
Detection of Microscopic Fungi and Yeast in Clinical Samples Using
Fluorescence Microscopy and Deep Learning
Jakub Paplh
´
am
1
, Vojt
ˇ
ech Franc
1
and Daniela L
ˇ
zi
ˇ
ca
ˇ
rov
´
a
2
1
Department of Cybernetics, Czech Technical University in Prague, Prague, Czech Republic
2
Second Faculty of Medicine, Charles University, Prague, Czech Republic
Keywords:
Filamentous Fungi, Yeast, Automated Detection, Fluorescence Staining, Poisson Image Editing.
Abstract:
Early detection of yeast and filamentous fungi in clinical samples is critical in treating patients predisposed to
severe infections caused by these organisms. The patients undergo regular screening, and the gathered samples
are manually examined by trained personnel. This work uses deep neural networks to detect filamentous fungi
and yeast in the clinical samples to simplify the work of the human operator by filtering out samples that are
clearly negative and presenting the operator with only samples suspected of containing the contaminant. We
propose data augmentation with Poisson inpainting and compare the model performance against expert and
beginner-level humans. The method achieves human-level performance, theoretically reducing the amount of
manual labor by 87%, given a true positive rate of 99% and incidence rate of 10%.
1 INTRODUCTION
Early detection of yeast and filamentous fungi in clin-
ical samples is critical in treating patients predisposed
to severe infections caused by these organisms. Fluo-
rescence microscopy is a suitable method for this de-
tection, where after application to a slide, the mate-
rial is stained with a fluorescent dye (e.g., Calcofluor
White), which binds to chitin contained in the fun-
gal cell wall. This staining process is non-specific,
as other structures that may occur accidentally in the
sample (e.g., dust, pollen, arthropods) can also bind
the dye. Clinical samples commonly consist of res-
piratory secretions or non-invasive tissue biopsy sam-
ples; the aforementioned foreign bodies are therefore
routinely present.
Severe infections caused by filamentous fungi are
sporadic but severe. Patients with a risk factor, there-
fore, undergo regular screening. It follows that a con-
siderable number of slides must be carefully exam-
ined, most of which do not contain yeast or filamen-
tous fungi.
This work was done in collaboration with Motol
University Hospital in Prague, whose staff amassed
a unique dataset of fluorescence microscopy images
over a period of several years. The goal of the col-
laboration is to develop an automated system, which
filters out samples that are easily distinguishable as
negative and presents the remaining potentially posi-
tive samples to an expert for verification.
Currently, fungi and yeast cells are detected manu-
ally by trained personnel. Laboratory staff then spend
a significant amount of time examining negative sam-
ples, which leads to job dissatisfaction, and the devel-
opment of musculoskeletal disorders caused by repet-
itive stress injuries. Deployment of the system would
therefore lead to reduction of the amount of manual
labor and an increase in the quality of work.
This paper represents a feasibility study for au-
tomation and uses mostly standard deep learning
methods. In actual deployment, the detector will be
applied to a sequence of images obtained from an au-
tomated microscope rather than a single image.
Our main contributions are the following: (i) a
detector based on convolutional neural networks
(CNNs) is trained on a unique dataset,, (ii) a data aug-
mentation technique specific to the detection task is
proposed,, or (iii) performance of the model is eval-
uated and compared against expert and novice level
humans.
2 RELATED WORKS
Recently, automated slide scanners were used for
slide imaging of clinical samples and CNNs for eval-
uation of the data. This allows for fully automatic de-
Paplhám, J., Franc, V. and Lži
ˇ
ca
ˇ
rová, D.
Detection of Microscopic Fungi and Yeast in Clinical Samples Using Fluorescence Microscopy and Deep Learning.
DOI: 10.5220/0011616100003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 4: VISAPP, pages
777-784
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
777

tection and classification of microscopic organisms.
The approach has been successfully used for the
detection of bacteria in Gram stains of blood culture,
(Smith et al., 2018), and detection of intestinal pro-
tozoa in trichrome stained stool samples, (Mathison
et al., 2020). Gram staining dyes were also applied
to samples containing yeast and yeast-like fungi, al-
lowing their successful classification, (Zieli
´
nski et al.,
2020). Perhaps most similar to the goals of this work,
(Gao et al., 2021) fully automate the process of scan-
ning and classifying fluorescent dye-stained skin sam-
ples containing fungi.
All of the listed methods utilize the industry stan-
dard technique of fine-tuning a pre-trained CNN.
They employ an automated scanner to obtain a large
number of images from each sample, classify the im-
ages separately, then aggregate the result over the im-
ages to classify the sample. An identical approach can
be observed in the entire field of automatic evaluation
of digital microscopy.
The methods achieve human-level performance,
motivating future large-scale deployment. Some nov-
elty can be observed in the use of non-standard clas-
sifier heads, e.g., (Zieli
´
nski et al., 2020) replace the
linear classification head with bag-of-words encoding
followed by a support vector machine (SVM) classi-
fier. To our knowledge, no notable domain-specific
modifications of the standard techniques were used in
these works.
