
Datasets," in IEEE Access, vol. 7, pp. 129617-129626,
2019, doi: 10.1109/ACCESS.2019.2940115.
T. Li, X. Zhang, F. Luo, F. -X. Wu and J. Wang,
"MultiMotifMaker: A Multi-Thread Tool for
Identifying DNA Methylation Motifs from Pacbio
Reads," in IEEE/ACM Transactions on Computational
Biology and Bioinformatics, vol. 17, no. 1, pp. 220-
225, 1 Jan.-Feb. 2020, doi: 10.1109/TCBB.2018.2
861399.
H. Zhang, L. Zhu and D. -S. Huang, "DiscMLA: An
Efficient Discriminative Motif Learning Algorithm
over High-Throughput Datasets," in IEEE/ACM
Transactions on Computational Biology and
Bioinformatics, vol. 15, no. 6, pp. 1810-1820, 1 Nov.-
Dec. 2018, doi: 10.1109/TCBB.2016.2561930.
H. Zhao, X. Xu, Y. Song, D. L. Lee, Z. Chen and H. Gao,
"Ranking Users in Social Networks with Motif-Based
PageRank," in IEEE Transactions on Knowledge and
Data Engineering, vol. 33, no. 5, pp. 2179-2192, 1
May 2021, doi: 10.1109/TKDE.2019.2953264.
X. Sun, Y. Tan, Q. Wu, B. Chen and C. Shen, "TM-Miner:
TFS-Based Algorithm for Mining Temporal Motifs in
Large Temporal Network," in IEEE Access, vol. 7, pp.
49778-49789, 2019, doi: 10.1109/ACCESS.2019.
2911181.
Z. Xing and S. Tu, "A Graph Neural Network Assisted
Monte Carlo Tree Search Approach to Traveling
Salesman Problem," in IEEE Access, vol. 8, pp.
108418-108428, 2020, doi: 10.1109/ACCESS.
2020.3000236.
S. Yu, F. Xia, Y. Sun, T. Tang, X. Yan and I. Lee,
"Detecting Outlier Patterns With Query-Based
Artificially Generated Searching Conditions," in IEEE
Transactions on Computational Social Systems, vol. 8,
no. 1, pp. 134-147, Feb. 2021, doi: 10.1109/
TCSS.2020.2977958.
M. U. Chaudhry and J. -H. Lee, "Feature Selection for
High Dimensional Data Using Monte Carlo Tree
Search," in IEEE Access, vol. 6, pp. 76036-76048,
2018, doi: 10.1109/ACCESS.2018.2883537.
Nicolae, M., Rajasekaran, S. qPMS9: An Efficient
Algorithm for Quorum Planted Motif Search. Sci Rep
5, 7813(2015). https://doi.org/10.1038/srep07813
Yu, Qiang & Zhang, Xiao. (2019). A New Efficient
Algorithm for Quorum Planted Motif Search on Large
DNA Datasets. IEEE Access. PP. 1-1. 10.1109/
ACCESS.2019.2940115.
Shrimankar, D.D. High performance computing
approachfor DNA motif discovery. CSIT 7, 295–297
(2019). https://doi.org/10.1007/s40012-019-00235-w
Peng Xiao, Xingyu Cai, and Sanguthevar Rajasekaran.
2021. EMS3: An Improved Algorithm for Finding Edit-
Distance Based Motifs. IEEE/ACM Trans. Comput.
Biol. Bioinformatics 18, 1 (Jan.-Feb. 2021)27,
37.https://doi.org/10.1109/TCBB.2020.3024222
Schultz, C.J., Wu, Y. & Baumann, U. A targeted
bioinformatics approach identifies highly variable cell
surface proteins that are unique to Glomeromycotina.
Mycorrhiza 32, 45–66(2022). https://doi.org/10.1007/
s00572-021-01066-x
J. Merlin and H. Dinh,"Poster: Randomized algorithms for
planted Motif Search," in 2013 IEEE 3rd International
Conference on Computational Advances in Bio and
medical Sciences (ICCABS), Las Vegas, NV, USA,
2012 pp. 1. doi: 10.1109/ICCABS.2012.6182654
Yu, Qiang & Huo, Hongwei & Vitter, Jeffrey & Huan, Jun
& Nekrich, Yakov. (2015). An Efficient Exact
MRQPMS: Design of a Map Reduce Bioinspired Model for Solving Quorum Planted Motif Search for High-Speed Deployments
129

Algorithm for the Motif Stem Search Problem over
Large Alphabets. Computational Biology and
Bioinformatics, IEEE/ACM Transactions on. 12. 384-
397. 10.1109/TCBB.2014.2361668.
EMS1: An Elegant Algorithm for Edit Distance Based
Motif Search, Sudipta Pathak (United States),
SANGUTHEVAR RAJASEKARAN (United States),
and MARIUS NICOLAE (United States),
International Journal of Foundations of Computer
Science 2013 24:04, 473-486
Reddy, U. Srinivasulu & Michael, Arock & A.V.Reddy,.
(2010). Planted (l, d) - Motif Finding using Particle
Swarm Optimization. International Journal of
Computer Applications. ecot. 10.5120/1541-144.
Kashiwabara, André Yoshiaki & Garbelini, Jader &
Sanches, Danilo. (2018). Sequence motif finder using
memetic algorithm. BMC Bioinformatics. 19.
10.1186/s12859-017-2005-1.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
130
