
Condition for Sustained Oscillations in Repressilator Based on a Hybrid
Modeling of Gene Regulatory Networks
Honglu Sun
1 a
, Jean-Paul Comet
2 b
, Maxime Folschette
3 c
and Morgan Magnin
1 d
1
Nantes Universit
´
e,
´
Ecole Centrale Nantes, CNRS, LS2N, UMR 6004, F-44000 Nantes, France
2
University C
ˆ
ote d’Azur, I3S Laboratory, UMR CNRS 7271, CS 40121, 06903 Sophia Antipolis Cedex, France
3
Univ. Lille, CNRS, Centrale Lille, UMR 9189 CRIStAL, F-59000 Lille, France
Keywords:
Hybrid Modeling, Repressilator, Sustained Oscillation, Gene Regulatory Network, Synthetic Biology.
Abstract:
In this work, we study the existence of sustained oscillations in the “canonical” repressilator, a basic synthetic
circuit of 3 genes leading to sustained oscillations. Previous works mostly used differential equations to study
the repressilator. In our work, a pre-existing hybrid modeling framework of gene regulatory networks called
HGRN is used to model this system. Compared to differential equations, dynamical properties of HGRNs
are easier to prove theoretically due to its lower dynamical complexity. The objective of this work is to
find conditions for the existence of sustained oscillations described by separable constraints on parameters.
With such separable constraints, each parameter is constrained individually by an interval, which can provide
useful information for the design of synthetic circuits. Our two major contributions are the following: firstly,
we develop, by using the Poincar
´
e map, a sufficient and necessary condition for the existence of sustained
oscillations; then, based on this condition, we give a method using the range enclosure property of Bernstein
coefficients to compute compatible separable constraints. By applying this method, we successfully obtain
sets of conditions for the existence of sustained oscillations described as separable constraints.
1 INTRODUCTION
With the developments in the field of synthetic biol-
ogy in the recent years, the construction of synthetic
gene regulatory networks in living cells which satisfy
certain properties becomes possible. Stable and con-
trollable synthetic circuits could have potential med-
ical applications. Mathematical modeling is one of
the ways to guide the design of these synthetic cir-
cuits. The modeling of many different synthetic cir-
cuits have been studied in the literature (Chaves and
Jong, 2021; Firippi and Chaves, 2020; Chaves and
Gouz
´
e, 2011; Bus¸e et al., 2010).
In this work, we focus on one class of synthetic
circuit: the repressilators, which are gene regulatory
networks consisting of at least one feedback loop, in
which the expression of each gene inhibits the expres-
sion of the next gene in the loop leading to an oscilla-
a
https://orcid.org/0000-0002-8265-0984
b
https://orcid.org/0000-0002-6681-3501
c
https://orcid.org/0000-0002-3727-2320
d
https://orcid.org/0000-0001-5443-0506
tory behavior. Among different repressilators, in this
work, we focus on the canonical repressilator with
three components (see Figure 1). Our objective is
to study the conditions for the existence of sustained
oscillations in this network. Long term perspective
application of sustained oscillations in a repressilator
could be the treatment of diseases related to circadian
rhythms, for instance, by allowing drug delivery at a
particular pace.
Figure 1: The influence graph of the canonical repressilator.
The existence of sustained oscillations in the
repressilator has been studied both mathematically
(Bus¸e et al., 2009; Bus¸e et al., 2010) and biologically
(Potvin-Trottier et al., 2016; Elowitz and Leibler,
2000). In particular, the first biological implemen-
tation of the repressilator uses three natural repressor
Sun, H., Comet, J., Folschette, M. and Magnin, M.
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks.
DOI: 10.5220/0011614300003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 29-40
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
29

proteins, the TetR, LacI and CI repressors (Elowitz
and Leibler, 2000).
In this work, we analyze mathematically the exis-
tence of sustained oscillations based on a pre-existing
formalism, which has not yet been used for model-
ing the repressilator. In fact, how to biologically im-
plement a repressilator with sustained oscillations is
still an open question, particularly in eukaryotic cells,
therefore exploring new models to search for condi-
tions for sustained oscillations is of high interest.
Previous works about mathematical analysis of
oscillations in the repressilator are mainly based on
differential equations. Many models of the repressila-
tor with three components are developed from a dif-
ferential equation model using 6 variables (Elowitz
and Leibler, 2000), with 3 variables for repressor-
protein concentrations and 3 variables for correspond-
ing mRNA concentrations. This 6-variable model
can also be reduced to a 3-variable model with only
repressor-protein variables under certain assumptions
(Bus¸e et al., 2010). These models and their variations
are extensively studied in the literature (Dukari
´
c et al.,
2019; Dil
˜
ao, 2014; Kuznetsov and Afraimovich,
2012; Bus¸e et al., 2009; M
¨
uller et al., 2006; El Samad
et al., 2005). One major limit about differential equa-
tions is that some dynamical properties are hard to an-
alyze. Finally, let us mention some pre-existing tools
that analyze dynamical properties on models such as
GNA (De Jong et al., 2003) and RoVerGeNe (Batt
et al., 2007), which are based mostly on qualitative
properties.
In this work, a class of hybrid model called
hybrid gene regulatory network (HGRN) (Behaegel
et al., 2016; Cornillon et al., 2016) is used to
model the repressilator. The HGRN is an extension
of Thomas’ discrete modeling framework (Thomas,
1973; Thomas, 1991). In HGRNs, the state space
is separated into several discrete states, as for dis-
crete models, and in each discrete state, the tempo-
ral derivative of the system is described by a constant
vector making the system evolve continuously over
time, as for differential equations. The most impor-
tant property of HGRNs is that the “sliding mode” is
allowed, which means that when a trajectory reaches
a black wall (a boundary of the discrete state which
cannot be crossed by trajectories) it is forced to move
along the black wall.
There are two major reasons why we choose
HGRNs to study synthetic circuits. Firstly, the dy-
namical complexity of HGRNs is lower than differ-
ential equations, making some dynamical properties
of HGRNs easier to prove theoretically, such as the
stability of limit cycles (Sun et al., 2022). Secondly,
from the parameters of a HGRN, we can easily derive
the time required for the expression value of one gene
to move from one threshold to another under certain
regulation, and this information can be useful for the
biological design of synthetic circuits.
The objective of this work is to find separable
constraints on the parameters of a HGRN of the re-
pressilator to ensure the existence of sustained os-
cillations. “Separable constraints on the parameters”
means that the constraints are separable in a conjunc-
tion of constraints which cover a unique parameter
each: each parameter is included individually in an in-
terval. Thus we are looking for constraints which can
be evaluated variable by variable. When all these con-
straints are satisfied, the global system shows the de-
sired behavior. The reason why we choose constraints
of separable form is that they can be easily interpreted
and used. If such separable constraints can be found
and if the individual intervals are not degenerated, the
measure of the solution space is not null leading to
a not null chance to be able to implement it in bio-
logical cells. These separable constraints represent a
bounding box in the parameter space.
This work has the following contributions:
• It is the first study of sustained oscillations in the
canonical repressilator based on HGRN.
• Similarly to the work (Sun et al., 2022), the
Poincar
´
e map is also used to analyze the stabil-
ity of a certain cycle in HGRN. But contrary to
(Sun et al., 2022) where the values of parameters
are known, in this work, the Poincar
´
e map is ana-
lyzed symbolically. By doing so, a sufficient and
necessary condition for the existence of sustained
oscillations in a HGRN of the canonical repressi-
lator is proposed for the first time.
• An intermediate result implies some new control
strategies for sustained oscillations in this HGRN
of the canonical repressilator: controlling certain
parameters such that their absolute values are suf-
ficiently small compared to others.
• The range enclosure property of Bernstein coef-
ficients, which can be used to over-approximate
the image of a polynomial function on a bounding
box, is adapted for the first time in this work to
find bounding boxes in which all models satisfy
certain conditions. Based on this method, some
bounding boxes which only contain models with
sustained oscillations are obtained.
The paper is organized as follows. In Section 2,
the HGRN framework is defined and a HGRN of the
canonical repressilator is introduced. In Section 3, we
discuss different qualitative properties of this HGRN.
