Visualizing Grassmannians via Poincare Embeddings
Huanran Li
1
and Daniel Pimentel-Alarcón
2
1
Department of Electrical Engineering, Wisconsin Institute for Discovery University of Wisconsin-Madison, U.S.A.
2
Department of Biostatistics, Wisconsin Institute for Discovery University of Wisconsin-Madison, U.S.A.
Keywords:
Grassmannian, Manifold Learning, Poincare Disk, t-SNE, High-Dimensional Data and Dimensionality
Reduction.
Abstract:
This paper introduces an embedding to visualize high-dimensional Grassmannians on the Poincaré disk,
obtained by minimizing the KL-divergence of the geodesics on each manifold. Our main theoretical result
bounds the loss of our embedding by a log-factor of the number of subspaces, and a term that depends on the
distribution of the subspaces in the Grassmannian. This term will be smaller if the subspaces form well-defined
clusters, and larger if the subspaces have no structure whatsoever. We complement our theory with synthetic and
real data experiments showing that our embedding can provide a more accurate visualization of Grassmannians
than existing representations.
1 INTRODUCTION
Subspaces are a cornerstone of data analysis, with ap-
plications ranging from linear regression to principal
component analysis (PCA) Knudsen (2001); Jansson
and Wahlberg (1996); Vaswani et al. (2018), low-rank
matrix completion (LRMC) Dai et al. (2011); Vidal
and Favaro (2014), computer vision Cao et al. (2016);
Chen and Lerman (2009); Hong et al. (2006); Lu and
Vidal (2006), recommender systems Koohi and Kiani
(2017); Ullah et al. (2014); Zhang et al. (2021), classifi-
cation Sun et al. (2015); Ahmed and Khan (2009); Xia
et al. (2017), and more Van Overschee (1997); Mevel
et al. (1999). However, there exist few tools to visual-
ize the Grassmann manifold
G(m,r)
of
r
-dimensional
subspaces of
R
m
. Perhaps the most intuitive of such
visualizations is the representation of
G(3,1)
as the
closed half-sphere where each point in the hemisphere
represents the 1-dimensional subspace (line) in
R
3
that
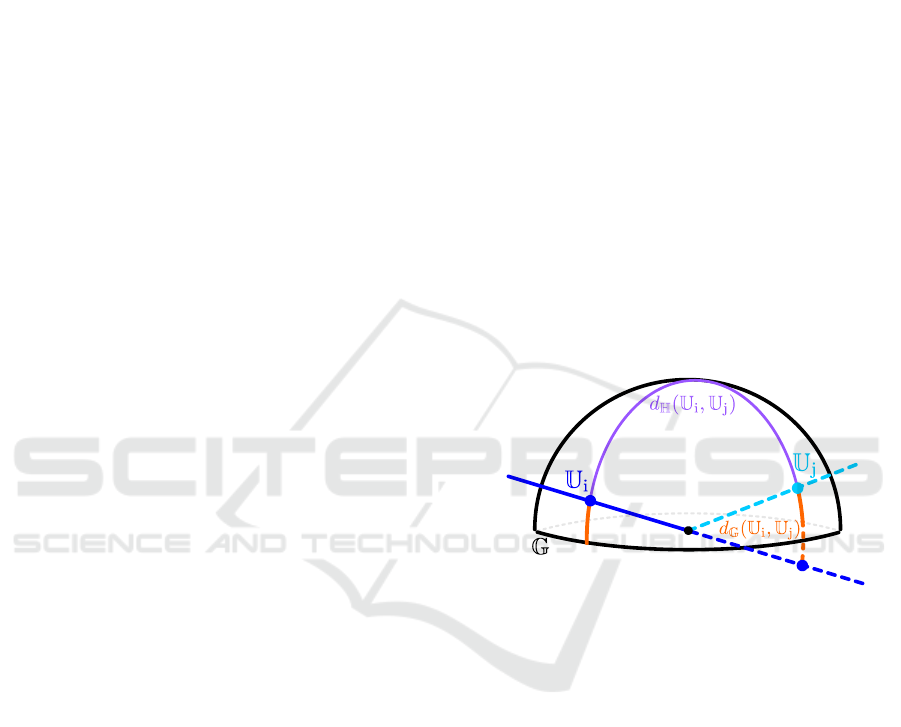
crosses that point and the origin (see Figure 1). While
intuitive, this visualization bears certain limitations.
First, this representation wraps around the edge, so
geodesic distances can be deceiving. For instance, two
points (subspaces) that may appear diametrically far
may in fact be arbitrarily close (see Figure 1). But
more importantly, the main caveat of this semi-sphere
representation is that it is unclear how to generalize it
to
m > 3
or
r > 1
, which makes it quite restrictive, spe-
cially for analysis of modern high-dimensional data.
Motivated by this gap, we propose visualizing a
collection of points in the Grassmannian (subspaces)
Figure 1: Classical 3D Representation of the Grassmannian
G(3,1)
. Each point represents the subspace
U
i
that connects
that point to the origin. This representation wraps around the
edge. Two points (subspaces) that appear diametrically far
w.r.t. the geodesic distance on the hemisphere (
d
H
(U
i
,U
j
)
)
are in fact close w.r.t. the geodesic distance on the Grassman-
nian (
d
G
(U
i
,U
j
)
; see
(1)
). An intuitive way to see this is to
extend the lines to the opposite side of the hemisphere and
compute their smallest angle.
through an embedding onto the Poincaré disk
D ⊂ R
2
.
This embedding, which we call GrassCaré, is inspired
by the well-known t-Distributed Stochastic Neighbor
Embedding (t-SNE), which is widely used to visualize
high-dimensional data (in
R
m
) on the
R
2
plane while
preserving Euclidean distances in
R
m
Van der Maaten
and Hinton (2008). The main difference between the
t-SNE and our GrassCaré embedding is that the latter
maps points in
G(m,r)
onto
D
while preserving the
geodesics on the Grassmannian as much as possible.
This allows to keep an accurate global representation
of the Grassmannian in a unit circle while at the same
time retaining any local structures. Our embedding
Li, H. and Pimentel-Alarcón, D.
Visualizing Grassmannians via Poincare Embeddings.
DOI: 10.5220/0011609400003417
In Proceedings of the 18th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2023) - Volume 3: IVAPP, pages 27-39
ISBN: 978-989-758-634-7; ISSN: 2184-4321
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
27

is obtained by minimizing the Kullback-Leibler (KL)
divergence between the geodesics on the Grassman-
nian and the Poincaré disk using Riemannian gradient
descent. Our main theoretical result shows that the
loss of our embedding (measured in terms of the KL-
divergence with respect to (w.r.t.) the Grassmannian
geodesics) is bounded by a log-factor of the number of
subspaces, and a term that depends on the distribution
of the subspaces in the Grassmannian. This term will
be smaller if the subspaces form well-defined clusters,
and it will be larger if the subspaces have no structure
whatsoever. In words, this result shows that under
reasonable assumptions, our embedding can be an ac-
curate representation of the Grassmannian. Equipped
with this result, we believe that the GrassCaré em-
bedding can be a powerful tool for subspace tracking,
classification, multi-dataset analysis, and any applica-
tion where there is an interest in visualizing subspaces.
This paper should be understood as a first introduction
of our embbedding method, a development of funda-
mental theory, and an exploration of its performance
on canonical datasets.
Paper Organization. In Section 2 we discuss several
applications of our GrassCaré embedding. Section
3 briefly summarizes related work. In Section 4 we
introduce the main formulation that determines our
embedding, together with the gradient steps for the
optimization. Section 5 presents our main theorem,
bounding the loss of our embedding, followed by its
proof. Finally, in Section 6 and 7 we demonstrate the
applicability of our GrassCaré embedding on real and
synthetic data, we compare it to naive alternatives, and
we discuss its advantages and limitations.
2 APPLICATIONS
Our GrassCaré embedding could be a valuable tool in
the following applications:
Subspace Clustering: aims to cluster a collection of
data points
x ∈ R
m
lying near a union of subspaces
Parsons et al. (2004). Equivalently, the goal is to
find a union of subspaces that approximates a high-
dimensional dataset. This method has applications in
motion segmentation Yang et al. (2008); Vidal et al.
(2008), face clustering Elhamifar and Vidal (2013),
data mining Agrawal et al. (1998), time series Ba-
hadori et al. (2015), and more. As we show in our
experiments, our GrassCaré embedding can aid ana-
lyzing the results of a subspace clustering algorithm
beyond a simple accuracy metric, providing insights
and summaries about the clusters characteristics and
relationships. It can also be a valuable tool for debug-
ging and understanding algorithmic performance. In
fact, analyzing and understanding a subspace cluster-
ing algorithm is what initially motivated this paper.
Low-Rank Matrix Completion: aims to recover the
missing entries of a low-rank matrix
X
Recht (2011).
This is equivalent to finding the low-dimensional row
and column spaces of
X
. Some applications of LRMC
include recommender systems Kang et al. (2016), im-
age processing Ji et al. (2010), drug discovery Zhang
et al. (2019), and electronic health records (EHR) Lee
et al. (2010). As we show in our experiments, our
GrassCaré embedding can help analyze the algorith-
mic behavior of LRMC methods as they make opti-
mization steps to complete
X
, showing proximity to
the target, convergence, step sizes, and patterns as they
move through the Grassmannian.