Our work focuses on a different domain, namely
fluorescent microscopy of human secretions. Further,
we are presented with only a single image per sample
and demonstrate that the method achieves sufficient
performance for deployment even in this setting.
3 METHODS
3.1 Dataset
The dataset contains high-resolution images of sam-
ples collected by staff of Motol University Hospital in
Prague from January 2018 to April 2021. The images
are a priori assumed to be negative and are considered
positive only when structures specific to microscopic
yeast or fungi are detected, even if low-resolution and
present only in a small portion of the image. An ex-
ample of such a case, where a single yeast cell is
present and covers only a very small portion of the
image, is shown in Figure 3.
The dataset contains a total of 1244 high-
resolution images. The pixel dimensions of the im-
ages are not identical, see Table 1. However, the
aspect ratio
Width
Height
≈ 1.33 is constant throughout the
Figure 1: Showcase of randomly selected positive samples
from the dataset.
Figure 2: Showcase of randomly selected negative samples
from the dataset.
dataset, and each image captures the same field of
view. The images can therefore be resized to a
uniform shape, which allows for mini-batching of
the data, improving the training efficiency. Annota-
tions are given in the form of the binary label (pos-
itive/negative) for each image. In other words, the
annotation provides no information on the location or
size of the specimen within the image.
The main challenges associated with the dataset
are twofold: (i) the amount of available data is rela-
tively low, and (ii) positive and negative images have
a high degree of similarity. Examples of positive and
negative samples are shown in Figure 1 and Figure 2,
respectively.
Table 1: Dimensions of images in the dataset.
Width × Height
Annotation
Positive Negative
4140 ×3096 374 546
2040 ×1536 77 3
1360 ×1024 231 13
Total 682 562
Sample Preparation. Clinical Material
1
was
smeared on a sterile slide and dried. The dried slides
were dyed with Calcofluor White mixed 1 : 1 with
20% potassium hydroxide solution and immediately
covered with cover slides and examined. Fluores-
cence microscopy was performed manually with the
1
Specifically (i) sputum, (ii) endotracheal or bronchial
aspirate, (iii) bronchoalveolar fluid or tissue, (iv) pleu-
ral fluid, (v) pericardial fluid, (vi) cerebrospinal fluid, or
(vii) liquid or solid contents of pathological cavities.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
778

Figure 3: Except for a single budding yeast cell, there is no
contaminant in the image. The yeast cell takes up only a
small portion of the image, but it is entirely responsible for
the final classification as a positive sample.
use of Olympus BX 53 fluorescence microscope, Up-
lanFLN 20x objective lens, FN 26,5. The entire slide
was examined, and a representative section of the
slide was selected. The image of the selected section
was captured using an Olympus DP72 microscope
digital camera.
3.2 Model
Formal Definition. Let us denote the common
pixel domain as D ⊂ Z
2
, and the monochromatic do-
main as M ⊂ {x | x ∈ R}, where lower and upper
bounds of the values and machine precision are ig-
nored for simplicity. The set of all possible grayscale
images is then I = M
D
.
The model is a binary image classifier, i.e., the
mapping h : X → Y , where X ⊂ I , Y = {+1, −1}.
Often the classifier h is further decomposed as
h = f ◦d, where f : X → R, and d : R → {+1,−1},
with the decoding mapping d defined as
d(x,θ) =
(
+1 for x ≥ θ,
−1 for x < θ,
for some fixed threshold θ ∈R. We further define an
ensemble of binary classifiers, as a binary classifier
where the score function f is defined as
f (x) =
∑
n
i=1
f
i
(x)
n
, f
i
: X → R.
(1)
Implementation. We choose ResNet-50 with a sin-
gle linear output layer as our baseline binary classifier
f and use the shallower variant, ResNet-18, to search
for an optimal training setup, e.g., data augmentations
and image preprocessing. We further train and com-
pare performance of (i) ResNet-50x1-V2, (He et al.,
2016), (ii) EfficientNet models, (Tan and Le, 2019),
from B0 to B4, (iii) EfficientNet-V2-S model, (Tan
and Le, 2021), and (iv) Vision Transformer ViT-B us-
ing 32 ×32 embeddings (Dosovitskiy et al., 2021).
All models we use are pretrained on ImageNet.
3.3 Saved Time Metric
Assuming that manual examination of each sample
takes constant time, the amount of human time saved
by the model is directly proportional to the number
of samples which need not be examined by human
personnel. If a sample is to be classified as positive,
manual confirmation is required. Therefore, the saved
time is proportional to the number of samples classi-
fied as negative by the model.
The maximal value of such a metric can be
achieved by classifying all samples as negative. This
is clearly undesirable. Therefore, we define the saved
time metric as the portion of samples classified as
negative while guaranteeing that the true positive rate
is higher than a specified level. The metric represents
an alternative to the standard ROC curve. It directly
measures the clinical utility of the model and can eas-
ily be explained to medical staff.
Formal Definition. We evaluate the prediction
rule h: X → {+1, −1} in terms of two metrics.