In Section 4, the Poincar
´
e map is used to compute a
sufficient and necessary condition for the existence of
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
30

sustained oscillations. In Section 5, a method based
on the range enclosure property of Bernstein coeffi-
cients is proposed to find separable constraints on pa-
rameters under which this HGRN has sustained oscil-
lations. Finally, in Section 6, we make a conclusion
and discuss our future works.
2 MODELING REPRESSILATOR
WITH HGRN
This section first defines a hybrid gene regulatory net-
work (HGRN). Then a HGRN of the repressilator is
introduced.
2.1 Hybrid Gene Regulatory Network
(HGRN)
Consider a gene regulatory network with N genes,
the i
th
gene has n
i
+ 1 discrete levels which are
represented by integers:
{
0,1,2,..., n
i
}
. A dis-
crete state s is obtained by attributing a valuation
for each gene among its discrete levels. We de-
note d
s
the integer vector which describes the
discrete levels of all genes in s in order; in the
following, for simplicity, we also call d
s
a dis-
crete state. The set of all discrete states is E
d
=
d
s
∈ N
N
| ∀i ∈
{
1,2, ...,N
}
,d
i
s
∈
{
0,1, ...,n
i
}
,
where d
i
s
is the i
th
component of d
s
. Based on
the notion of discrete state, HGRNs are defined as
follows:
Definition 1 (Hybrid gene regulatory network
(HGRN)). A hybrid gene regulatory network (HGRN)
is noted H = (E
d
,c) where E
d
is the set of all discrete
states and c is a function from E
d
to R
N
. For each
d
s
∈ E
d
, c(s), also noted c
s
, is called the celerity of
discrete state d
s
and describes the temporal deriva-
tive of the system in d
s
.
In HGRNs, a hybrid state is used to fully describe
the state of the system: it contains the discrete state
in which the system currently is, and a fractional part
that represents the (normalized) position of each vari-
able inside this discrete state.
Definition 2 (Hybrid state of a HGRN). A hybrid
state of a HGRN is a couple h = (π,d
s
) containing a
fractional part π, which is a real vector in [0,1]
N
, and
a discrete state d
s
in E
d
. E
h
is the set of all hybrid
states.
Unless there is an ambiguity, a hybrid state will be
called simply a state. Based on this notion of state, a
trajectory and a boundary are defined as follow.
Definition 3 ((Hybrid) trajectory). A (hybrid) tra-
jectory τ is a function from a time interval [0,t
0
] to
E
τ
= E
h
∪ E
sh
, where t
0
∈ R
+
∪ {∞}, E
h
is the set of
all states, and E
sh
is the set of all sequences of states
(E
sh
=
(h
0
,h
1
,..., h
m
) ∈ (E
h
)
m+1
| m ∈ N ∪ {∞}
).
A trajectory represents a simulation of the system
over time. Consider a trajectory τ on [0,t
0
]. For any
t ∈ [0,t
0
], if τ(t) = (h
0
,h
1
,..., h
m
) ∈ E
sh
, this means
that there is a sequence of instant transitions at t,
which begins from h
0
, reaches h
1
at first, then reaches
h
2
,..., and finally reaches h
m
; otherwise, if τ(t) ∈ E
h
,
then the trajectory in t is made of a regular point. See
below for an illustration.
Definition 4 (Boundary). A boundary in a discrete
state d
s
is a set of states defined by e(i,π
0
,d
s
) =
(π,d
s
) ∈ E
h
| π
i
= π
0
,
, where i ∈
{
1,2, ...,N
}
,d
s
∈
E
d
and π
0
∈
{
0,1
}
. The boundary e(i,π
0
,d
s
) is in-
side the discrete state d
s
. In the rest of this paper, we
simply use e to represent a boundary.
A toy example of HGRN, not based on any real-
world biological system, is shown in Figure 2. This
example is related to a negative feedback loop with
two genes: A (first dimension) and B (second dimen-
sion), where A activates B and B inhibits A. Each
gene has two discrete levels, so there are four discrete
states in this system. Black arrows represent the celer-
ities of each discrete state and red arrows represent a
possible trajectory of this system, which happens, in
this particular case, to be a closed trajectory.
The state h
M
= ((π
1
M
,1), (1,1)) of point M be-
longs to the upper boundary e
1
in the second dimen-
sion of the discrete state 11, which is a shorthand no-
tation of (1,1). Since there is no other discrete state
on the other side of e
1
, the trajectory from h
M
cannot
cross e
1
and has to slide along e
1
. Boundaries like
e
1
, which can be reached by trajectories but cannot be
crossed, are defined as attractive boundaries. If there
was another discrete state on the other side of e
1
, in
which the celerity is negative in the second dimen-
sion (towards the boundary), then the trajectory from
h
M
could still not cross it, and in this case e
1
would
also be an attractive boundary.
The state h
P
= ((π
1
P
,0), (0,1)) of point P belongs
to the lower boundary e
2
in the second dimension of
the discrete state 01. The trajectory from h
P
will reach
instantly h
Q
= ((π
1
Q
,1), (0,0)), which belongs to the
upper boundary e
3
in the second dimension of discrete
state 00, because the celerities on both sides allow this
(instant) discrete transition. e
2
is defined as an output
boundary of 01 and e
3
is defined as an input boundary
of 00.
The trajectory in Figure 2 only reaches one new
boundary at a time, however generally a trajectory
can reach several new boundaries at the same time.
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks
31
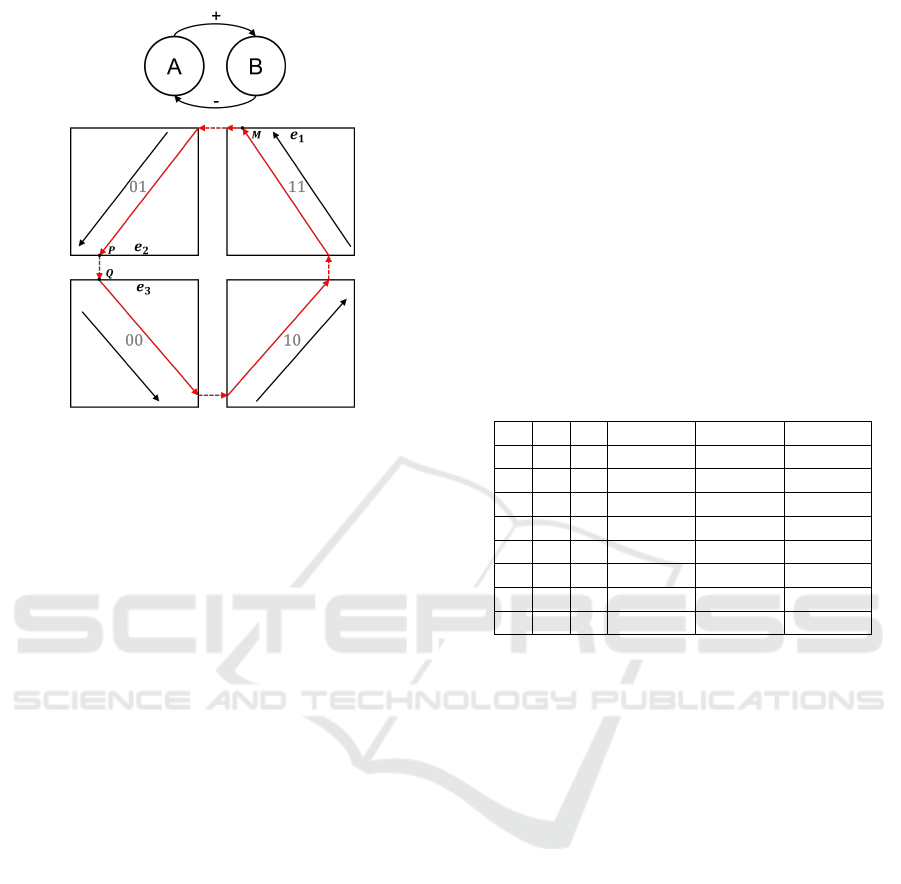
Figure 2: Example of a HGRN of negative feedback loop
with 2 genes: gene A and gene B, where A activates B and
B inhibits A. Abscissa represents the first gene (gene A) and
ordinate represents the second gene (gene B).
When a trajectory reaches several output boundaries
at the same time, it can cross any of them but can
only cross one boundary at a time, which causes non-
deterministic behaviors.