Subspace Tracking: aims to constantly estimate a
subspace
U
t
that changes over time (moves in the
Grassmannian), based on iterative observations
x
t
∈U
t
Vaswani et al. (2018); He et al. (2011); Xu et al. (2013).
This model has applications in signal processing Stew-
art (1998), low-rank matrix completion Balzano et al.
(2010), and computer vision He et al. (2011), where,
for example, one may want to estimate the subspace
corresponding to the moving background of a video.
Here our GrassCaré embedding can be used to track
the subspace path as it moves through the Grassman-
nian. This could provide insights about the subspaces’
behavior: moving speed and distance, zig-zag or cy-
cling patterns, etc.
Multi-Dataset Analysis. Principal Component Analy-
sis (PCA) is arguably the most widely used dimension-
ality reduction technique, with applications ranging
from EHR Lee et al. (2010) to genomics Novembre
et al. (2008); Song et al. (2019) to vehicle detection
Wu and Zhang (2001); Wu et al. (2001). In a nutshell,
PCA identifies the low-dimensional subspace that best
approximates a high-dimensional dataset. In modern
situations, several of these datasets may be distributed
or related in some way. For instance, the EHRs of
a population of certain location could be tightly re-
lated to those of another. However, due to privacy
concerns, security, size, proprietorship, and logistics,
exchange of information like this could prove chal-
lenging if not impossible. The principal subspaces,
however, could be efficiently shared without many of
these concerns, potentially providing new informative
insights. Our GrassCaré embedding could provide a
visualization tool to analyze the relationships between
related datasets like these, potentially revealing simi-
larities, clusters and patterns.
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
28

3 PRIOR WORK
To the best of our knowledge, general visualiza-
tions of Grassmannians have been studied using Self-
Organizing Mappings (SOM) Kirby and Peterson
(2017), which were introduced first for general dimen-
sionality reduction Kohonen (1982, 1990, 1998, 2013).
The extension of SOM to Grassmannians iteratively
updates points on a 2D index space to find the best
arrangement, such that points that are neighbors in the
Grassmannian are still close in the embedding. How-
ever, SOM present several limitations. For instance,
like most neural networks, they require large datasets,
which may not always be available in practice. They
also suffer of large parameter spaces, and are quite
difficult to analyze, making it hard to derive theoreti-
cal guarantees about the accuracy of their embeddings.
It is worth mentioning that there are numerous meth-
ods for general high-dimensional data visualization,
including umapMcInnes et al. (2018), LargeVisTang
et al. (2016), Laplacian eigenmaps Belkin and Niyogi
(2001, 2003), isomap Tenenbaum et al. (2000), and
more Liu et al. (2016); Engel et al. (2012); Ashokku-
mar and Don (2017); Kiefer et al. (2021). However,
since these embeddings are not compact, they are not
appropriate to represent the Grassmannian.
Another more suitable alternative are the Grass-
mannian Diffusion Maps (GDMaps) dos Santos et al.
(2020) introduced as an extension of Diffusion Maps
Coifman et al. (2005). GDMaps consist of two seper-
ate stages. The first stage projects the given data point
(i.e. vector, matrix, tensor) onto the Grassmannian us-
ing a singular value decomposition. The second stage
uses diffusion maps to identify the subspace structures
on the projected Grassmannian. Although the embed-
ding can be quickly generated, it is unfortunately less
accurate than other methods in this paper. On the other
hand, Stochastic Neighbor Embeddings (SNE) were
first presented by Hinton and Roweis in Hinton and
Roweis (2002). It formed the basis for t-SNE, which
was introduced later by Maaten and Hinton in Van der
Maaten and Hinton (2008). Both algorithms minimize
the KL-divergence between the distributions represent-
ing the probability of choosing the nearest neighbor on
the high and low dimensional spaces. These embed-
dings have become some of the most practical tools
to visualize high dimensional data on Euclidean space.
However, Euclidean distances are poor estimators of
geodesics of Grassmannians, so a direct application of
these methods would result in an inaccurate represen-
tation of subspaces arrangements.
Motivated by these issues we decided to explore the
use of the Poincaré disk, which has recently received
increasing attention for high-dimensional embeddings
Nickel and Kiela (2017); Klimovskaia et al. (2020).
Intuitively, the Poincaré disk is a 2D hyperbolic geo-
metric model, usually displayed as a unit circle where
the geodesic distance between two points in the disk
is represented as the circular arc orthogonal to the unit
circle Goodman-Strauss (2001), which corresponds to
the projection of the hyperbolic arc of their geodesic
(see Figure 2 to build some intuition). This unique
feature brings several advantages for serving as the
embedding space for Grassmannian. First, since these
hyperbolic arcs get larger (tending to infinity) as points
approach the disk boundary, the Poincaré disk is an
effective model to accurately represent the global struc-
ture of complex hierarchical data while retaining its
local structures. Specifically, the Poincaré disk can
be viewed as a continuous embedding of tree nodes
from the top of the tree structure, where the root node
is at the origin, and the leaves are distributed near
the boundary. So, it is naturally suited to represent
hierarchical structures. This is suitable to represent
structured clusters, where the points from the same
cluster can be regarded a branch of the tree, because
they share a similar distance to other clusters. Sec-
ond, the hyperbolic disk has a Riemannian manifold
structure that allow us to perform gradient-based opti-
mization, which is crucial to derive convergence guar-
antees, and for parallel training of large-scale dataset
models. Finally, our main result showing the accuracy
of our embedding enables efficient clustering using the
Poincaré low-dimensional representation. That is, in-
stead of clustering subspaces on the high-dimensional
dataset, the clustering method can be performed on the
mutual distances acquired from the embedding, with
the knowledge that the embedding would represent the
high-dimensional subspace accurately enough.
4 SETUP AND FORMULATION
In this section we present the mathematical formula-
tion of our GrassCaré embedding. To this end let us
first introduce some terminology. Recall that we use
G(m,r)
to denote the Grassmann manifold that con-
tains all the
r
-dimensional subspaces of
R
m
. For any
two subspaces
U
1
,U
2
∈G(m,r)
, the geodesic distance
between them is defined as:
d
G
(U
i
,U
j
) :=
s
r
∑
ℓ=1
arccos
2
σ
ℓ
(U
T
i
U
j
), (1)
where
U
i
,U
j
∈ R
m×r
are orthonormal bases of
U
i
,U
j
,
and
σ
ℓ
(·)
denotes the
ℓ
th
largest singular value. As for
the embedding space, recall that the Poincaré disk
D
is
the Riemannian manifold defined as the open unit ball
Visualizing Grassmannians via Poincare Embeddings
29
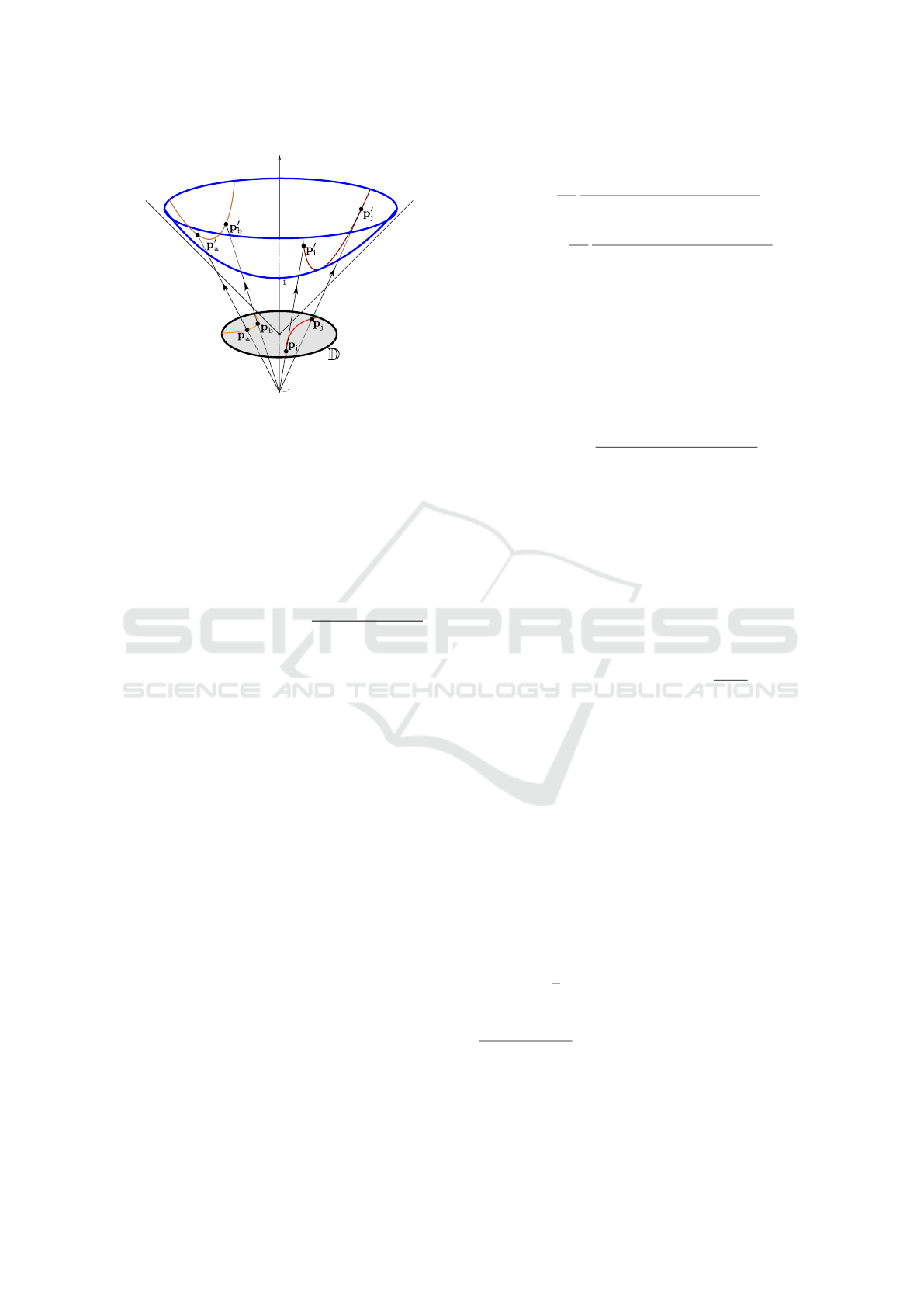
Figure 2: Geodesics in the Poincaré disk
D
. The geodesic
distance
d
D
(p
i
,p
j
)
is given by the Euclidean length of the
hyperbolic arc between
p
′
i
and
p
′
j
, and is often depicted in
the disk by the arc between
p
i
and
p
j
(and similarly for
d
D
(p
a
,p
b
)
). Points closer to the disk’s boundary will be
projected higher on the hyperbolic space, resulting in larger
distances (see
(2)
). In words, distances near the edge are
larger than they appear.