First, the true positive rate (a.k.a. sensitiv-
ity) TPR(h) = E
x∼p(x|y=+1)
[[h(x) = +1]], which is
the probability that a positive sample is correctly
classified as positive. Secondly, the saved time
ST(h) = E
x∼p(x)
[[h(x) = −1]], equal to the probabil-
ity that any input sample is classified as negative. It is
useful to rewrite the saved time as
ST(h) =
1 −p(y = +1)
·
1 −FPR(h)
+ p(y = +1) ·
1 −TPR(h)
,
(2)
where p(y = +1) is the prior probability of the posi-
tive class, and FPR(h) = E
x∼p(x|y=−1)
[[h(x) = +1]] is
the false positive rate, i.e., the probability that a neg-
ative sample is incorrectly classified as positive. The
equation (2) shows that the two metrics, TPR(h) and
ST(h), are antagonistic, i.e., increasing one leads to a
decrease of the other and vice versa.
Evaluating the Metric. As defined in Section 3.2,
the model is a binary image classifier of the form
h(x;θ) =
(
+1 for f (x) ≥ θ,
−1 for f (x) < θ,
(3)
where f : X → R is a score function trained from ex-
amples and θ ∈ R is a decision threshold used to tune
Detection of Microscopic Fungi and Yeast in Clinical Samples Using Fluorescence Microscopy and Deep Learning
779

the operating point of the model. With a slight abuse
of notation, we use TPR(θ) and ST(θ) as a shortcut
for TPR(h(·;θ)) and ST(h(·;θ)), respectively.
The number of positive and negative samples in
the available test set is approximately the same, which
does not match the real distribution at the deploy-
ment time. According to Motol University Hospi-
tal’s staff, approximately 9 out of 10 samples that ar-
rive at the laboratory for examination are negative.
Therefore, when evaluating the detector we assume
that the incidence rate is p(y = +1) = 0.1. How-
ever, the methodology as a whole is general and,
if necessary, can be applied to any incidence rate.
When evaluating the metric, we resolve the men-
tioned distribution mismatch as follows. Given a test
set {(x
i
,y
i
) ∈ X ×{+1,−1}| i = 1, ..., n}, we com-
pute the empirical estimates of TPR(θ) and FPR(θ),
d
TPR(θ) =
1
n
+
n
∑
i=1
[[h(x
i
;θ) = +1 ∧y
i
= +1]], (4)
d
FPR(θ) =
1
n
−
n
∑
i=1
[[h(x
i
;θ) = +1 ∧y
i
= −1]], (5)
where n
+
=
∑
n
i=1
[[y
i
= +1]] and n
−
=
∑
n
i=1
[[y
i
= −1]].
Then, we fix the positive class prior to the expert
estimate of the incidence rate, p(y = +1) = 0.1,
and compute the empirical estimate of the saved
time
c
ST(θ) by substituting
d
TPR(θ) and
d
FPR(θ)
into equation (2). We evaluate the predictor h
by a curve
n
d
TPR(θ) ,
c
ST(θ)
| θ ∈ (−∞,∞)
o
which summarizes the entire space of achievable
true positive rates and saved times. As a ref-
erence, we also plot the best achievable saved
time curve as a function of TPR, i.e., we plot
the curve
{
(TPR,ST
∗
(TPR)) | TPR ∈(0,1)
}
where
ST
∗
(TPR) = p(y = +1) ·[1 −TPR] + [1 − p(y = +1)],
which is obtained from equation (2) when assuming
an ideal predictor with zero FPR(h).
In case we need to evaluate the predictor by a sin-
gle scalar, e.g., when ranking different models, we
report the saved time at desired true positive rate τ,
which is defined as
c
ST
τ
= max
θ∈(−∞,∞)
c
ST(θ) subject
to
d
TPR(θ) ≥ τ .
3.4 Domain-Specific Data
Augmentation
To enlarge the number of samples used for train-
ing, we utilize standard image data augmentations,
namely (i) horizontal flip, (ii) vertical flip, (iii) rota-
tion, and (iv) crop and resize. We also implement and
evaluate the effects of a custom augmentation method
described in the following text.
Motivation & Overview. While obtaining negative
samples is simple, getting positive samples is com-
paratively complex and expensive. We propose an
augmentation method specific to the detection task,
where any negative image becomes positive if the
contaminant (e.g., yeast or fungi) is introduced. The
technique takes advantage of a large number of nega-
tive images and uses them to generate synthetic pos-
itive images by inpainting the positive contaminant
into a negative background. The contaminant can fur-
ther be rotated and shifted to create practically an un-
limited number of positive samples.
To generate additional positive samples, we locate
the fungi or yeast within the image either by (i) gradi-
ent-based localization, Grad-CAM (Selvaraju et al.,
2017), which is a broadly applicable method with
minimal prerequisites, or (ii) by exploiting the fluo-
rescent staining process. We then augment the image
by inpainting the located yeast and fungi into a nega-
tive background using Poisson image editing, (P
´
erez
et al., 2003).
We also generate synthetic negative samples, to
keep the augmentation symmetrical with respect to
classes; preventing the model from associating poten-
tial inpainting artifacts with the positive class. In neg-
ative samples, we inpaint structures that are visually
similar to the yeast and fungi, localized using Grad-
CAM, (Selvaraju et al., 2017).