2.2 HGRN of the Repressilator
Here we only focus on the influence graph of the
canonical repressilator, see Figure 1. We assume that
each gene has one threshold when it influences one
another gene. Although, a priori, one gene can have
multiple thresholds for one another gene, in this work
we only consider the simplest case. Based on this as-
sumption, since each gene only influences one other
gene in this influence graph, it has only two discrete
levels separated by only one threshold.
The parameters (celerities) of this HGRN of the
repressilator are shown symbolically in Table 1. Each
parameter in Table 1 is strictly positive and is denoted
by C
xyiz j
, which represents the absolute value of the
celerity of variable x when the discrete level of vari-
able y is i and the discrete level of variable z is j. Con-
sider the influence of A on B: when the discrete level
of A is 1, meaning that the expression of A is above
the threshold to inhibit B, then the temporal derivative
of B is always negative, no matter the discrete level of
B (0 or 1) which corresponds to the negative values
−C
ba1b0
and −C
ba1b1
. On the other hand, when the
expression of A is below the threshold to inhibit B,
the temporal derivative of B is always positive, cor-
responding to parameters C
ba0b0
and C
ba0b1
. From
the parameters in this table, we can also see that the
number of different parameters (12) is smaller than
the multiplication of the number of dimensions by the
number of discrete states (24), because some discrete
states have celerities in common (same regulation on
some variables).
In addition to the threshold which separates the
discrete levels 0 and 1, each gene also has a maximal
value and a minimal value. For example, when A in-
hibits B (see Figure 1), B will continue to decrease
until it reaches the minimal value (most of the time
this minimal value is 0) which is related to the lower
boundary in the second dimension (the dimension of
gene B) of discrete state 10∗, where ∗ can be 0 or 1.
Similarly, when A does not inhibit B, B will continue
to increase until it reaches the upper boundary in the
second dimension of 01∗.
Table 1: Parameters of the HGRN of the repressilator.
A B C C
A
C
B
C
C
0 0 0 C
ac0a0
C
ba0b0
C
cb0c0
0 0 1 −C
ac1a0
C
ba0b0
C
cb0c1
0 1 0 C
ac0a0
C
ba0b1
−C
cb1c0
0 1 1 −C
ac1a0
C
ba0b1
−C
cb1c1
1 0 0 C
ac0a1
−C
ba1b0
C
cb0c0
1 0 1 −C
ac1a1
−C
ba1b0
C
cb0c1
1 1 0 C
ac0a1
−C
ba1b1
−C
cb1c0
1 1 1 −C
ac1a1
−C
ba1b1
−C
cb1c1
Figure 3 gives two simulations with two different
choices of parameters. The simulation on the left rep-
resents a sustained oscillation while the simulation on
the right represents a damped oscillation. In the sim-
ulation on the left, gene C continues to increase from
t = 0 until it reaches the maximal value, which also
means that the trajectory reaches an attractive bound-
ary, then the trajectory will slide along this boundary
(called the sliding mode) so that the value of gene C
stays unchanged for some time. The sliding mode is
an important property of HGRNs.
3 QUALITATIVE BEHAVIORS IN
THIS HGRN OF THE
REPRESSILATOR
In this section, we discuss different qualitative prop-
erties of this HGRN. To analyze dynamical proper-
ties of a HGRN, we need to firstly analyze the transi-
tion graph of discrete states, which is determined by
the signs of celerities. The signs of celerities of this
HGRN of the repressilator can be found in Table 1,
based on which the transition graph of discrete states
can be constructed using the classical discrete asyc-
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
32
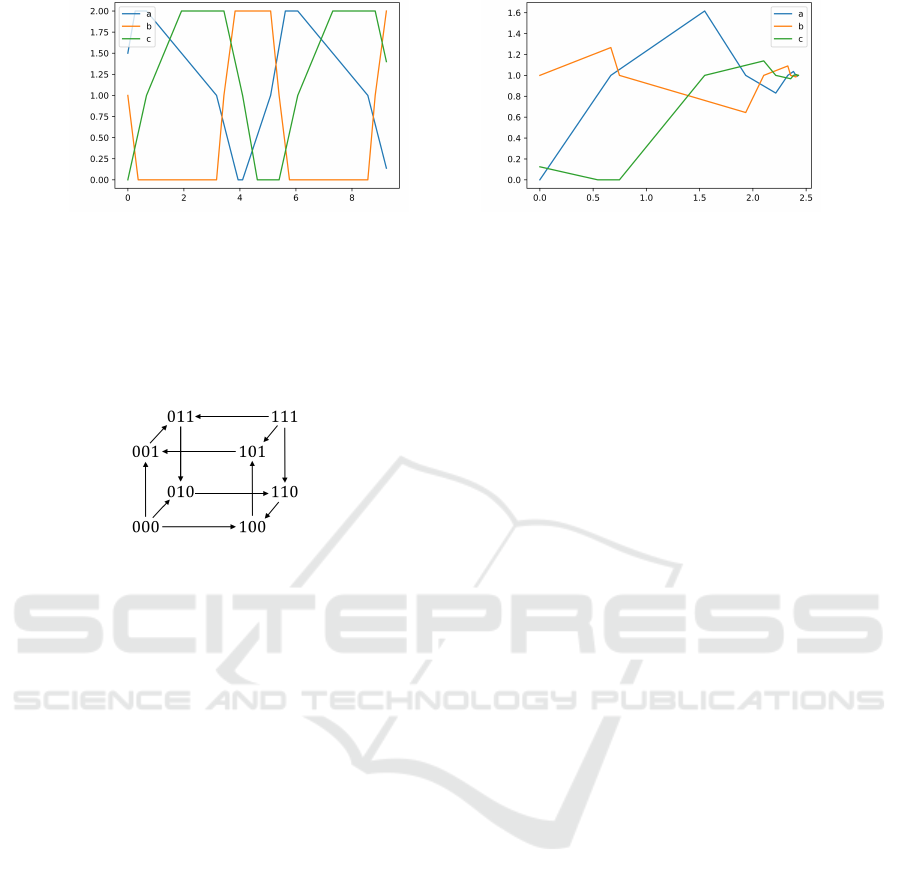
Figure 3: Simulations of the HGRN of the repressilator with two different choices of parameters (Abscissa represents time
and ordinate represents the sum of the fractional part and the discrete state of each gene). Parameters of the model on
the left: C
ac0a0
= 1, C
ac0a1
= 1.9, C
ac1a0
= 1.3, C
ac1a1
= 0.4, C
ba0b0
= 3.8, C
ba0b1
= 2.5, C
ba1b0
= 2.7, C
ba1b1
= 3.3,
C
cb0c0
= 1.5, C
cb0c1
= 0.8, C
cb1c0
= 1.9, C
cb1c1
= 1.5. Parameters of the model on the right: C
ac0a0
= 1.5, C
ac0a1
= 0.7,
C
ac1a0
= 0.6, C
ac1a1
= 1.6, C
ba0b0
= 2.1, C
ba0b1
= 0.4, C
ba1b0
= 0.3, C
ba1b1
= 3.3, C
cb0c0
= 1.25, C
cb0c1
= 0.25, C
cb1c0
= 0.23,
C
cb1c1
= 1.23.
nhronous semantics, see Figure 4.
Figure 4: Transition graph of discrete states of the HGRN
of the repressilator.
From the transition graph, we can see that there
is a unique cycle of discrete states, which is 001 −→
011 −→ 010 −→ 110 −→ 100 −→ 101 −→ 001. This cycle
is a global attractor of discrete states (the unique ter-
minal strongly connected component), which means
that any trajectory in this HGRN will finally enter this
cycle.