in
R
2
equipped with the following distance function
between two points p
i
,p
j
∈ D:
d
D
(p
i
,p
j
) := arcosh
1 + 2
∥p
i
−p
j
∥
2
(1−∥p
i
∥
2
)(1−∥p
j
∥
2
)
.
(2)
Notice from
(2)
that the geodesic distance in the disk
is amplified smoothly as
p
i
or
p
j
move away from
the origin. Intuitively, this means that an arc of the
same Euclidean length in the disk represents a larger
geodesic distance (tending to infinity) as it approaches
the edge of the disk. In other words, distances near the
edge of the disk are larger than they appear (see Figure
3 to build some intuition). Conversely, distances at
the center of the disk are smaller than they appear.
This allows to plot denser regions of the Grassmannian
with higher granularity (thus retaining local structure)
while at the same time keeping an accurate global
representation of the Grassmannian inside an open
circle.
To find our embedding, we will mimic the sym-
metric SNE approach in Van der Maaten and Hinton
(2008). That is, we will first compute a probability
matrix
P
G
∈ [0,1]
N×N
whose
(i,j)
th
entry represents
the probability that
U
i
is chosen as a nearest neighbor
of
U
j
, which is equal to zero if
i = j
, and for
i ̸= j
is
given by:
[P
G
]
ij
:=
1
2N
exp(−d
G
(U
i
,U
j
)
2
/2γ
2
i
)
∑
k̸=i
exp(−d
G
(U
i
,U
k
)
2
/2γ
2
i
)
+
1
2N
exp(−d
G
(U
j
,U
i
)
2
/2γ
2
j
)
∑
k̸=j
exp(−d
G
(U
j
,U
k
)
2
/2γ
2
j
)
, (3)
where
γ
i
is adapted to the data density: smaller values
for denser regions of the data space. In our experi-
ments, we choose it to be the variance of distances
from point
i
to other points. Next we create the proba-
bility matrix
P
D
∈ [0, 1]
N×N
, whose
(i,j)
th
entry repre-
sents the probability that point
p
i
in our embedding
D
is chosen as a nearest neighbor of point
p
j
∈ D
, which
is equal to zero if i = j, and for i ̸= j is given by:
[P
D
]
ij
:=
exp(−d
D
(p
i
,p
j
)
2
/β)
∑
k̸=l
exp(−d
D
(p
k
,p
l
)
2
/β)
(4)
where
β > 0
(usually set to
1
or
2
) controls the em-
bedding’s scattering Klimovskaia et al. (2020). The
larger
β
, the smaller variance in the probability matrix
P
D
. In practice, we did not notice much variability
in our results as a function of this parameter. Thus,
following standard practice Klimovskaia et al. (2020),
we pick
β = 1
. Our goal to obtain the embedding is
to maximize the similarity between the two distribu-
tions
P
G
and
P
D
, which we do by minimizing their
Kullback-Leibler (KL) divergence:
KL(P
G
||P
D
) =
∑
i,j
[P
G
]
ij
log
[P
G
]
ij
[P
D
]
ij
.
Since
P
G
is a constant given
{U
i
}
, this is the same as
minimizing the following loss
L = −
∑
i,j
[P
G
]
ij
log[P
D
]
ij
.
To minimize this loss over the Poincaré disk
D
we
will use Riemannian Stochastic Gradient Descent
Bonnabel (2013), which updates p
t+1
i
according to:
p
t+1
i
← R(p
t
i
−η∇
i
L), (5)
where
η > 0
is the step size (set as
η = 1
in the im-
plementation),
∇
i
L
denotes the Riemannian gradient
of
L
w.r.t.
p
i
, and
R
denotes a retraction
1
from the
tangent space of p
i
onto D. It is easy to see that
∇
i
L =
4
β
∑
j
([P
G
]
ij
−[P
D
]
ij
)(1 + d
D
(p
i
,p
j
)
2
)
−1
·d
D
(p
i
,p
j
)∇
i
d
D
(p
i
,p
j
), (6)
1
A mapping
R
from the tangent bundle
T M
to the mani-
fold
M
such that its restriction to the tangent space of
M
at
p
i
satisfies a local rigidity condition which preserves gradi-
ents at
p
i
; see Chapters 3 and 4 of Absil et al. (2009) for a
more careful treatment of these definitions.
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
30
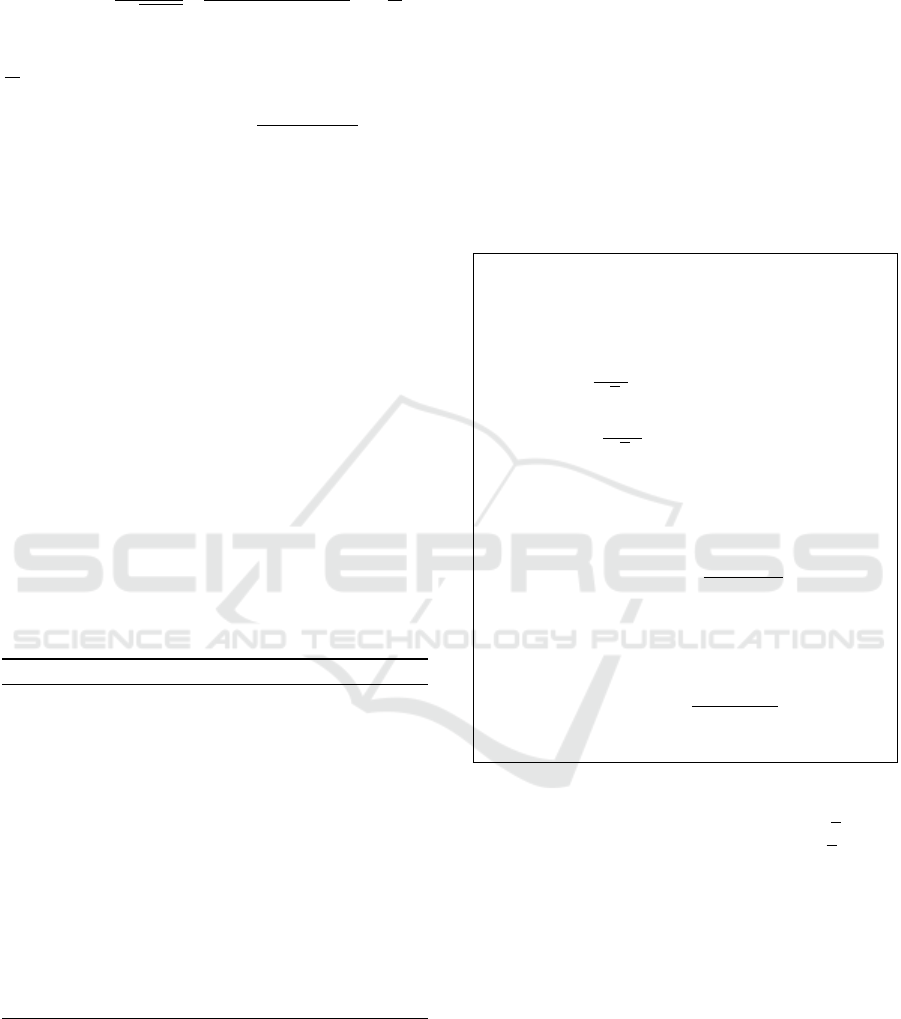
where the gradient of d
D
w.r.t. p
i
is given by:
∇
i
d
D
(p
i
,p
j
) =
4
b
√
c
2
−1
||p
j
||
2
−2⟨p
i
,p
j
⟩+ 1
a
2
p
i
−
p
j
a
!