Overview of Poisson Inpainting. Consider the task
of inpainting a portion of a source image s into a back-
ground b to form a resulting image r . The naive ap-
proach is to directly copy the pixel values from the
source s to the background b. This, however, cre-
ates visible edges between the inpainted region and
the background. Instead of copying values of the pix-
els, Poisson image editing, (P
´
erez et al., 2003), copies
the gradient. A comparison with the naive procedure
is shown in Figure 6.
To inpaint a region of the source into the back-
ground using Poisson inpainting, (i) pixels on the bor-
der of the source region are set to match the neighbor-
ing pixels in the background, and (ii) the remaining
pixel values of the inpainted source region are found
by solving the Poisson equation with the condition of
preserving the gradient of the source image. I.e., the
color of the inpainted region is modified to match the
color of the background, but the relative color differ-
ence between pixels is preserved.
Formal Definition of Poisson Inpainting. Here,
we briefly review our usage of the method famously
introduced by (P
´
erez et al., 2003). Let us denote a
background image, a source image, and a resulting
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
780

image as b,s,r ∈ I , respectively. By Ω ⊂ D, let us de-
note a region within the source image s which is to be
inpainted into the background b to form the resulting
image r. Let us further denote by p ∈ D a pixel po-
sition and by s
p
, b
p
, r
p
values of the pixel within the
source, background, and result images, respectively.
To seamlessly inpaint the region Ω of the source
image into the background image, we solve the opti-
mization problem
min
r
∑
⟨p,q⟩∩Ω̸=
/
0
(r
p
−r
q
−s
p
+ s
q
)
2
, (6)
subject to
r
p
= b
p
, ∀p ∈ δΩ, (7)
where ⟨p,q⟩ is a pixel neighbor pair.
→
Figure 4: Thresholding a fluorescent stained positive sam-
ple to obtain the Ω regions for Poisson inpainting.
→
Figure 5: Synthetic sample created by inpainting region
containing the positive class into a negative background.
Figure 6: Comparison of the inpainting methods. The in-
painted region has a distinct background color when the
value of pixels is copied directly. The Poisson image editing
ensures that the cutout seamlessly blends into the negative
image, creating a more realistic result.
Generating Synthetic Positive Samples. The aug-
mentation procedure requires (i) a positive source im-
age s, (ii) a localization mask defining the region Ω,
and (iii) a negative background image b. To local-
ize the fungi and yeast, we exploit the fact that due
to the fluorescent staining process, the pixel values of
the yeast and fungi are always greater than those of
the background. Therefore, we can obtain a rough lo-
calization mask by thresholding the pixel values, e.g.,
by employing Yen’s non-parametric thresholding al-
gorithm, (Yen et al., 1995). It must be noted that the
fluorescent dye is non-specific, however, and other
structures, such as dust, pollen, or arthropods, may
also bind the dye. Yeast or fungi is therefore always
inpainted, but some miscellaneous structures are in-
advertently inpainted as well. This, however, does not
harm the creation of new positive examples.
If the localization mask contains multiple con-
nected components, we interpret each connected com-
ponent as a region Ω and inpaint it separately with
random rotation and random position. This modifica-
tion is especially suited for yeast.
The following steps summarize the augmentation:
(i) Localize regions Ω of a positive source image s
which contain yeast or fungi., (ii) Select a negative
background image b at random., or (iii) Inpaint each
region of s discovered in step (i) into the negative im-
age b to form the resulting image. The position and
rotation of the inpainted region within the result are
selected at random.
Generating Synthetic Negative Samples. The
augmentation procedure requires (i) a negative im-
age, serving as both a source s and background b, and
(ii) a localization mask defining the region Ω. We
use the gradient-based Grad-CAM localization, (Sel-
varaju et al., 2017), to discover the Ω regions. Grad-
CAM provides a course heatmap from which we gen-
erate a binary mask by thresholding. It must be men-
tioned that the heatmap specifies the relative magni-
tude of activations of the learned convolutional filters.
If there are no structures in the image that are similar
to fungi or yeast, the activations across the entire im-
age are of similar magnitude and the resulting mask
covers the entire image. We, therefore, do not aug-
ment the sample in such a case. The following steps
summarize the augmentation: (i) Localize regions Ω
of a negative image that are visually similar to yeast
or fungi., or (ii) Inpaint each region discovered in step
(i) into the image to form the resulting image. The po-
sition and rotation of the inpainted region within the
result are selected at random. This step can be re-
peated multiple times.