Following the same sequence of discrete
states, there is a special hybrid trajectory:
((1,1, 0),(0, 0,1)) −→ ((1,0, 0),(0, 1,1)) −→
((1,0, 1),(0, 1,0)) −→ ((0,0, 1),(1, 1,0)) −→
((0,1, 1),(1, 0,0)) −→ ((0,1, 0),(1, 0,1)) −→
((1,1, 0),(0, 0,1)). It is illustrated in Figure 5
by green arrows. This trajectory contains 6 different
states (the last and the first one are identical). From
each state in this trajectory, there is an instant tran-
sition (transition which crosses boundaries between
discrete states and takes no time), which reaches the
next state. This trajectory represents also a closed
trajectory, meaning that beginning from each of
these 6 states, the trajectory will return to the initial
state. When representing trajectories, we often use
an embedding of hybrid states in R
N
: a state (π,d
s
)
is represented in R
N
by summing its discrete and
fractional parts: π + d
s
. We say that the hybrid state
(π,d
s
) and the point π + d
s
∈ R
N
are related. When
doing so, the six previous hybrid states are embedded
in the same point H
0
in R
3
: H
0
= (1.0,1.0, 1.0). H
0
is called a characteristic state of this HGRN. The
characteristic state is formally defined as follows:
Definition 5 (Characteristic state). A characteristic
state of a HGRN is a state H in the continuous space
such that: for any hybrid state h
0
related to H, all
trajectories from h
0
will never reach a hybrid state
which is not related to H, and there exist oscillations
in any small neighborhood of H.
In this paper, we say that a trajectory of a HGRN
is an oscillation if it is related to an oscillation in the
continuous space. Likewise, the nature of an oscilla-
tion of a HGRN (damped or sustained) and the rela-
tion between an oscillation and a characteristic state
(converging to the characteristic state, moving away
from it, etc.) are determined by the related oscilla-
tion in the continuous space. A neighborhood of a
characteristic state H is defined as a set N (H) =
{
(π,d
s
) ∈ E
h
| ∥π + d
s
− H∥ < r
}
where r ∈ R is the
radius of the neighborhood.
In the continuous space, a characteristic state is a
fixed point, because from any hybrid state h related
to a characteristic state, all trajectories can only reach
hybrid states related to this characteristic state.
We can easily prove that H
0
is a characteristic
state: apart from the 6 hybrid states on this closed tra-
jectory, there are two other hybrid states which are re-
lated to H
0
: ((1,1, 1),(0,0,0)) and ((0,0,0), (1,1, 1))
From each of these two other states, all trajectories
reach directly the closed trajectory, and once they do,
they can never leave it. Finally, we can find oscilla-
tions which follow the unique cycle of discrete states
in any neighborhood of H
0
. In this HGRN, there is
only one characteristic state. This can be proved by
verifying all “corners” of discrete states.
All trajectories in this HGRN will oscillate in this
unique cycle of discrete states, except some special
trajectories from discrete state 000 or 111 which can
reach directly a hybrid state related to the characteris-
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks
33
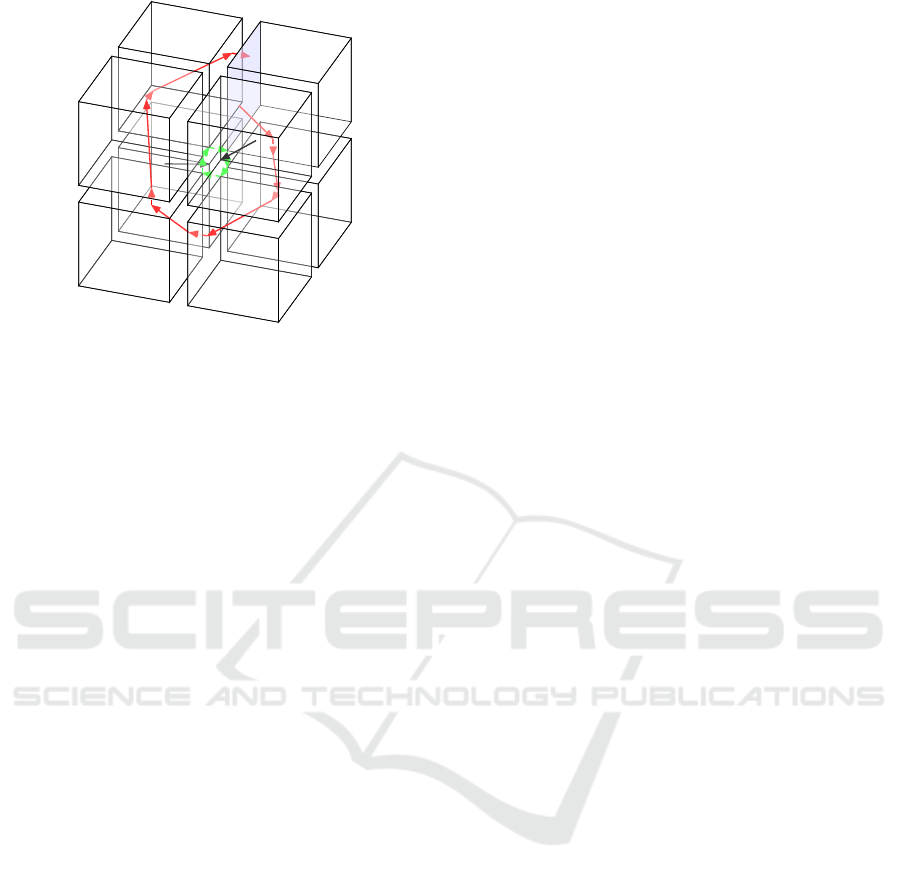
000
100
t
4
010
t
2
001
t
6
s
7
011
s
1
t
1
101
t
5
110
t
3
111
Figure 5: Illustration of the closed trajectory with only
instant transitions (green arrows), two special trajectories
which can reach directly the characteristic state (black ar-
rows), and a trajectory without sliding mode (red arrows) in
this HGRN of the canonical repressilator.
tic state, see for example the black arrows in Figure 5.
These oscillations can have several different dynami-
cal properties.
To better illustrate the possible dynamical prop-
erties in this repressilator, different qualitative behav-
iors of a HGRN of negative feedback loop in 2 dimen-
sions are illustrated in Figure 6, where black arrows
represent celerities in each discrete state and red ar-
rows represent some trajectories. The three figures
are obtained by choosing three different parameteri-
sations, which represent the three qualitative behav-
iors of this HGRN. The closed trajectory with states
P,M,N, Q contains only instant transitions. The state
in continuous space that is related to states P,M,N, Q
is a characteristic state, and is the only characteris-
tic state in this HGRN. On the left, the characteristic
state is stable as all trajectories tend to converge to it.
In this case, there is no sustained oscillation. On the
right, the characteristic state is unstable as all trajec-
tories from a small neighborhood of the characteristic
state will move away from it and will finally reach a
stable limit cycle containing at least one sliding mode,
like the closed trajectory in Figure 2. Between these
two cases, in the middle, there is a third possibility in
which all trajectories circle around the characteristic
state without getting closer or moving away, which we
call parallel cycles and can be considered as a special
case of sustained oscillations.
The HGRN of the repressilator in 3 dimensions
(Figure 1) is more complicated than the example
HGRN of negative feedback loop in 2 dimensions
(Figures 2 and 6), but we think that it also has three
similar qualitative behaviors, since each discrete state
in the only cycle of discrete states has only one suc-
cessor (Figure 4). However, we have no proof for the
non-existence of other possibilities yet, for example
chaos or the co-existence of sustained oscillations and
a stable fixed point. So in this work, we make Hypoth-
esis 1.
Hypothesis 1. In this HGRN of canonical repressila-
tor (see Figure 1 and Table 1), either the characteris-
tic state is stable and all oscillations are damped, or
the characteristic state is unstable and all oscillations
are sustained.
In this HGRN of the canonical repressilator, the
characteristic state is said stable if we can find a small
neighborhood around the characteristic state such that
all oscillations which begin from this neighborhood
converge to the characteristic state, and the character-
istic state is said unstable if there is no damped oscil-
lation which converges to it.
Now, based on Hypothesis 1, we can use the sta-
bility of the characteristic state to determine the ex-
istence of sustained oscillations in this HGRN of the
repressilator: all oscillations are sustained if and only
if the characteristic state is unstable. This is the main
idea used in this work to find conditions for the ex-
istence of sustained oscillations. A similar idea was
used in other works with differential models, see for
example (Page and Perez-Carrasco, 2018; Wang et al.,
2006; El Samad et al., 2005).
The problem now is how to analyze the stability of
the characteristic state. To do so, we apply a method
based on the Poincar
´
e map.
4 ANALYSIS OF THE POINCAR
´
E
MAP
In this section, a method based on the Poincar
´
e map is
proposed to compute a sufficient and necessary con-
dition for the existence of sustained oscillations.