.
Here
a = 1 − ||p
i
||
2
,
b = 1 − ||p
j
||
2
, and
c = 1 +
2
ab
||p
i
−p
j
||
2
. Finally, the retraction step is given by
R(p
i
−η∇
i
L) = proj
p
i
−η
(1 −||p
i
||
2
)
2
4
∇
i
L
,
where
proj(p
i
) =
(
p
i
/(||p
i
||+ ε) i f ||p
i
|| ≥ 1
p
i
otherwise,
and
ε
is a small constant number; in our experiments
we set this to 10
−5
.
In our implementation we use random initialization
for the points in the embedding. We point out that ini-
tialization is crucial for t-SNE. This is because t-SNE
is generally used to embed points in the Euclidean
space, which is open. In contrast, the Grassmannian
is spherical and compact, and hence, we observed that
varying initialization resulted in similar/equivalent em-
beddings of the Grassmannian, observed from differ-
ent angles. This is further demonstrated in our exper-
iments section (Figure 5), where the average loss of
GrassCaré over 100 trials varies very little in compari-
son to all other embeddings, showing that besides this
point-of-view difference, our results do not depend
heavily on the initialization. The entire embedding
procedure is summarized in Algorithm 1.
Algorithm 1: GrassCaré.
Input: A collection of subspaces
{U
1
,U
2
,...,U
N
} ∈
G(m,r).
Output: A collection of points
{p
1
,p
2
,...,p
N
}
in the
Poincaré disk D.
Parameter: β ∈ [1,2],ε < 10
−5
Construct P
G
according to (3)
Randomly initialize p
1
,p
2
,...,p
N
∈ D
repeat
Construct P
D
according to (4)
for i ∈ [1, N] do
Compute KL-loss gradient ∇
i
L as in (6)
Update point p
i
according to (5)
end for
until Converge
5
MAIN THEORETICAL RESULTS
AND PROOFS
First observe that convergence of our embedding fol-
lows directly by now-standard results in Riemannian
optimization (see e.g. Proposition in Adams et al.
(1996)). In fact, local convergence of our embedding
follows directly because our Riemannian steps are
gradient-related Adams et al. (1996). Our main theo-
retical result goes one step further, bounding the loss
of our embedding by a log-factor of the number of
subspaces, and a term that depends on the arrange-
ment of the subspaces in the Grassmannian. This term
will be smaller if the subspaces form well-defined
clusters, and larger if the subspaces have no structure
whatsoever. Intuitively, this result shows that under
reasonable assumptions, our embedding can provide
an accurate representation of Grassmannians.
Theorem 1. Suppose
N > 3
. Define
γ := min
i
γ
i
and
Γ := max
i
γ
i
. Let
{U
1
,...,U
K
}
be a partition
of
{U
1
,...,U
N
}
such that
|U
k
|≥n
K
> 1 ∀ k
. Let
δ :=
1
√
2γ
max
k
max
U
i
,U
j
∈U
k
d
G
(U
i
,U
j
),
∆ :=
1
√
2Γ
min
U
i
∈U
k
,U
j
∈U
ℓ
:
k̸=ℓ
d
G
(U
i
,U
j
).
Then the optimal loss of GrassCaré is bounded
by:
L
⋆
< log D +
5e
δ
2
−∆
2
β(n
K
−1)
,
where
D := N(n
K
−1) + N(N −n
K
)
·exp
−arcosh
2
1 +
2sin(π/K)
0.75
2
/β
. (7)
In words, Theorem 1 requires that the subspaces
can be arranged into clusters of size
n
k
> 1
such that
the intra-cluster distances are smaller than
√
2δγ
, and
the outer-cluster distances are larger than
√
2∆Γ
(see
Figure 3). Notice that this can always be done as long
as
N > 3
. However, depending on the arrangement,
δ
could be too large or
∆
too small, resulting in a loose
bound. Ideally we want a small
δ
and a large
∆
, so that
the subspaces form well-defined clusters and
e
δ
2
−∆
2
is
small, resulting in a tighter bound.
Proof.
Theorem 1 follows by a similar strategy as in
Shaham and Steinerberger (2017), which essentially
bounds the optimal loss by that of an artificial embed-
ding. In our case we will use an embedding that maps
{U
1
,...,U
K
}
to
K
points uniformly distributed in the
circle of radius
1/2
, i.e.,
p
i
= p
j
for every
U
i
,U
j
∈ U
k
(see Figure 3). This way, for any subspaces
U
i
,U
j
in
Visualizing Grassmannians via Poincare Embeddings
31

Figure 3: Left: Theorem 1 requires that the intra-cluster dis-
tances are smaller than
√
2δγ
, and the outer-cluster distances
are larger than
√
2∆Γ
. Right: Example of the artificial em-
bedding (with
K = 5
) in the proof of Theorem 1, which maps
all subspaces in cluster
U
k
to the same point in the circle of
radius 1/2.
different clusters
U
k
,U
ℓ
, the geodesic distance of their
embeddings on the Poincaré disk is upper and lower
bounded by
2.2 >
1 +
2
0.75
2
≥ d
D
(p
i
,p
j
)
≥ arcosh
1 +
2sin(π/K)
0.75
2
=: Φ.
It follows that the (i,j)
th
entry of P
D
is bounded by
[P
D
]
ij
:=
exp(−d
D
(p
i
,p
j
)
2
/β)
∑
k̸=l
exp(−d
D
(p
k
,p
l
)
2
/β)
≥
exp(−d
D
(p
i
,p
j
)
2
/β)
N(n
K
−1) + N(N −n
K
)exp(−Φ
2
/β)
, (8)
where the denominator is precisely
D
as defined in
(7)
.
Now, if
U
i
and
U
j
are in the same cluster
U
k
, the bound
in
(8)
simplifies to
1/D
. Otherwise, it simplifies to
exp(−2.2
2
/β)/D
. Plugging these bounds in the loss,
we see that:
L
∗
<
∑
i,j in same cluster
[P
G
]
ij
logD
+
∑
i,j in different clusters
[P
G
]
ij
(2.2
2
/β + log D)
≤ log D +
∑
i,j in different clusters
[P
G
]
ij
(2.2
2
/β). (9)
Next notice that if
U
i
,U
j
are not in the same cluster,
[P
G
]
ij
:=
1
2N
exp(−d
G
(U
i
,U
j
)
2
/2γ
2
i
)
∑
k̸=i
exp(−d
G
(U
i
,U
k
)
2
/2γ
2
i
)
+
1
2N
exp(−d
G
(U
j
,U
i
)
2
/2γ
2
j
)
∑
k̸=j
exp(−d
G
(U
j
,U
k
)
2
/2γ
2
j
)
≤
1
2N
e
−∆
2
∑
k̸=i
exp(−d
G
(U
i
,U
k
)
2
/2γ
2
i
)
+
1
2N
e
−∆
2
∑
k̸=j
exp(−d
G
(U
j
,U
k
)
2
/2γ
2
j
)
≤
1
N
e
−∆
2
N(n
k
−1)e
−δ
2
.
Plugging this into
(9)
we obtain the desired result.
6 EXPERIMENTS
Recall that the main motivation of this paper is to de-
velop a novel method to visualize Grassmannians of
high ambient dimension. Our bound above describes
the theoretical accuracy of our embedding. We now
present a series of experiments on real and synthetic
datasets to analyze its practical performance. In par-
ticular, we will test on normal simulated data, and
one canonical datasetTron and Vidal (2007). These
datasets have moderately high ambient dimension (i.e.,
many features), but low intrinsic dimension (i.e., lie
in a low-dimensional subspace). In other words, these
datasets would fit in high-dimensional Grassmannians
of low-dimensional subspaces. We believe that these
well-studied datasets are a perfect fit for our setting,
and convenient for an initial exploration and compari-
son against existing baselines.
Comparison Baseline. To evaluate the effectiveness
of our method we used t-SNEVan der Maaten and Hin-
ton (2008), GDMaps dos Santos et al. (2020), and a
naive visualization based on the most common dimen-
sionality reduction technique: Principal Components
Analysis (PCA). To this end we first vectorize (stack
the columns of) each orthonormal basis
U
i
into a vec-
tor
u
i
∈ R
mr
. Next we concatenate all vectors
u
i
into
a matrix of size
mr ×N
, on which we apply PCA. In
this naive PCA (nPCA) visualization, the subspace
U
i
is represented in the
(x,y)
plane by
v
i
∈ R
2
, the
coefficients of u
i
w.r.t. the leading principal plane V.