4 EXPERIMENTS & RESULTS
Setup. Unless explicitly stated otherwise, we train
and evaluate the models using 30-fold cross-
validation with training, validation, and test sets con-
taining 80%, 10%, and 10% of the total data set, re-
Detection of Microscopic Fungi and Yeast in Clinical Samples Using Fluorescence Microscopy and Deep Learning
781
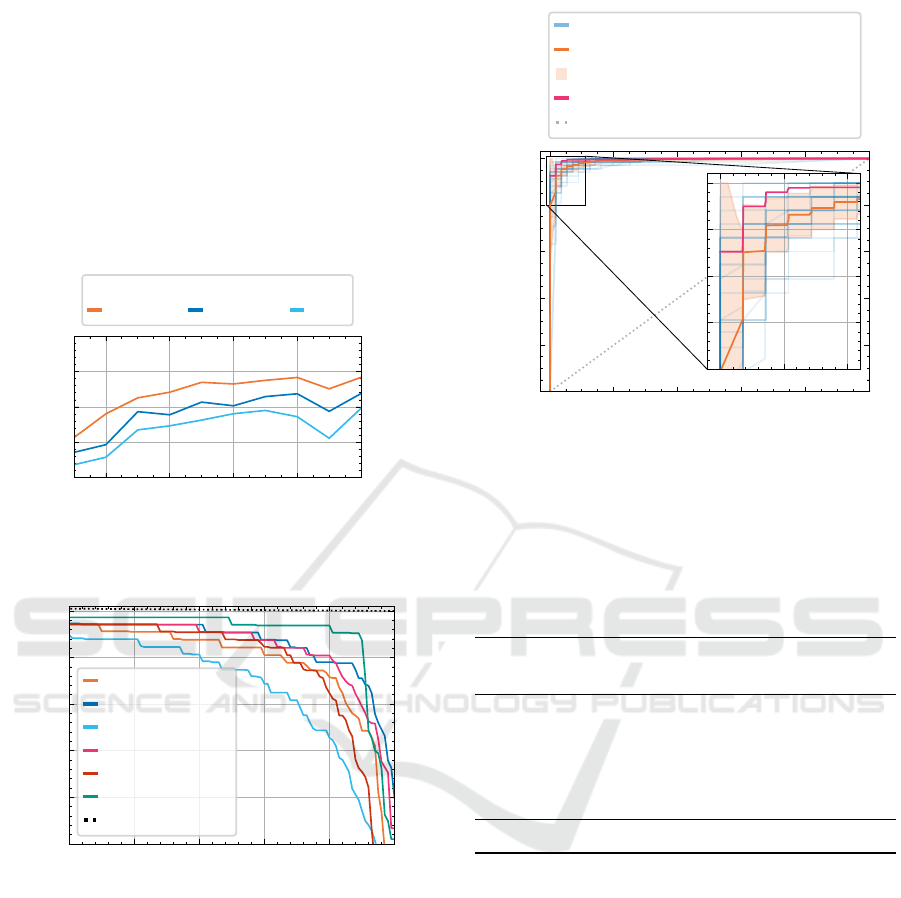
spectively. We use the SGD optimizer with 0.9 Nes-
terov momentum. The initial learning rate is set to
0.001 and reduced by a factor of 3 upon reaching 33%
and 66% of the 150 total training epochs. We train
using a batch size of 10 images due to hardware limi-
tations. We use images of a uniform size of 952×716
pixels. We show the effects of image size on perfor-
mance of a ResNet-18 model in Figure 7. A larger im-
age size results in better performance; however, an in-
crease over 680 ×512 yields only marginal improve-
ments.
272 × 205
544 × 409
816 × 614
1088 × 819
1360 × 1024
Image Size
0.00
0.25
0.50
0.75
1.00
Saved Time
True Positive Rate
0.98 0.99 0.995
Figure 7: Dependence of saved time metric on image size.
0.95 0.96 0.97 0.98 0.99 1.00
True Positive Rate
0.4
0.5
0.6
0.7
0.8
0.9
Saved Time
ResNet-50
EfficientNet-B2
ViT-B-32
EfficientNet-V2-S
ResNet-50-V2
Ensemble
Upper Bound
Figure 8: Comparison of different model architectures.
4.1 Model Architecture
We train multiple state-of-the-art models of similar
complexity as ResNet-50. It is well known that en-
sembling techniques result in improved performance
at the cost of increased computational complexity.
We, therefore, also produce an ensemble of the ar-
chitectures by averaging their predictions.
The achieved saved time metric values are dis-
played in Table 2 and the curve of achievable val-
ues is shown in Figure 8. The performance of all
the models is within a 5% margin, except for an
outlier, the Vision Transformer, which performs sig-
0.0 0.2 0.4 0.6 0.8 1.0
True Negative Rate
0.0
0.2
0.4
0.6
0.8
1.0
True Positive Rate
ResNet-50 Instance
Mean (AUC = 0.991)
± 1 Standard Deviation
Ensemble (AUC = 0.997)
Random Classifier
0.00 0.05 0.10
0.80
0.85
0.90
0.95
1.00
Figure 9: ROC curve (receiver operating characteristic) of
ResNet-50 models. The orange curve shows the mean ROC
over all folds, with the light orange area marking the stan-
dard deviation between folds. The pink curve shows the
ROC for an ensemble of different model architectures.
Table 2: Saved time metric comparison for different model
architectures. The first value indicates the mean saved time
metric; the second is the standard deviation between folds.