4.1 Poincar
´
e Map in HGRNs
The Poincar
´
e map was initially proposed to study
limit cycles of nonlinear dynamical systems and has
also been used later to study limit cycles of hybrid
systems (Belgacem et al., 2020; Firippi and Chaves,
2020; Znegui et al., 2020; Flieller et al., 2006; Ed-
wards and Glass, 2005; Girard, 2003; Hiskens, 2001;
Edwards, 2000; Mestl et al., 1996). The Poincar
´
e map
is the intersection of a periodic orbit with a lower di-
mension subspace which is called the Poincar
´
e sec-
tion. The properties of the periodic orbit can be de-
rived from the Poincar
´
e map. In the case of the re-
pressilator studied in this paper, the blue boundary in
Figure 5 could be chosen as a Poincar
´
e section.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
34

Figure 6: Illustration of different qualitative behaviors in a HGRN of negative feedback loop in 2 dimensions. The three
subfigures represent three different choices of parameters.
A method based on the Poincar
´
e map was pro-
posed in (Sun et al., 2022) to analyze the stability of
limit cycles in HGRNs. To illustrate this method, con-
sider another example in 2 dimensions, with an addi-
tional discrete level in the first dimension, as shown
in Figure 7. In this example, we assume that all pa-
rameters (celerities) are known, that is, their real val-
ues can directly be used. The upper boundary in the
second dimension of discrete state 00 is chosen as
Poincar
´
e section. We can see that there is a closed tra-
jectory which intersects this Poincar
´
e section at state
P = (π
P
,(0, 0)), where π
P
∈ [0, 1]
2
is the fractional
part of state P. We note that in this particular case,
π
P
= (x,1) where x ∈ [0,1], thus there is only one
varying dimension to study in this Poincar
´
e section.
According to the properties of HGRNs, we can find
on the Poincar
´
e section a neighborhood N around P,
such that trajectory from any state M = (π
M
,(0, 0))
from N will return to the Poincar
´
e section at state
M
′
= (π
M
′
,(0, 0)) and the relation between π
M
and
π
M
′
can be described by an affine matrix application:
π
M
′
= G(π
M
) = Aπ
M
+ b (1)
where A is a matrix and b is a vector. We note that
the form of Equation (1) is general and applies to any
case, including those of dimension higher than 2. At
this point, the stability of the closed trajectory de-
pends on the eigenvalues of A. For more details, see
(Sun et al., 2022).
In this work, we adapt this method based on the
Poincar
´
e map to analyze the stability of the character-
istic state, which is a special case of closed trajectory,
in order to find conditions for sustained oscillations.
As opposed to (Sun et al., 2022) where parameters are
assumed to be known, here we compute and analyze
symbolically the Poincar
´
e map.
4.2 Symbolic Poincar
´
e Map
In order to analyze the stability of the characteristic
state, we only need to consider trajectories without
sliding mode around this state. Indeed, the charac-
teristic state is stable if we can find a small neigh-
borhood around the characteristic state such that all
Figure 7: Illustration of the Poincar
´
e map in a HGRN.
oscillations which begin from this neighborhood con-
verge to the characteristic state. Such a trajectory ex-
ists because otherwise, the celerities would prevent
the characteristic state from existing. Without loss
of generality, we choose the lower boundary in the
second dimension of discrete state 011 as Poincar
´
e
section; see the blue boundary in Figure 5. Now, we
consider any trajectory τ which begins from a state
s
1
= ((x
1
,0, z
1
),(0, 1,1)) on the Poincar
´
e section and
returns to the Poincar
´
e section for the first time at
s
7
= ((x
7
,0, z
7
),(0, 1,1)) without sliding mode; such
a trajectory is illustrated in red in Figure 5. Thus,
the Poincar
´
e map is an affine application describing
the relation between (x
1
,0, z
1
) and (x
7
,0, z
7
). Since
s
1
is on an input boundary of the discrete state 011,
from s
1
, τ will first (continuously) cross the discrete
state 011 and reach a state ((x
2
,y
2
,0), (0,1, 1)) on the
lower boundary in the third dimension of 011 which is
the output boundary of this discrete state towards 010.
We name this output boundary e
1
. Then, it crosses in-
stantly e
1
and reaches an input boundary of 010 in
state ((x
2
,y
2
,1), (0,1, 0)). The duration of crossing in
discrete state 011 is:
t
1
=
0 − z
1
−C
cb1c1
(2)
It should be noted that, to ensure that this trajec-
tory τ has no sliding mode in 011, we also need to
ensure that the lower boundary in the first dimension
and the upper boundary in the second dimension of
011 are not reached before e
1
, which gives us two ad-
ditional inequalities:
t
1
<
0 − x
1
−C
ac1a0
(3)
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks
35

t
1
<
1 − 0
C
ba0b1
(4)
In fact, these inequalities can always be satisfied if τ
is sufficiently close to the characteristic state, which
is the case we consider here. Therefore, in the rest
of this section, we do not consider these additional
constraints.
Based on the duration t
1
and the fact that there
is no sliding mode, we can get the relation between
(x
1
,0, z
1
) and (x
2
,y
2
,1):
x
2
= x
1
−C
ac1a0
×t
1
(5)
y
2
= 0 +C
ba0b1
×t
1
(6)
Following the same process, we can get the du-
ration of τ in each discrete state and the relations be-
tween states from one input boundary to another input
boundary:
t
2
=
1 − x
2
C
ac0a0
t
3
=
0 − y
3
−C
ba1b1
t
4
=
1 − z
4
C
cb0c0
(7)
t
5
=
0 − x
5
−C
ac1a1
t
6
=
1 − y
6
C
ba0b0
(8)
y
3
= y
2
+C
ba0b1
×t
2
z
3
= 1 −C
cb1c0
×t
2
(9)
x
4
= 0 +C
ac0a1
×t
3
z
4
= z
3
−C
cb1c0
×t
3
(10)
x
5
= x
4
+C
ac0a1
×t
4
y
5
= 1 −C
ba1b0
×t
4
(11)
y
6
= y
5
−C
ba1b0
×t
5
z
6
= 0 +C
cb0c1
×t
5
(12)
x
7
= 1 −C
ac1a0
×t
6
z
7
= z
6
+C
cb0c1
×t
6
(13)
where t
2
,t
3
,t
4
,t
5
,t
6
are the durations of τ in dis-
crete states 010, 110, 100, 101, 001 respectively, and
(0,y
3
,z
3
),(x
4
,1, z
4
),(x
5
,y
5
,0), (1,y
6
,z
6
),(x
7
,0, z
7
)
are the fractional parts of the states when τ first
reaches 110, 100, 101, 001, 011 respectively.
Based on the above equations, the Poincar
´
e map
can be calculated to describe the relation between
(x
1
,0, z
1
) and (x
7
,0, z
7
) as follows. One dimension is
missing in the matrix below; indeed, this dimension
is useless in the computation of the stability, for more
details see (Sun et al., 2022).
x
7
z
7
=
b
1
c
1
b
2
c
2
x
1
z
1
+
a
1
a
2
(14)
In the above equation, a
1
, a
2
, b
1
, b
2
, c
1
, c
2
are nonlin-
ear combinations of the celerity values. Their expres-
sions are given in Supplementary Material Section 1.
We can easily derive that b
1
and c
2
are strictly posi-
tive, while b
2
and c
1
are strictly negative.
4.3 Analysis of Eigenvalues
The stability of the characteristic state depends on the
two eigenvalues of
b
1
c
1
b
2
c
2
, which are:
λ
1
=
b
1
+ c
2
+
p
(b
1
− c
2
)
2
+ 4c
1
b
2
2
(15)
λ
2
=
b
1
+ c
2
−
p
(b
1
− c
2
)
2
+ 4c
1
b
2
2
(16)
Property 1. These two eigenvalues are real and
strictly positive.
The proof of Property 1 is given in Supplementary
Material Section 3.
Suppose that two eigenvectors which are related
to λ
1
and λ
2
respectively are v
1
= (v
1
1
,v
2
1
) and v
2
=
(v
1
2
,v
2
2
). We have the following property on v
1
and v
2
.
Property 2. v
1
1
× v
2
1
< 0 and v
1
2
× v
2
2
> 0.
The proof of Property 2 is given in Supplementary
Material Section 4.
Based on these properties, we develop the follow-
ing theorem to verify the stability of the characteristic
state.