Clustering Synthetic Data. In our synthetic experi-
ments we study our embedding when the subspaces are
uniformly distributed among
K
clusters. To this end we
first generated
K
centers in the Grassmannian
G(m,r)
,
each defined by a
m ×r
matrix
C
k
with i.i.e. standard
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
32

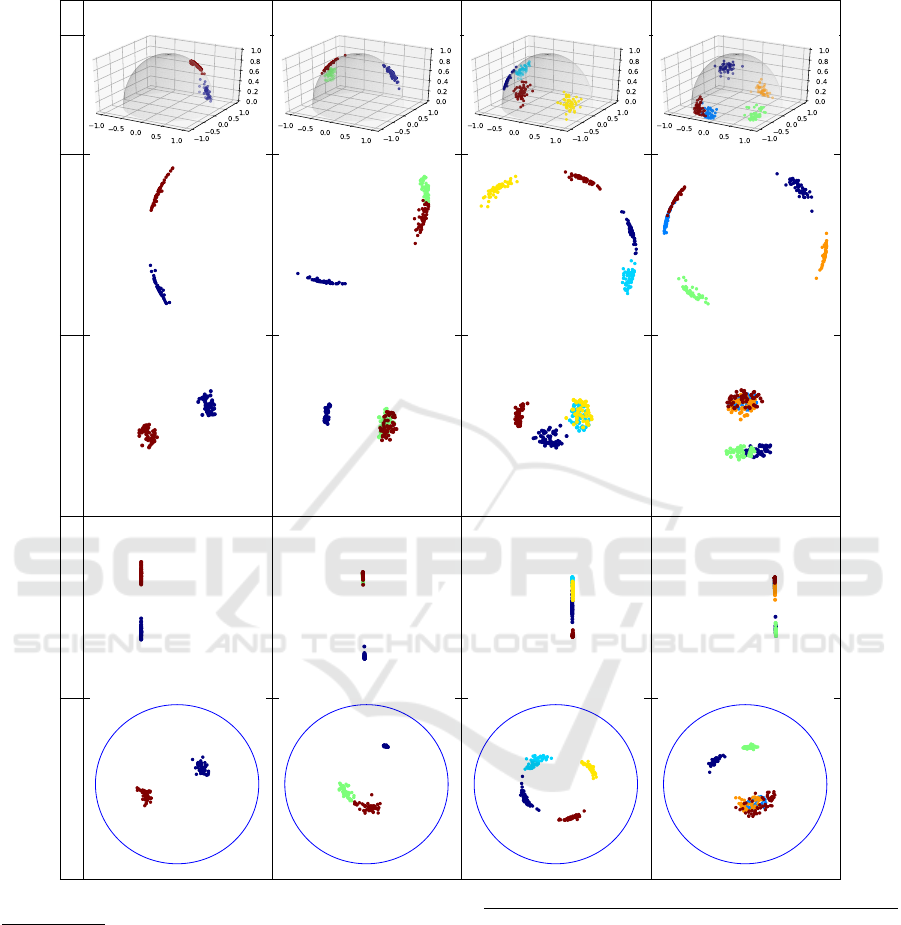
Classic 3D nPCA GDMaps t-SNE GrassCaré
Figure 4: Visualizations of clusters in
G(3,1)
with 4 clusters.
GrassCaré produces a more accurate
representation of the Grassmannian
. nPCA and even the 3D representation display Clusters 1 and 2 (cyan and yel-
low) nearly diametrically apart. In reality they are quite close, as depicted by GrassCaré.
normal entries whose columns are later orthonormal-
ized. Then for each
k
we independently generate
n
k
subspaces, each spanned by a basis
U
i
whose entries
are equal to those of
C
k
plus i.i.d. normal random vari-
ables with variance
σ
2
. This will produce
K
clusters
in
G(m,r)
, each with
n
k
subspaces. The smaller
σ
, the
closer the subspaces in the same cluster will be to one
another, and vice versa.
In our first experiment we study a controlled setting
where we can actually visualize the low-dimensional
Grassmannian
G(3,1)
, and compare it with our em-
bedding on the Poincaré disk. We hope that this ex-
periment provides a visual intuition of how points are
embedded in higher-dimensional cases. To this end
we generated
n
k
= 50
subspaces per cluster (
m = 3
,
r = 1
), and we set
σ = 0.1
, which produced visually
well-defined cluster clouds. Figure 4 shows some re-
sults for
K = 4
clusters (see Figure 9 in the Appendix
for additional values of
K
). At first glance it might
appear like our GrassCaré embedding is not too dif-
ferent from the other approaches, especially as t-SNE
and nPCA seems to be doing a decent job displaying
the clusters. However, a more careful look reveals that
t-SNE clearly agglomerates several pairs of clusters,
while the GrassCaré can separate them nicely.
In particular, notice that in Figure 4, both nPCA
and even the classic 3D representation fail to show the
true local structure of the Grassmannian that the Grass-
Caré plot reveals. To see this pay special attention to
the cyan and yellow clusters. Based on the first two
rows (classic 3D representation and nPCA) these clus-
ters would appear to be nearly diametrically apart (in
the 3D representation, the cyan cluster is in the back
side of the hemisphere). However, computing their
geodesics one can verify that the subspaces that they
represent are in fact quite close in the Grassmannian.
An intuitive way to see this is to extend the lines to
the opposite side of the hemisphere and compute their
smallest angle, or to remember that in the 3D repre-
sentation, the hemisphere wraps around the edge (see
Figure 3). In contrast, our GrassCaré plot accurately
displays the true global structure of the Grassman-
nian, mapping these two clusters close to one another.
Also notice that the embeddings are plotted with equal
scale on horizontal and vertical axis. The GrassCaré
makes a better use of the visual space, spreading all
data more broadly while at the same time keeping the
clusters well-defined. In contrast, GDMaps has much
less range on the horizontal axis, which makes it look
like a straight line and not be able to display the full
information. More examples for different values of K
are presented in the Appendix.
The previous experiment shows the qualitative su-
periority of the GrassCaré embedding over alternative
embeddings in the low-dimensional case
m = 3
and
r = 1
(where no vectorization is needed for nPCA). In
our next experiment we will show in a more quanti-
tative way that the advantages of GrassCaré are even
more evident in higher dimensional cases. First notice
that the classic 3D representation only applies to the
case
m = 3
,
r = 1
, and there is no clear way how to
extend it to higher dimensions. On the other hand, re-
call that for
r > 1
, nPCA requires vectorizing the bases
U
i
, which will naturally interfere even more with the
structure of the Grassmannian. To see this consider:
U =
1 0
0 1
0 0
and U
′
=
0 1
1 0
0 0
.