True positive rate
Model 98% 99% 99.5%
RN-50 0.81 (0.12) 0.76 (0.18) 0.64 (0.20)
EN-B2 0.84 (0.05) 0.79 (0.10) 0.76 (0.10)
ViT-B-32 0.74 (0.12) 0.63 (0.18) 0.47 (0.22)
EN-V2-S 0.84 (0.05) 0.81 (0.12) 0.70 (0.12)
RN-50-V2 0.82 (0.08) 0.72 (0.19) 0.56 (0.28)
Ensemble 0.87 (0.03) 0.87 (0.16) 0.84 (0.16)
nificantly worse. The best performance is achieved
by EfficientNet-B2 models, reaching both the highest
value of the saved time metric and the lowest stan-
dard deviation between folds, i.e., the architecture
performs the best consistently. From the EfficientNet
family of models, we only report the best performer,
EfficientNet-B2. By ensembling, the performance
can further be improved, resulting in a theoretical re-
duction of manual labor (saved time) of 87%, given a
true positive rate of 99%. We show the ROC curve in
Figure 9. The ensemble comprises (i) ResNet-50, (ii)
EfficientNet-B2, (iii) ViT-B-32, (iv) EfficientNet-V2-
S, (v) ResNet-50-V2. Score function of the ensem-
ble is computed as the mean of score functions of the
component models.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
782
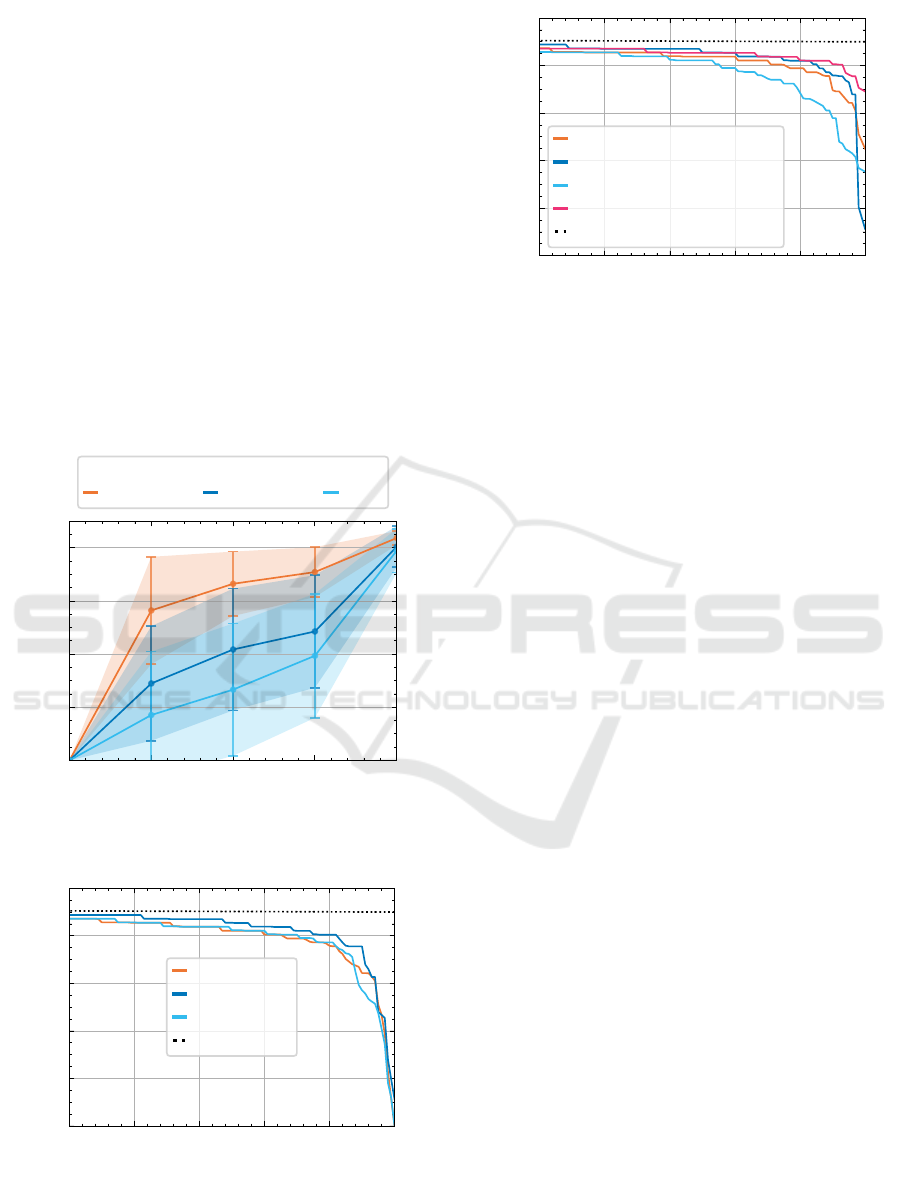
4.2 Poisson Augmentation
We construct a learning curve, shown in Figure 10,
to verify that the model can benefit from a larger
dataset. The performance steeply improves with ad-
ditional data, motivating further data augmentations
beyond the standard techniques.