Theorem 1. The characteristic state is unstable if
and only if λ
1
≥ 1.
The proof of Theorem 1 is given in Supplementary
Material Section 5.
Based on Hypothesis 1 and Theorem 1, the condi-
tion λ
1
≥ 1 is a sufficient and necessary condition for
the existence of sustained oscillations in this HGRN
of canonical repressilator.
Since our final objective is to provide practical in-
formation for the construction of synthetic networks,
conditions like λ
1
≥ 1 might not be a good result,
because the set of models under this constraint is
not easy to figure. In the next section, a method to
compute separable constraints based on the condition
λ
1
≥ 1 is presented.
5 COMPUTATION OF
SUFFICIENT SEPARABLE
CONSTRAINTS ON
PARAMETERS
In this section, we propose a method to compute sepa-
rable constraints on parameters based on the condition
λ
1
≥ 1 which is developed in the previous section.
In this paper, separable constraints mean constraints
with separable form: each parameter is constrained
by an interval, for example C
ac0a0
∈ [C
ac0a0
,C
ac0a0
].
In other words, these separable constraints represent
a n-dimensional bounding box in the space of param-
eters (celerities). What we want to do is to find such a
n-dimensional box so that any model in this bounding
box satisfies the condition λ
1
≥ 1, which means that
any model in this box has sustained oscillations.
Firstly, we present a simplification of the condi-
tion λ
1
≥ 1. Secondly, we introduce a method to ver-
ify if all models in a given bounding box satisfy this
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
36

simplified condition. At last, using the method in the
second part, we propose a search algorithm to find
some separable constraints.
5.1 Condition Simplification
The condition:
λ
1
=
b
1
+ c
2
+
p
(b
1
− c
2
)
2
+ 4c
1
b
2
2
≥ 1 (17)
can be reformulated as:
b
1
+ c
2
− 2 ≥ −
q
(b
1
− c
2
)
2
+ 4c
1
b
2
(18)
which is equivalent to:
(b
1
+ c
2
− 2 ≥ 0) ∨
((b
1
+ c
2
− 2 < 0) ∧
((b
1
+ c
2
− 2)
2
≤ (b
1
− c
2
)
2
+ 4c
1
b
2
))
(19)
or:
(b
1
+ c
2
− 2 ≥ 0) ∨ (b
1
c
2
− c
1
b
2
− b
1
− c
2
+ 1 ≤ 0)
(20)
This last condition is equivalent to (P
1
≥ 0)∨(P
2
≥ 0)
where P
1
and P
2
are polynomials on parameters. The
expressions of P
1
and P
2
are given in Supplementary
Material Section 2.
Condition (P
1
≥ 0) ∨ (P
2
≥ 0) seems preferable
to λ
1
≥ 1 because it only contains polynomials. In
fact, one can easily prove that solutions for (P
1
≥
0) ∨ (P
2
≥ 0) exist. For example, by only consider-
ing Cac0a0 and Cac0a1, which are two parameters
describing the derivative of gene A when gene C does
not inhibit gene A, P
1
and P
2
can be expressed by:
P
1
=p
11
×C
ac0a0
+ p
12
×C
ac0a1
+ p
13
×C
ac0a0
×C
ac0a1
+ p
14
(21)
P
2
=p
21
×C
ac0a0
+ p
22
×C
ac0a1
+ p
23
×C
ac0a0
×C
ac0a1
+ p
24
(22)
where p
i j
(with i ∈
{
1,2
}
, j ∈
{
1,2, 3,4
}
) are expres-
sions of parameters which do not include C
ac0a0
and
C
ac0a1
. We can see that if C
ac0a0
and C
ac0a1
converge
to 0 while other parameters remain unchanged, P
1
and P
2
converge to p
14
and p
24
respectively, which
are both positive. This indicates that the solutions
of (P
1
≥ 0) ∨ (P
2
≥ 0) exist and also implies a new
control strategy for the existence of sustained oscil-
lations: controlling the derivatives of gene A when
A is not inhibited by C such that these derivatives are
sufficiently small, while keeping other parameters un-
changed.
5.2 Satisfiability Under Separable
Constraints
In this subsection, we adapt the range enclosure prop-
erty of Bernstein coefficients to verify if all models
in a given bounding box satisfy the condition (P
1
≥
0) ∨ (P
2
≥ 0). The Bernstein coefficients have been
used in the literature to, for example, compute images
for polynomial dynamical system (Dang and Salinas,
2009; Dang and Testylier, 2012), or compute affine
lower bound functions for polynomials (Garloff and
Smith, 2003), etc.
Before introducing the Bernstein coefficients, we
firstly introduce the notion of multi-indice. A multi-
indice is a vector of non-negative integers. Given two
multi-indice i = (i
1
,i
2
,..., i
n
) and j = ( j
1
, j
2
,..., j
n
),
we write i ≤ j if ∀k ∈
{
1,2, ...,n
}
, i
k
≤ j
k
. We
also write
i
j
for (
i
1
j
1
,
i
2
j
2
,...,
i
n
j
n
) and
j
i
for
j
1
i
1
j
2
i
2
...
j
n
i
n
which is the multiplication of all bi-
nomial coefficient
j
k
i
k
, k ∈
{
1,2, ...,n
}
.
Using the multi-indice, a polynomial f : R
n
→ R
can be represented as follows:
f (x) =
∑
i∈I
d
a
i
x
i
(23)
where a
i
∈ R, i and d are multi-indices, I
d
is set of
all multi-indices i such that i ≤ d, and x
i
= x
i
1
1
x
i
2
2
...x
i
n
n
which is the product of all x
i
j
j
, where x
j
is the j
th
vari-
able of polynomial f .
f can also be expressed by Bernstein expansion as
follows:
f (x) =
∑
i∈I
d
b
i
B
d,i
(x) (24)
where
B
d,i
(x) = β
d
1
,i
1
(x
1
)...β
d
n
,i
n
(x
n
) (25)
β
d
k
,i
k
(x
k
) =
d
k
i
k
x
i
k
k
(1 − x
k
)
d
k
−i
k
(26)
b
i
=
∑
j≤i
i
j
d
j
a
j
(27)
where d and i are multi-indices and k ∈
{
1,2, ...,n
}
.
The values b
i
, for i ∈ I
d
are called Bernstein coeffi-
cients.
One fundamental property of Bernstein coeffi-
cients for our approach is the range enclosure prop-
erty, which can be derived from the convex hull prop-
erty. The convex hull of a set S, noted Conv(S), is the
smallest convex set that contains S.
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks
37
Lemma 1 (Convex hull property).
Conv
{
(x, f (x)) | x ∈ B
}
⊆ Conv
{
(i/d, b
i
) | i ∈ I
d
}
.
Lemma 2 (Range enclosure property).
min
{
b
i
| i ∈ I
d
}
≤ f (x) ≤ max
{
b
i
| i ∈ I
d
}
, ∀x ∈ B,
where B = [0, 1]
n
is the unit box.
The range enclosure property over-approximates
the range of image of f on B and can be used to verify
if all models in a given bounding box satisfy the con-
dition (P
1
≥ 0) ∨(P
2
≥ 0). To do so, we need to firstly
make a change of variables of polynomials P
1
and P
2
,
such that all variables are included in [0,1]. For exam-
ple, the variable C
ac0a0
∈ [C
ac0a0
,C
ac0a0
] is replaced
by C
ac0a0
= (C
ac0a0
−C
ac0a0
)× X
ac0a0
+C
ac0a0
, where
X
ac0a0
∈ [0,1]. By doing so, we get two new polyno-
mials P
′
1
and P
′
2
.
So now, to verify if (P
1
≥ 0) ∨ (P
2
≥ 0) is always
true in a given bounding box, we only need to ver-
ify if (P
′
1
≥ 0) ∨ (P
′
2
≥ 0) is always true in the unit
box. To do so, we compute the Bernstein coeffi-
cients
{
b
1,i
}
and
{
b
2,i
}
(where i ∈ I
d
) of P
′
1
and P
′
2
respectively. A sufficient condition for the condition
“(P
′
1
≥ 0) ∨ (P
′
2
≥ 0) is always true in the unit box”
(condition1) is “(∀i ∈ I
d
, b
1,i
≥ 0) ∨ (∀i ∈ I
d
, b
2,i
≥ 0)”
(condition2), according to the range enclosure prop-
erty. In fact, since the minimum value of the image
of P
′
1
on the unit box is always larger or equal to the
minimum value of
{
b
1,i
}
, “
{
b
1,i
}
are not negative”
(∀i ∈ I
d
, b
1,i
≥ 0) indicates that “P
′
1
are not negative on
the unit box”, and the same holds for P
′
2
. Since there is
a finite number of Bernstein coefficients, condition2
can be verified. Therefore, in this work, condition2 is
used to verify if all models in a given bounding box
always have sustained oscillations.