While both span the same subspace in
G(3,2)
, the Eu-
clidean distance of their vectorizations is large, which
would result in distant points in the nPCA embed-
ding. t-SNE and GDMaps present similar inaccuracy
behavior. To verify this we generated subspaces in
the exact same way as described before (with
K = 3
,
n
k
= 17
, and different values of
m
and
r
), except this
time we measured the quality of the visualization in
terms of the representation error, which we define
as the Frobenius difference between the (normalized)
distance matrices produced by the subspaces in the
Grassmannian and the points in each embedding. In
the case of GrassCaré, distances in the embedding are
measured according to the Poincaré geodesics, so the
representation error of the GrassCaré embedding will
Visualizing Grassmannians via Poincare Embeddings
33
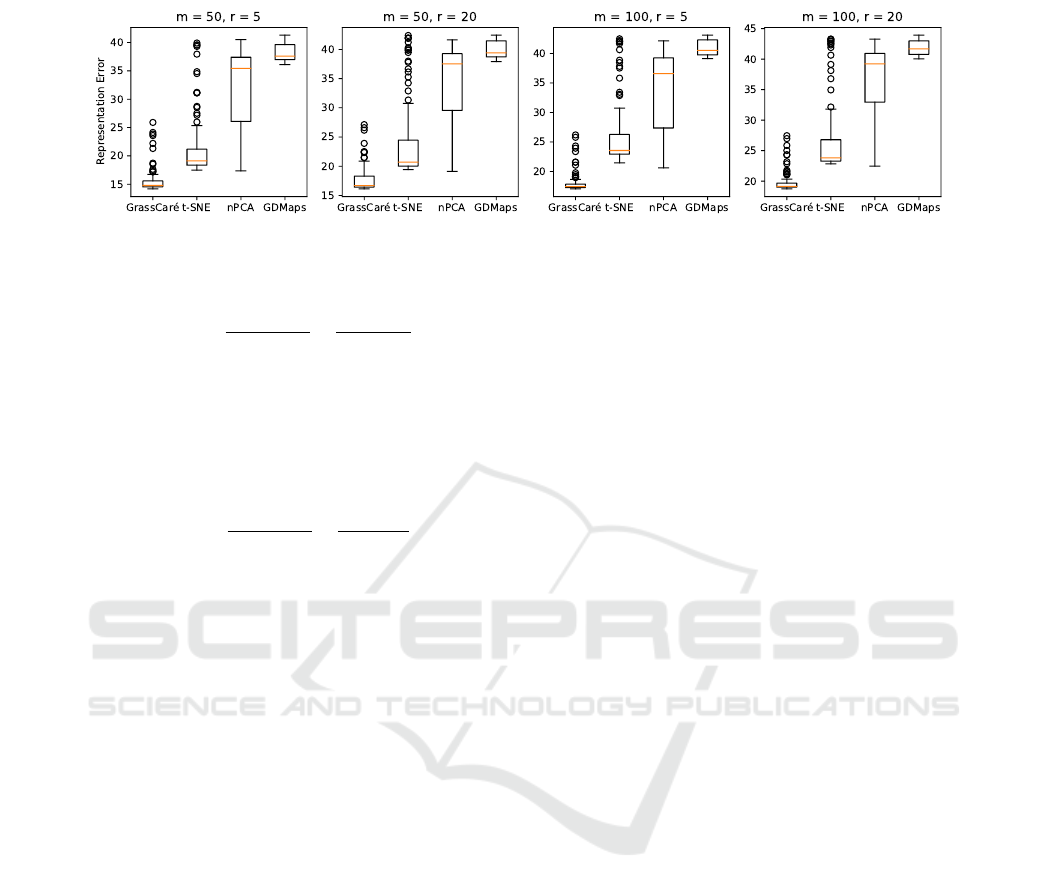
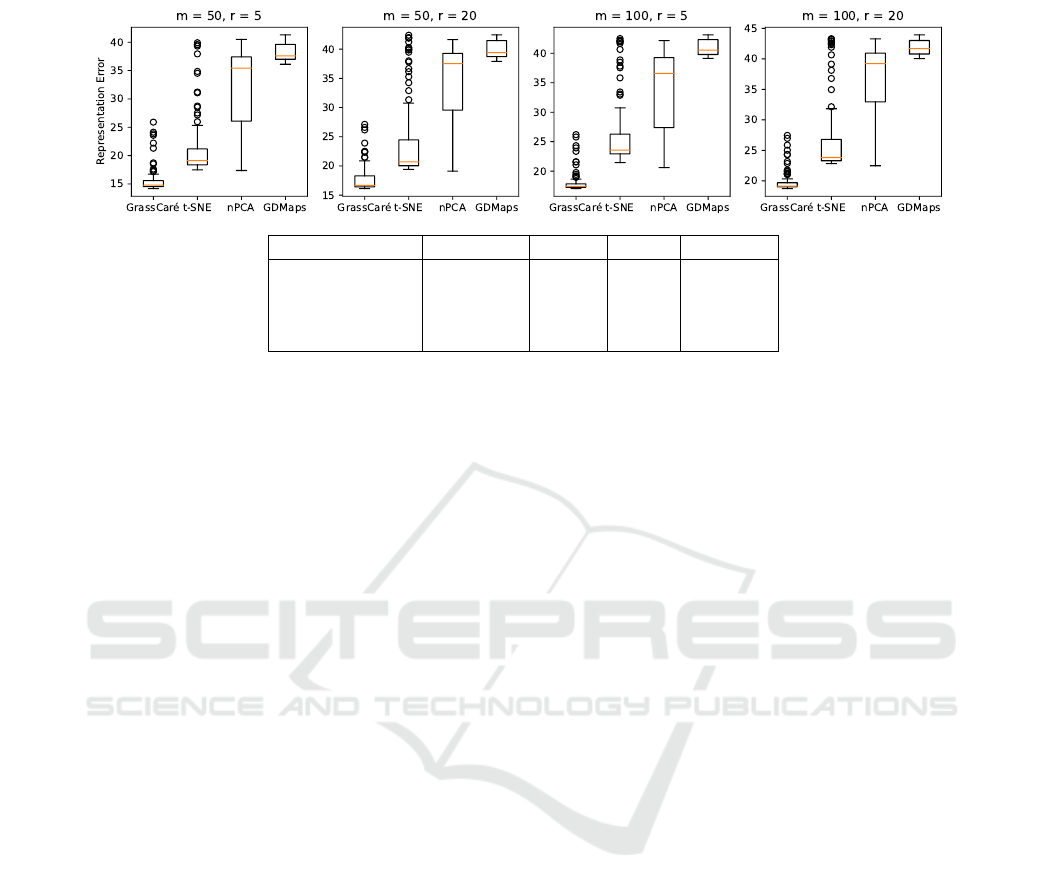
Figure 5: Representation error of GrassCaré (this paper) and other methods for high-dimensional Grassmannians G(m,r).
be measured as:
ε
2
(D) =
∑
i,j
d
G
(U
i
,U
j
)
Z
G
−
d
D
(p
i
,p
j
)
Z
D
2
, (10)
where
Z
2
G
=
∑
i,j
d
2
G
(U
i
,U
j
)
and
Z
2
D
=
∑
i,j
d
2
D
(p
i
,p
j
)
are
normalization terms. Similarly, since the distance that
all other embeddings aim to minimize is Euclidean,
the representation error of the other embeddings will
be measured as:
ε
2
(V) =
∑
i,j
d
G
(U
i
,U
j
)
Z
G
−
∥v
i
−v
j
∥
Z
V
2
, (11)
where
Z
2
V
=
∑
i,j
∥v
i
−v
j
∥
2
is a normalization term.
The results of 100 trials are summarized in Figure 5,
which confirms the superiority of our GrassCaré em-
bedding, and the loss of structure of the naive approach.
The computation time is also summarized in Figure 10
in the Appendix.
Subspace Estimation from Incomplete Data. In our
next experiment we apply our GrassCaré embedding
to visualize the path of subspaces produced by the
subspace estimation algorithm known as GROUSE
(Grassmannian Rank-One Update Subspace Estima-
tion) Balzano et al. (2010). The applicability of this
algorithm ranges from online video analysis (to track
the subspace of the background in real time) to sub-
space clustering Parsons et al. (2004) and low-rank
matrix completion (LRMC) Balzano et al. (2010). In
the latter, the algorithm receives a subset of the entries
of a data matrix
X ∈ R
m×n
whose columns lie in an
unknown subspace
U
⋆
∈ G(m, r)
, and the goal is to
estimate
U
⋆
. To this end GROUSE starts with a sub-
space estimate
U
0
∈ G(m, r)
, and iteratively tilts it in
the direction of a column of
X
, producing a sequence
of subspaces U
1
,...,U
N
.
To emulate this setup we first generate true and
initial subspaces with bases
U
⋆
,U
0
∈ R
m×r
with
i.i.d. standard normal entries. Next we generate
a coefficient matrix
Θ ∈ R
r×n
with i.i.d. standard
normal entries, so that
X = U
⋆
Θ
is rank-
r
. Then
we run GROUSE using a fraction
Ω
of the en-
tries of
X
, selected uniformly at random. We store
each of GROUSE’s steps, and visualize their path
U
0
,U
1
,...,U
N
(together with the target
U
⋆
) using our
GrassCaré embedding, nPCA, GDMaps, and t-SNE.
Figure 6 shows sample plots when
m = 200
,
r = 5
,
Ω = 0.7
(corresponding to 30% missing data; both
cases share the same initialization), and
n = N = 50
(corresponding to the case where GROUSE only it-
erates once over each column). Here once again
the GrassCaré plot shows a richer depiction of the
subspaces and a better usage of the available visual
space. From the GrassCaré plot we can clearly vi-
sualize each separate path, and see that, as expected,
the full-data estimate gets much closer to the target
than the missing-data estimate (and much faster). In
contrast, the paths in the nPCA plot are hardly dis-
tinguishable and misleading, showing the opposite of
the truth: an incomplete-data estimate much closer to
the target than the full-data estimate. To verify once
again (beyond our visual interpretation) that the Grass-
Caré embedding is much more representative of the
true distribution of subspaces in the Grassmannian, we
measured the distance of each iterate to the target, in
the Grassmannian and in each embedding. The nor-
malized results are in figure 7. They show that the
trajectories in the GrassCaré embedding mimic closely
those in the Grassmannian. In contrast, the nPCA em-
bedding can be quite misleading, showing in fact an
opposite representation, with distances in the embed-
ding growing over iterations, while in reality they are
decreasing in the Grassmannian.