We train ResNet-50 with additional synthetic pos-
itive samples, which were created either (i) by Pois-
son inpainting, or (ii) by directly copying the pixel
values. The results are shown in Figure 11 and
demonstrate that the Poisson inpainting is crucial, as
the naive technique does not result in any perfor-
mance improvements.
We, therefore, train ResNet-50 with additional
synthetic samples, both positive and negative, created
with Poisson inpainting. Models trained using the
augmentation of both classes consistently outperform
the baseline. The results are shown in Figure 12.
0 25 50 75 100
Dataset Size [%]
0.0
0.2
0.4
0.6
0.8
Saved Time
True Positive Rate
0.98 0.99 0.995
Figure 10: Learning curve for ResNet-50. The transparent
area shows ±1 standard deviation. Result on 11 folds.
0.95 0.96 0.97 0.98 0.99 1.00
True Positive Rate
0.0
0.2
0.4
0.6
0.8
1.0
Saved Time
Baseline
Poisson
Naive Copy
Upper Bound
Figure 11: Training with synthetic positive samples created
with Poisson inpainting results in better performance than
naively copied pixel values. Result on 20 folds.
0.95 0.96 0.97 0.98 0.99 1.00
True Positive Rate
0.0
0.2
0.4
0.6
0.8
1.0
Saved Time
Baseline
Positive Poisson
Negative Poisson
Positive & Negative Poisson
Upper Bound
Figure 12: Training with both, positive and negative, syn-
thetic samples results in significant increase in performance.
Result on 15 folds.
4.3 Human-Machine Comparison
To assess the difficulty of the task and further eval-
uate the model performance, we select 100 positive
and 100 negative images from the dataset at random
and compare the performance of the model with the
performance of humans on the classification.
The images were shown to 4 expert microbiol-
ogists. They were prompted to classify the images
as either positive or negative. A group of 3 begin-
ners was also shown the images after a brief training
session that included a showcase of 20 representative
positive and 20 negative samples. The task was ver-
bally explained, and special attention was given to the
specific structures of the contaminants.
For each of the images, the automated classifica-
tion was produced by an ensemble model, which con-
tained the given image in its test set.
All expert microbiologists perform similarly,
achieving a true positive rate of 89%, 89%, 90% and
94% with a saved time of 91.1%, 91.1%, 89.2% and
90.6% respectively. The experts achieve values of the
saved time at the theoretical upper bound, i.e., the
experts achieve a false positive rate of FPR ≈ 0. It
should be noted that the presented task is significantly
different from the standard operating procedure of the
experts. In the usual setting, the expert is presented
with an entire slide and can freely move between por-
tions of the slide and search for the contaminant. In
this experiment, the view is locked, and the expert is
presented with only a single image.
The beginner-level humans perform significantly
worse than the automated model. They either (i) do
not achieve sufficient true positive rate, or (ii) achieve
sufficient true positive rate, but a low value of the
saved time metric. The ensemble of models performs
at the same level or a better level than the expert hu-
mans. The result is shown in Figure 13.
Detection of Microscopic Fungi and Yeast in Clinical Samples Using Fluorescence Microscopy and Deep Learning
783
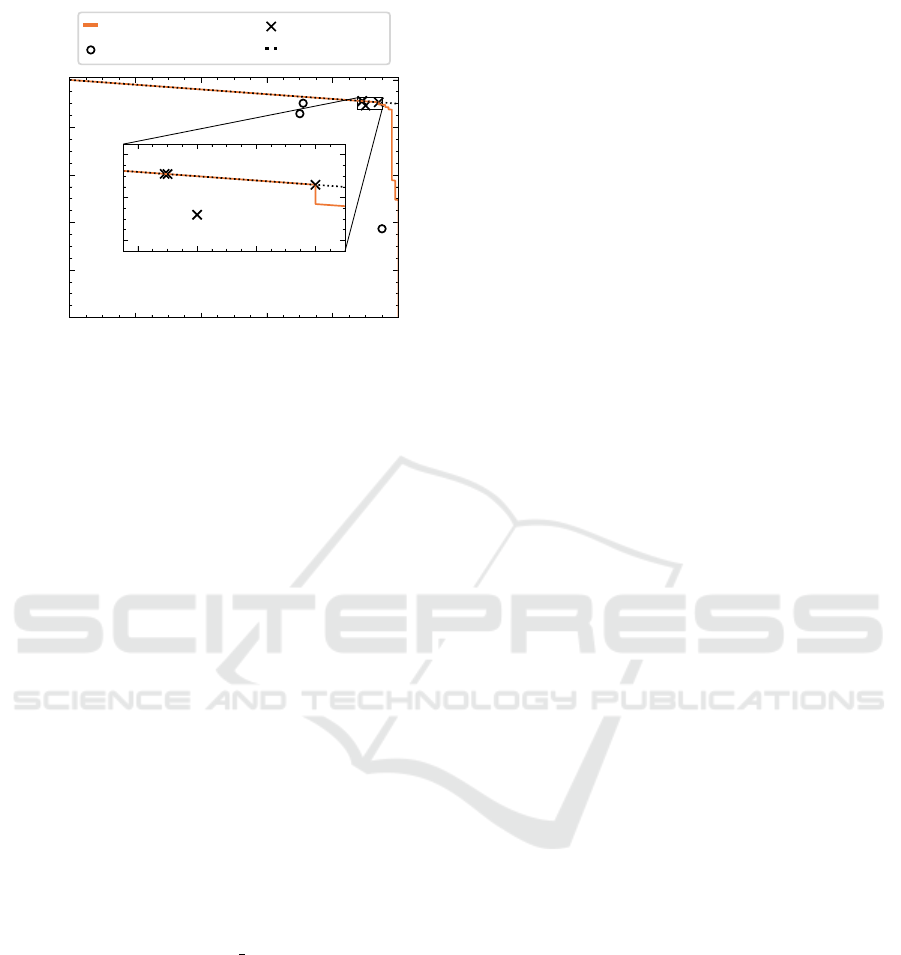
0.0 0.2 0.4 0.6 0.8 1.0
True Positive Rate
0.0
0.2
0.4
0.6
0.8
1.0
Saved Time
Ensemble
Beginner
Expert
Upper Bound
0.88 0.90 0.92 0.94
0.88
0.90
0.92
Figure 13: Human-machine performance comparison. Re-
sults on a randomly selected set of 100 positive and 100 neg-
ative samples. Medical experts perform significantly better
than beginner humans, who are outperformed by the auto-
mated model by a significant margin.