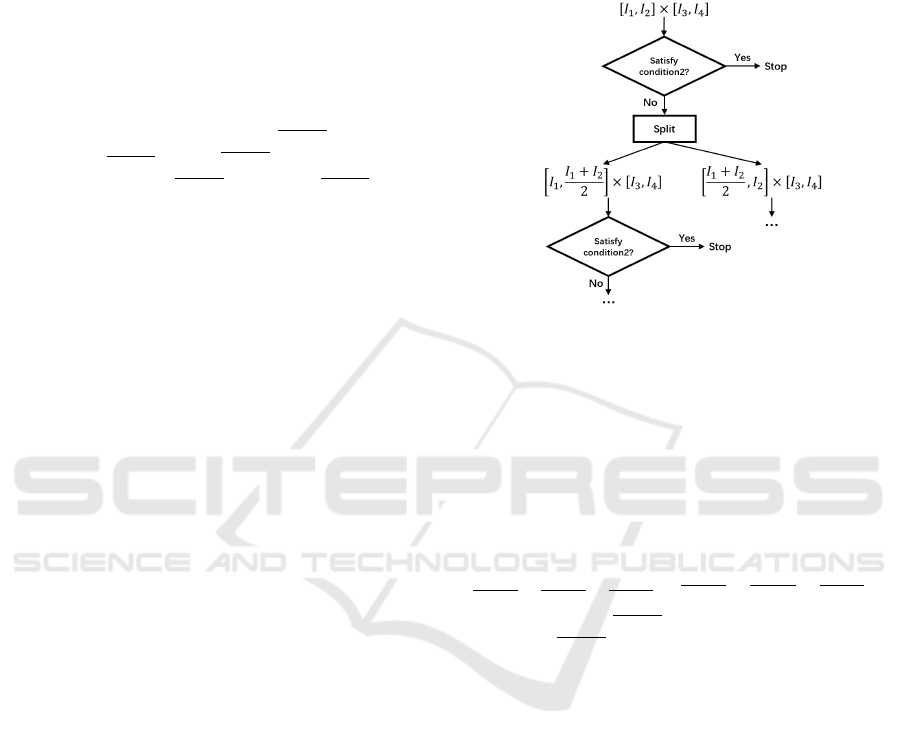
5.3 Search of Separable Constraints
Based on the method introduced in the previous sub-
section, we propose a depth first algorithm to find
some bounding boxes which satisfy condition2. This
algorithm is illustrated in Figure 8. Initially, each pa-
rameter is included in an interval. In this implementa-
tion, without loss of generality, we assume that each
parameter is included initially in [0, 1]. Then, we ver-
ify if condition2 is satisfied for this bounding box us-
ing the method proposed in the previous subsection.
If it is satisfied, then it is a bounding box such that all
models in it have sustained oscillations. If condition2
is not satisfied, then there might be some models in
this bounding box which do not have sustained oscil-
lations, in this case the bounding box is split into two
smaller bounding boxes (by splitting the largest in-
terval into two) which have the same volume and the
process is repeated on each of these two new bound-
ing boxes. Each path in this algorithm will stop, ei-
ther when a bounding box which satisfies condition2
is found or when the length of the largest interval is
smaller than a certain threshold. In fact, similar ideas
are widely used to find solution sets under non-linear
constraints (Ziat et al., 2019; Pelleau et al., 2013).
Figure 8: Illustration of the algorithm to search for some
bounding boxes.
Since the HGRN of the canonical repressilator has
12 parameters (see Table 1), and if we assume that
the number of possible smallest intervals for each pa-
rameter are the same, noted m, then there are at most
m
12
smallest bounding boxes to check. Verifying all
these boxes can be time consuming. In our imple-
mentation, we assume that the intervals of these three
genes are identical, which means that we search for
bounding boxes such that C
acia j
= C
baib j
= C
cbic j
and
C
acia j
= C
baib j
= C
cbic j
for any i, j ∈
{
0,1
}
, where, for
example, [C
ac0a0
,C
ac0a0
] is the interval of the param-
eter C
ac0a0
, so that we only need to consider 4 inde-
pendent intervals when searching for bounding boxes.
This assumption is only applied here to decrease the
number of possible bounding boxes. Similar assump-
tion about the symmetry between these three genes
was also made in works based on differential equa-
tions, see for example (Bus¸e et al., 2010). We also
assume that the minimal length of interval is greater
or equal to 0.5. A value smaller than 0.5 could natu-
rally be chosen, but this might exponentially increase
the number of possible bounding boxes, which could
also exponentially increase the execution time. With
these assumptions, we obtain 5 bounding boxes which
satisfy the condition2. In the results below, (y,x) rep-
resents (c, a), (a,b) and (b,c), where y inhibits x, for
example C
xy0x0
presents C
ac0a0
, C
ba0b0
and C
cb0c0
.
Bounding box 1: C
xy0x0
∈ [0,0.5],C
xy0x1
∈
[0,0.5],C
xy1x0
∈ [0.5, 1],C
xy1x1
∈ [0, 1]
Bounding box 2: C
xy0x0
∈ [0,0.5],C
xy0x1
∈
[0.5,1],C
xy1x0
∈ [0, 0.5],C
xy1x1
∈ [0, 0.5]
Bounding box 3: C
xy0x0
∈ [0,0.5],C
xy0x1
∈
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
38

[0.5,1],C
xy1x0
∈ [0.5, 1],C
xy1x1
∈ [0, 1]
Bounding box 4: C
xy0x0
∈ [0.5,1],C
xy0x1
∈
[0.5,1],C
xy1x0
∈ [0, 0.5],C
xy1x1
∈ [0, 0.5]
Bounding box 5: C
xy0x0
∈ [0.5,1],C
xy0x1
∈
[0.5,1],C
xy1x0
∈ [0.5, 1],C
xy1x1
∈ [0, 1]
We can see that these constraints are easy to in-
terpret and some intuitive results can be derived from
them: for instance, from Bounding box 2, we can get
that sustained oscillations exist if the values of C
xy0x1
are close to each other and are sufficiently larger than
any C
xy0x0
, C
xy1x0
and C
xy1x1
.
6 CONCLUSIONS
In this work, a HGRN of the canonical repressila-
tor is constructed. By computing and analyzing an-
alytically a Poincar
´
e map and based on a hypothe-
sis, a sufficient and necessary condition for the ex-
istence of sustained oscillations is developed. Then
the range enclosure property of Bernstein coefficients
is adapted to find some bounding boxes in parame-
ters space which satisfy this sufficient and necessary
condition. These bounding boxes (intervals of param-
eters) can provide useful information for the design
of synthetic circuits. Moreover, an intermediate re-
sult implies some new control strategies for sustained
oscillations: controlling the absolute values of the
derivatives of one gene under certain regulation such
that these values are sufficiently small, while keeping
other parameters of the system unchanged.
Naive assumptions about the bounding boxes are
made in this work. For example, we assume that
the influence between these three genes are symmetri-
cal. In other words, some groups of parameters (such
as C
ac0a0
, C
ba0b0
and C
cb0c0
) are constrained by the
same intervals. These assumptions could be replaced
by more realistic ones with access to more biologi-
cal knowledge. For instance, knowing the intervals of
possible values for some parameters would allow to
speed up the enumeration of the bounding boxes.
Only the canonical repressilator with three com-
ponents is considered in this work. This method could
be extended for more complex influence graphs that
exist in the literature (Page, 2019; Perez-Carrasco
et al., 2018; Goh et al., 2008). However, for more
complex influence graphs, conditions for the exis-
tence of sustained oscillations can be harder to de-
velop, as there could be several cycles of discrete
states around one characteristic state: in such a case,
one would have to consider the disjunction of the con-
ditions associated with each cycle. It is also possible
to extend this work to find condition for other dynam-
ical properties expressed with temporal logics.