Motion Segmentation. In our final experiment we
use our GrassCaré plot to visualize the subspaces
describing moving objects in videos from the Hopkins
155 dataset Tron and Vidal (2007). This dataset
contains the locations over time of landmarks
of several moving objects (e.g., cars, buses, or
checkerboards) in 155 video sequences. Recall that
the stacked landmarks of each rigid object over time
approximately lie in a 4-dimensional subspace Tomasi
and Kanade (1992); Kanatani (2001). So for our
experiment we split all landmarks of the same object
in groups of 5 (if at any point there were fewer than
5
landmarks left, they were discarded), and for each
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
34
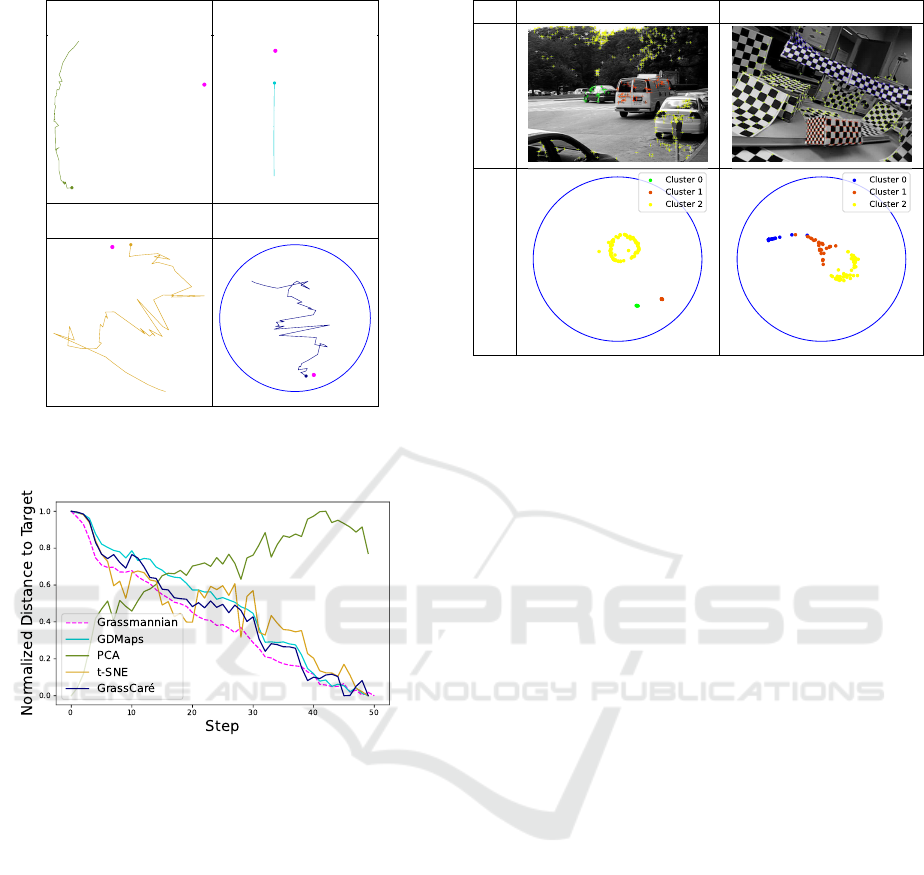
nPCA GDMaps
t-SNE GrassCaré
Figure 6: Visualization of the path generated by GROUSE
using nPCA, GDMaps, t-SNE, and GrassCaré (this paper).
Figure 7: Distance to target (in the Grassmannian and its
embeddings) of the sequence generated by GROUSE.
group we performed a singular value decomposition
to identify its 4-dimensional principal subspace
U
i
.
Figure 8 shows the embedding of the subspaces of all
groups, color-coded by object. Notice that GrassCaré
displays the subspaces of the same object nearby.
This is consistent with theory, as they represent
slightly noisy versions of the subspace describing the
object’s trajectory. Notice the higher variance in the
yellow cluster, which is consistent with its landmarks,
corresponding to several trees, cars, and pavement, as
opposed to just one rigid object. But not only that.
From the GrassCaré embedding we can also analyze
the trajectories themselves, and their relationships.
For instance, in the traffic plot we can see that the
green and red clusters (corresponding to the moving
car and van) are close to one another, indicating that
their trajectories resemble each other. In contrast,
these clusters are farther from the yellow one, which
matches our observation that the trajectories of the
cars3 2RT3RCR
Sample Frame
GrassCaré
Figure 8: GrassCaré embedding for two motion sequences
of the Hopkins155 dataset.
moving car and van are quite different from
the nearly static background.
7 CONCLUSIONS AND
LIMITATIONS
This paper presents an embedding method of Grass-
mannian points on a 2-d disk with lower representation
error and a more effective distribution over the visual
space. We believe GrassCaré will be a powerful tool
for visualizing subspaces extracted from high dimen-
sional real-world data, and that it will help researchers
analyze both local and global structures (e.g., paths and
clusters). In our experiments, GrassCaré is marginally
slower than t-SNE and GDMaps. This is because com-
puting distances in the Poincaré disk requires slightly
more calculations than in Euclidean space. However,
we believe this price is worth it for two reasons. First,
GrassCaré outputs a more accurate visual representa-
tion. In fact, as shown by our main theoretical result,
the representation loss of GrassCaré is lower bounded
under mild assumptions. This can be verified in Fig-
ure 5. Second, GrassCaré make a better use of space
within the unit circle, which eliminates the visual mis-
leading effect of different axis scales, like GDMaps
does.
REFERENCES
Absil, P.-A., Mahony, R., and Sepulchre, R. (2009). Opti-
mization algorithms on matrix manifolds. Princeton
University Press.
Adams, L. M., Nazareth, J. L., et al. (1996). Linear and non-
Visualizing Grassmannians via Poincare Embeddings
35

linear conjugate gradient-related methods, volume 85.
Siam.
Agrawal, R., Gehrke, J., Gunopulos, D., and Raghavan, P.
(1998). Automatic subspace clustering of high dimen-
sional data for data mining applications. In Proceed-
ings of the 1998 ACM SIGMOD international confer-
ence on Management of data, pages 94–105.
Ahmed, M. S. and Khan, L. (2009). Sisc: A text classifica-
tion approach using semi supervised subspace cluster-
ing. In 2009 IEEE International Conference on Data
Mining Workshops, pages 1–6. IEEE.
Ashokkumar, P. and Don, S. (2017). High dimensional data
visualization: A survey. Journal of Advanced Research
in Dynamical and Control Systems, 9(12):851–866.
Bahadori, M. T., Kale, D., Fan, Y., and Liu, Y. (2015). Func-
tional subspace clustering with application to time se-
ries. In International conference on machine learning,
pages 228–237. PMLR.
Balzano, L., Nowak, R., and Recht, B. (2010). Online
identification and tracking of subspaces from highly
incomplete information. In 2010 48th Annual allerton
conference on communication, control, and computing
(Allerton), pages 704–711. IEEE.
Belkin, M. and Niyogi, P. (2001). Laplacian eigenmaps
and spectral techniques for embedding and clustering.
Advances in neural information processing systems,
14.
Belkin, M. and Niyogi, P. (2003). Laplacian eigenmaps
for dimensionality reduction and data representation.
Neural computation, 15(6):1373–1396.
Bonnabel, S. (2013). Stochastic gradient descent on rie-
mannian manifolds. IEEE Transactions on Automatic
Control, 58(9):2217–2229.
Cao, J., Zhang, K., Luo, M., Yin, C., and Lai, X. (2016).
Extreme learning machine and adaptive sparse repre-
sentation for image classification. Neural networks,
81:91–102.
Chen, G. and Lerman, G. (2009). Spectral curvature cluster-
ing (scc). International Journal of Computer Vision,
81(3):317–330.
Coifman, R. R., Lafon, S., Lee, A. B., Maggioni, M., Nadler,
B., Warner, F., and Zucker, S. W. (2005). Geometric
diffusions as a tool for harmonic analysis and structure
definition of data: Diffusion maps. Proceedings of the
national academy of sciences, 102(21):7426–7431.
Dai, W., Milenkovic, O., and Kerman, E. (2011). Sub-
space evolution and transfer (set) for low-rank matrix
completion. IEEE Transactions on Signal Processing,
59(7):3120–3132.
dos Santos, K. R., Giovanis, D. G., and Shields, M. D. (2020).
Grassmannian diffusion maps based dimension reduc-
tion and classification for high-dimensional data. arXiv
preprint arXiv:2009.07547.
Elhamifar, E. and Vidal, R. (2013). Sparse subspace cluster-
ing: Algorithm, theory, and applications. IEEE trans-
actions on pattern analysis and machine intelligence,
35(11):2765–2781.
Engel, D., Hüttenberger, L., and Hamann, B. (2012). A
survey of dimension reduction methods for high-
dimensional data analysis and visualization. In Vi-
sualization of Large and Unstructured Data Sets:
Applications in Geospatial Planning, Modeling and
Engineering-Proceedings of IRTG 1131 Workshop
2011. Schloss Dagstuhl-Leibniz-Zentrum fuer Infor-
matik.
Goodman-Strauss, C. (2001). Compass and straightedge
in the poincaré disk. The American Mathematical
Monthly, 108(1):38–49.
He, J., Balzano, L., and Lui, J. (2011). Online robust sub-
space tracking from partial information. arXiv preprint
arXiv:1109.3827.
Hinton, G. and Roweis, S. T. (2002). Stochastic neighbor
embedding. In NIPS, volume 15, pages 833–840. Cite-
seer.
Hong, W., Wright, J., Huang, K., and Ma, Y. (2006). Mul-
tiscale hybrid linear models for lossy image repre-
sentation. IEEE Transactions on Image Processing,
15(12):3655–3671.
Jansson, M. and Wahlberg, B. (1996). A linear regression
approach to state-space subspace system identification.
Signal Processing, 52(2):103–129.
Ji, H., Liu, C., Shen, Z., and Xu, Y. (2010). Robust video
denoising using low rank matrix completion. In 2010
IEEE Computer Society Conference on Computer Vi-
sion and Pattern Recognition, pages 1791–1798. IEEE.
Kanatani, K.-i. (2001). Motion segmentation by subspace
separation and model selection. In Proceedings Eighth
IEEE International Conference on computer Vision.