5 CONCLUSION
The results indicate that the detection of microscopic
yeast and fungi in clinical samples can be tackled by
standard deep-learning methods, employing an en-
semble of convolutional neural networks. The de-
veloped model consistently performs on par or better
than a human expert and, if deployed, should reduce
the amount of manual labor by approximately 87%
when operating at a true positive rate of 99%. The re-
sults are achieved with annotations only on the image
level, i.e., the network was not instructed what part of
the image is responsible for the classification.
ACKNOWLEDGEMENTS
The authors acknowledge the support of the OP VVV
project CZ.02.1.01/0.0/0.0/16 019/0000765 Research
Center for Informatics. The paper also acknowledges
project No. VJ02010041 supported by the Ministry
of the Interior of the Czech Republic. Special thanks
belongs to MUDr. Daniela L
ˇ
zi
ˇ
ca
ˇ
rov
´
a for collecting
the dataset and to MUDr. Kamila Dundrov
´
a, MUDr.
Vanda Chrenkov
´
a, Bc. Karla Ka
ˇ
nkov
´
a, Vladim
´
ır
Kryll, RNDr. Pavl
´
ına Lyskov
´
a, Ph. D. for filling out
the human-machine comparison survey.
REFERENCES
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Min-
derer, M., Heigold, G., Gelly, S., Uszkoreit, J., and
Houlsby, N. (2021). An image is worth 16x16 words:
Transformers for image recognition at scale. ArXiv,
abs/2010.11929.
Gao, W., Li, M., Wu, R., Du, W., Zhang, S., Yin, S., Chen,
Z., and Huang, H. (2021). The design and appli-
cation of an automated microscope developed based
on deep learning for fungal detection in dermatology.
Mycoses, 64(3):245–251.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Identity
mappings in deep residual networks.
Mathison, B. A., Kohan, J. L., Walker, J. F., Smith,
R. B., Ardon, O., Couturier, M. R., and Pritt, B. S.
(2020). Detection of intestinal protozoa in trichrome-
stained stool specimens by use of a deep convolutional
neural network. Journal of Clinical Microbiology,
58(6):e02053–19.
P
´
erez, P., Gangnet, M., and Blake, A. (2003). Poisson im-
age editing. ACM Trans. Graph., 22(3):313–318.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. In 2017 IEEE International Conference
on Computer Vision (ICCV), pages 618–626.
Smith, K. P., Kang, A. D., and Kirby, J. E. (2018). Auto-
mated interpretation of blood culture gram stains by
use of a deep convolutional neural network. Journal
of Clinical Microbiology, 56(3):e01521–17.
Tan, M. and Le, Q. (2019). EfficientNet: Rethinking model
scaling for convolutional neural networks. In Chaud-
huri, K. and Salakhutdinov, R., editors, Proceedings of
the 36th International Conference on Machine Learn-
ing, volume 97 of Proceedings of Machine Learning
Research, pages 6105–6114. PMLR.
Tan, M. and Le, Q. V. (2021). Efficientnetv2: Smaller mod-
els and faster training. ArXiv, abs/2104.00298.
Yen, J.-C., Chang, F.-J., and Chang, S. (1995). A new
criterion for automatic multilevel thresholding. IEEE
Transactions on Image Processing, 4(3):370–378.
Zieli
´
nski, B., Sroka-Oleksiak, A., Rymarczyk, D., Piekar-
czyk, A., and Brzychczy-Włoch, M. (2020). Deep
learning approach to describe and classify fungi mi-
croscopic images. PLOS ONE, 15(6):1–16.
VISAPP 2023 - 18th International Conference on Computer Vision Theory and Applications
784