In this work, constraints are obtained based on
given dynamical properties. In future works, the con-
verse approach might be considered, that is, given a
bounding box of parameters, predict possible behav-
iors of the system.
ACKNOWLEDGEMENTS
We would like to thank Gilles Bernot and Thao Dang
for their fruitful discussions.
This work is partly supported by China Scholar-
ship Council.
ADDITIONAL INFORMATION
Supplementary Material is available at https://hal.
archives-ouvertes.fr/hal-03890505/document. The
code of this work is available at https://github.com/
Honglu42/HGRN repressilator.
REFERENCES
Batt, G., Belta, C., and Weiss, R. (2007). Model check-
ing genetic regulatory networks with parameter uncer-
tainty. In International Workshop on Hybrid Systems:
Computation and Control, pages 61–75. Springer.
Behaegel, J., Comet, J.-P., Bernot, G., Cornillon, E., and
Delaunay, F. (2016). A hybrid model of cell cycle in
mammals. Journal of bioinformatics and computa-
tional biology, 14(01):1640001.
Belgacem, I., Gouz
´
e, J.-L., and Edwards, R. (2020). Control
of negative feedback loops in genetic networks. In
2020 59th IEEE Conference on Decision and Control
(CDC), pages 5098–5105. IEEE.
Bus¸e, O., Kuznetsov, A., and P
´
erez, R. A. (2009). Ex-
istence of limit cycles in the repressilator equa-
tions. International Journal of Bifurcation and Chaos,
19(12):4097–4106.
Bus¸e, O., P
´
erez, R., and Kuznetsov, A. (2010). Dynamical
properties of the repressilator model. Physical Review
E, 81(6):066206.
Chaves, M. and Gouz
´
e, J.-L. (2011). Exact control of ge-
netic networks in a qualitative framework: the bistable
switch example. Automatica, 47(6):1105–1112.
Chaves, M. and Jong, H. d. (2021). Qualitative modeling,
analysis and control of synthetic regulatory circuits.
Synthetic Gene Circuits, pages 1–40.
Cornillon, E., Comet, J.-P., Bernot, G., and En
´
ee, G. (2016).
Hybrid gene networks: a new framework and a soft-
ware environment. advances in Systems and Synthetic
Biology.
Dang, T. and Salinas, D. (2009). Image computation for
polynomial dynamical systems using the bernstein ex-
Condition for Sustained Oscillations in Repressilator Based on a Hybrid Modeling of Gene Regulatory Networks
39

pansion. In International Conference on Computer
Aided Verification, pages 219–232. Springer.
Dang, T. and Testylier, R. (2012). Reachability analysis
for polynomial dynamical systems using the bernstein
expansion. Reliab. Comput., 17(2):128–152.
De Jong, H., Geiselmann, J., Hernandez, C., and Page, M.
(2003). Genetic network analyzer: qualitative simu-
lation of genetic regulatory networks. Bioinformatics,
19(3):336–344.
Dil
˜
ao, R. (2014). The regulation of gene expression in eu-
karyotes: bistability and oscillations in repressilator
models. Journal of theoretical biology, 340:199–208.
Dukari
´
c, M., Errami, H., Jerala, R., Lebar, T., Romanovski,
V. G., T
´
oth, J., and Weber, A. (2019). On three genetic
repressilator topologies. Reaction Kinetics, Mecha-
nisms and Catalysis, 126(1):3–30.
Edwards, R. (2000). Analysis of continuous-time switching
networks. Physica D: Nonlinear Phenomena, 146(1-
4):165–199.
Edwards, R. and Glass, L. (2005). A calculus for relating
the dynamics and structure of complex biological net-
works. Adventures in Chemical Physics: A Special
Volume of Advances in Chemical Physics, 132:151–
178.
El Samad, H., Del Vecchio, D., and Khammash, M. (2005).
Repressilators and promotilators: Loop dynamics in
synthetic gene networks. In Proceedings of the 2005,
American Control Conference, 2005., pages 4405–
4410. IEEE.
Elowitz, M. B. and Leibler, S. (2000). A synthetic oscil-
latory network of transcriptional regulators. Nature,
403(6767):335–338.
Firippi, E. and Chaves, M. (2020). Topology-induced
dynamics in a network of synthetic oscillators with
piecewise affine approximation. Chaos: An Interdisci-
plinary Journal of Nonlinear Science, 30(11):113128.
Flieller, D., Riedinger, P., and Louis, J.-P. (2006). Compu-
tation and stability of limit cycles in hybrid systems.
Nonlinear Analysis: Theory, Methods & Applications,
64(2):352–367.
Garloff, J. and Smith, A. P. (2003). A comparison of
methods for the computation of affine lower bound
functions for polynomials. In International Workshop
on Global Optimization and Constraint Satisfaction,
pages 71–85. Springer.
Girard, A. (2003). Computation and stability analysis of
limit cycles in piecewise linear hybrid systems. IFAC
Proceedings Volumes, 36(6):181–186.
Goh, K.-I., Kahng, B., and Cho, K.-H. (2008). Sustained
oscillations in extended genetic oscillatory systems.
Biophysical Journal, 94(11):4270–4276.
Hiskens, I. A. (2001). Stability of hybrid system limit cy-
cles: Application to the compass gait biped robot. In
Proceedings of the 40th IEEE Conference on Decision
and Control (Cat. No. 01CH37228), volume 1, pages
774–779. IEEE.
Kuznetsov, A. and Afraimovich, V. (2012). Heteroclinic
cycles in the repressilator model. Chaos, Solitons &
Fractals, 45(5):660–665.
Mestl, T., Lemay, C., and Glass, L. (1996). Chaos in high-
dimensional neural and gene networks. Physica D:
Nonlinear Phenomena, 98(1):33–52.
M
¨
uller, S., Hofbauer, J., Endler, L., Flamm, C., Widder,
S., and Schuster, P. (2006). A generalized model of
the repressilator. Journal of mathematical biology,
53(6):905–937.
Page, K. M. (2019). Oscillations in well-mixed, determinis-
tic feedback systems: Beyond ring oscillators. Journal
of Theoretical Biology, 481:44–53.
Page, K. M. and Perez-Carrasco, R. (2018). Degradation
rate uniformity determines success of oscillations in
repressive feedback regulatory networks. Journal of
the Royal Society Interface, 15(142):20180157.
Pelleau, M., Min
´
e, A., Truchet, C., and Benhamou, F.
(2013). A constraint solver based on abstract do-
mains. In International Workshop on Verification,
Model Checking, and Abstract Interpretation, pages
434–454. Springer.
Perez-Carrasco, R., Barnes, C. P., Schaerli, Y., Isalan, M.,
Briscoe, J., and Page, K. M. (2018). Combining a tog-
gle switch and a repressilator within the ac-dc circuit
generates distinct dynamical behaviors. Cell systems,
6(4):521–530.
Potvin-Trottier, L., Lord, N. D., Vinnicombe, G., and Pauls-
son, J. (2016). Synchronous long-term oscillations in
a synthetic gene circuit. Nature, 538(7626):514–517.
Sun, H., Folschette, M., and Magnin, M. (2022). Limit
cycle analysis of a class of hybrid gene regulatory
networks. In International Conference on Computa-
tional Methods in Systems Biology, pages 217–236.
Springer.
Thomas, R. (1973). Boolean formalization of genetic
control circuits. Journal of theoretical biology,
42(3):563–585.
Thomas, R. (1991). Regulatory networks seen as asyn-
chronous automata: a logical description. Journal of
theoretical biology, 153(1):1–23.
Wang, R., Chen, L., and Aihara, K. (2006). Construction
of genetic oscillators with interlocked feedback net-
works. Journal of theoretical biology, 242(2):454–
463.
Ziat, G., Mar
´
echal, A., Pelleau, M., Min
´
e, A., and Truchet,
C. (2019). Combination of boxes and polyhedra ab-
stractions for constraint solving. In International Sym-
posium on Formal Methods, pages 119–135. Springer.
Znegui, W., Gritli, H., and Belghith, S. (2020). Design of an
explicit expression of the poincar
´
e map for the passive
dynamic walking of the compass-gait biped model.
Chaos, Solitons & Fractals, 130:109436.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
40