ICCV 2001, volume 2, pages 586–591. IEEE.
Kang, Z., Peng, C., and Cheng, Q. (2016). Top-n recom-
mender system via matrix completion. In Proceedings
of the AAAI Conference on Artificial Intelligence, vol-
ume 30.
Kiefer, A., Rahman, M., et al. (2021). An analytical survey
on recent trends in high dimensional data visualization.
arXiv preprint arXiv:2107.01887.
Kirby, M. and Peterson, C. (2017). Visualizing data sets on
the grassmannian using self-organizing mappings. In
2017 12th international workshop on self-organizing
maps and learning vector quantization, clustering and
data visualization (WSOM), pages 1–6. IEEE.
Klimovskaia, A., Lopez-Paz, D., Bottou, L., and Nickel, M.
(2020). Poincaré maps for analyzing complex hier-
archies in single-cell data. Nature communications,
11(1):1–9.
Knudsen, T. (2001). Consistency analysis of subspace identi-
fication methods based on a linear regression approach.
Automatica, 37(1):81–89.
Kohonen, T. (1982). Self-organized formation of topolog-
ically correct feature maps. Biological cybernetics,
43(1):59–69.
Kohonen, T. (1990). The self-organizing map. Proceedings
of the IEEE, 78(9):1464–1480.
Kohonen, T. (1998). The self-organizing map. Neurocom-
puting, 21(1-3):1–6.
Kohonen, T. (2013). Essentials of the self-organizing map.
Neural networks, 37:52–65.
Koohi, H. and Kiani, K. (2017). A new method to find
neighbor users that improves the performance of col-
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
36

laborative filtering. Expert Systems with Applications,
83:30–39.
Lee, D., Kim, B. H., and Kim, K. J. (2010). Detecting
method on illegal use using pca under her environment.
In 2010 International Conference on Information Sci-
ence and Applications, pages 1–6. IEEE.
Liu, S., Maljovec, D., Wang, B., Bremer, P.-T., and Pas-
cucci, V. (2016). Visualizing high-dimensional data:
Advances in the past decade. IEEE transactions on vi-
sualization and computer graphics, 23(3):1249–1268.
Lu, L. and Vidal, R. (2006). Combined central and subspace
clustering for computer vision applications. pages 593–
600.
McInnes, L., Healy, J., and Melville, J. (2018). Umap:
Uniform manifold approximation and projection for di-
mension reduction. arXiv preprint arXiv:1802.03426.
Mevel, L., Hermans, L., and Van der Auweraer, H. (1999).
Application of a subspace-based fault detection method
to industrial structures. Mechanical Systems and Signal
Processing, 13(6):823–838.
Nickel, M. and Kiela, D. (2017). Poincaré embeddings
for learning hierarchical representations. Advances in
neural information processing systems, 30:6338–6347.
Novembre, J., Johnson, T., Bryc, K., Kutalik, Z., Boyko,
A. R., Auton, A., Indap, A., King, K. S., Bergmann, S.,
Nelson, M. R., et al. (2008). Genes mirror geography
within europe. Nature, 456(7218):98–101.
Parsons, L., Haque, E., and Liu, H. (2004). Subspace clus-
tering for high dimensional data: a review. Acm sigkdd
explorations newsletter, 6(1):90–105.
Recht, B. (2011). A simpler approach to matrix completion.
Journal of Machine Learning Research, 12(12).
Shaham, U. and Steinerberger, S. (2017). Stochastic neigh-
bor embedding separates well-separated clusters. arXiv
preprint arXiv:1702.02670.
Song, Y., Westerhuis, J. A., Aben, N., Michaut, M., Wessels,
L. F., and Smilde, A. K. (2019). Principal compo-
nent analysis of binary genomics data. Briefings in
bioinformatics, 20(1):317–329.
Stewart, G. W. (1998). An updating algorithm for subspace
tracking. Technical report.
Sun, W., Zhang, L., Du, B., Li, W., and Lai, Y. M. (2015).
Band selection using improved sparse subspace clus-
tering for hyperspectral imagery classification. IEEE
Journal of Selected Topics in Applied Earth Observa-
tions and Remote Sensing, 8(6):2784–2797.
Tang, J., Liu, J., Zhang, M., and Mei, Q. (2016). Visualizing
large-scale and high-dimensional data. In Proceedings
of the 25th international conference on world wide
web, pages 287–297.
Tenenbaum, J. B., Silva, V. d., and Langford, J. C. (2000). A
global geometric framework for nonlinear dimension-
ality reduction. science, 290(5500):2319–2323.
Tomasi, C. and Kanade, T. (1992). Shape and motion
from image streams under orthography: a factoriza-
tion method. International journal of computer vision,
9(2):137–154.
Tron, R. and Vidal, R. (2007). A benchmark for the com-
parison of 3-d motion segmentation algorithms. In
2007 IEEE conference on computer vision and pattern
recognition, pages 1–8. IEEE.
Ullah, F., Sarwar, G., and Lee, S. (2014). N-screen aware
multicriteria hybrid recommender system using weight
based subspace clustering. The Scientific World Jour-
nal, 2014.
Van der Maaten, L. and Hinton, G. (2008). Visualizing data
using t-sne. Journal of machine learning research,
9(11).
Van Overschee, P. (1997). Subspace identification: Theory,
implementation, application.
Vaswani, N., Bouwmans, T., Javed, S., and Narayanamurthy,
P. (2018). Robust subspace learning: Robust pca, ro-
bust subspace tracking, and robust subspace recovery.
IEEE signal processing magazine, 35(4):32–55.
Vidal, R. and Favaro, P. (2014). Low rank subspace cluster-
ing (lrsc). Pattern Recognition Letters, 43:47–61.
Vidal, R., Tron, R., and Hartley, R. (2008). Multiframe mo-
tion segmentation with missing data using powerfactor-
ization and gpca. International Journal of Computer
Vision, 79(1):85–105.
Wu, J. and Zhang, X. (2001). A pca classifier and its ap-
plication in vehicle detection. In IJCNN’01. Interna-
tional Joint Conference on Neural Networks. Proceed-
ings (Cat. No. 01CH37222), volume 1, pages 600–604.
IEEE.
Wu, J., Zhang, X., and Zhou, J. (2001). Vehicle detec-
tion in static road images with pca-and-wavelet-based
classifier. In ITSC 2001. 2001 IEEE Intelligent Trans-
portation Systems. Proceedings (Cat. No. 01TH8585),
pages 740–744. IEEE.
Xia, C.-Q., Han, K., Qi, Y., Zhang, Y., and Yu, D.-J. (2017).
A self-training subspace clustering algorithm under
low-rank representation for cancer classification on
gene expression data. IEEE/ACM transactions on
computational biology and bioinformatics, 15(4):1315–
1324.
Xu, J., Ithapu, V. K., Mukherjee, L., Rehg, J. M., and Singh,
V. (2013). Gosus: Grassmannian online subspace
updates with structured-sparsity. In Proceedings of
the IEEE international conference on computer vision,
pages 3376–3383.
Yang, A. Y., Wright, J., Ma, Y., and Sastry, S. S. (2008).
Unsupervised segmentation of natural images via lossy
data compression. Computer Vision and Image Under-
standing, 110(2):212–225.
Zhang, H., Ericksen, S. S., Lee, C.-p., Ananiev, G. E., Wlo-
darchak, N., Yu, P., Mitchell, J. C., Gitter, A., Wright,
S. J., Hoffmann, F. M., et al. (2019). Predicting kinase
inhibitors using bioactivity matrix derived informer
sets. PLoS computational biology, 15(8):e1006813.
Zhang, W., Wang, Q., Yoshida, T., and Li, J. (2021). Rp-
lgmc: rating prediction based on local and global in-
formation with matrix clustering. Computers & Oper-
ations Research, 129:105228.
Visualizing Grassmannians via Poincare Embeddings
37
APPENDIX
2 Clusters 3 Clusters 4 Clusters 5 Clusters
Classic 3DnPCAt-SNE
GDMaps
GrassCaré (this paper)
Figure 9: Alternative visualizations of clusters in
G(3,1)
.
GrassCaré produces a more accurate representation of the
Grassmannian
, e.g., the case of
K = 4
clusters, where nPCA and even the 3D representation display Clusters 1 and 2
(cyan and yellow) nearly diametrically apart. In reality they are quite close, as depicted by GrassCaré. See discussion for
details.
IVAPP 2023 - 14th International Conference on Information Visualization Theory and Applications
38
Parameter GrassCaré t-SNE nPCA GDMaps
m = 50;r = 5 59.71s 15.77s 0.04s 0.09s
m = 50;r = 20 59.40s 16.5s 0.03s 0.11s
m = 100;r = 5 60.03s 16.26s 0.03s 0.10s
m = 100;r = 20 77.16 23.10 0.05 0.16
Figure 10: Representation error of GrassCaré (this paper) and other methods for high-dimensional Grassmannians
G(m,r)
.
The experiments were performed on GoogleColab pro version with cpu.
Visualizing Grassmannians via Poincare Embeddings
39
