
Gene Editing of Saccharomyces Cerevisiae Using CRISPR
Chenyu Yang
1,*
and Yilin Li
2
1
Queen Mary College, Nanchang University, the Medical College, Block C, No. 999, Xuefu Avenue, Honggutan New
District, Nanchang, China
2
Biomedical Science Department, Southern University of Science and Technology, No. 1088, Xueyuan Avenue, Nanshan
District, Shenzhen, China
Keywords: CRISPR Cas9, PCR’ Gene Editing, Fluorescent Protein Gene.
Abstract: This experiment aims to integrate the mCherry gene into saccharomyces cerevisiae (S.cerevisiae) genome and
examine its expression in S.cerevisiae. This experiment is to detect whether the integration and mCherry gene
expression are successful. By applying the CRISPR/Cas9 and PCR techniques, we have shown that the
integration into S. cerevisiae is successful, and the mCherry gene is expressed perfectly. This experiment
shows the feasibility of the CRISPR/Cas9 technique, which can be applied in clinical treatments or mass
production of certain medical products.
1 INTRODUCTION
CRISPR/Cas9 is a mechanism of gene editing,
applied to form guide RNA (gRNA) plasmids and
integrate target gene into cells, in order to express the
target gene (L.S.Q, 2013).
CRISPR/Cas9 is categorized into two types: Non-
Homologous End Joining (NHEJ) and Homology
Directed Repair (HDR). NHEJ is a pathway that
repairs double-strand break mutations by insertions or
deletions, before knocking out the gene. HDR repairs
the template to complete gene editing.
The technique has the potential to affect scientific
research. It enables more precise gene editing, a
higher probability of successful transformation and a
purer transformation product. This technique
supports experts to express certain DNA and obtain
the target substances (Yan, 2022). CRISPR/Cas9 is a
perfect tool to transform cells, making them produce
certain substances, which can also be applied in
medical treatments, mass production of certain
medical products, and so on.
CRISPR/Cas9 employs the Cas9 Nickases from
BJ5464-p414-Cas9 as a catalyst. The gRNA binds
with and cuts the target sequence, which is the pre-
designed target gene (Wu, 2014). In this experiment,
the binding happens with the plasmids and transforms
the S.cerevisiae. The advantage of CRISPR/Cas9 is
its high efficiency, which increases its ability to
modify the gene accurately and efficiently. Nucleic
acid probe-based qPCR identifies the mutation site
rapidly. It can be used for genotyping to the
subsequent formation of a colony (
Vasu, 2021)
.
CRISPR/Cas9 is more accurate than the double
enzyme digestion technique, and it provides blunt
ends, which means the sequence cut by CRISPR/Cas9
will be less possible to adhere to other sequences. The
sequence cut by the technique is more likely to be
pure target fragments. The difficulty in purifying and
detecting the aim proteins (the expression of the
target gene) will decrease. However, as a pre-mature
technique, CRISPR/Cas9 still needs to be optimized.
The experiment aims to use S.cerevisiae to
express the fluorescent protein gene by the
CRISPR/Cas9 technology. The exogenous
expression box (YPet fluorescent protein) has already
been structured. In this experiment, we have activated
the S. cerevisiae BJ5464/ p414-TEF1p-Cas9-CYC1t
culture and made the competent cells to gain S.
cerevisiae host bacteria.
2 MATERIALS AND METHODS
2.1 Materials
S. cerevisiae BJ5464/ p414- TEF1p- Cas9- CYC1t,
E.coli.
PCR centrifuge tubes, 1.5ml centrifuge tubes.
PCR Machine, electrophoresis apparatus.
404
Yang, C. and Li, Y.
Gene Editing of Saccharomyces Cerevisiae Using CRISPR.
DOI: 10.5220/0012021800003633
In Proceedings of the 4th International Conference on Biotechnology and Biomedicine (ICBB 2022), pages 404-409
ISBN: 978-989-758-637-8
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
2.2 Methods
2.2.1 Design of gRNA Plasmids
Design the gRNA sequence to participate in the
reaction with the plasmids in the experiment. Anneal
the primer, bind the gRNA with the enzyme carrier
(p426- pSNR-gRNA- tSUP4) of E. coli, let the
transformed plasmids proliferate in the E. coli, and
then extract the plasmids.
1) Guide sequences
LEU- sgRNA target sequence:
TTTGTTGCCATCTGCGTCCT;
LEU- sgRNA- F: 5’ -
GATCTTTGTTGCCATCTGCGTCCTG- 3’;
LEU- sgRNA- R: 5’ -
AAAACAGGACGCAGATGGCAACAAA- 3’.
2) Primer annealing
Suspend the forward and reverse oligo in 100uM
HEPES (pH= 7.9) water.
3) Connection
Dilute the tenfold annealed double strands DNA
(0.1μL)
T4 DNA Ligase (0.25μL)
Buffer (0.5 μL)
Carrier (0.5 μL)
Up to 5 μL
Reaction conditions: 16℃, overnight.
4) E. coli transformation
Remove the competent cells (100μL) out from a
fridge at -80℃ and place them on ice for about 30
min. Add 10μL connection products and mix them,
and place them on ice for about 30 min.
Heat the competent cells with the connection
products in a water bath for 90 s, then place them on
ice for 2 min immediately. Add the competent cells
into 800μL LB medium without screening resistance
(preheated to 37℃), cultivate at 37℃, at a 180-rpm
table for about 1h.
Centrifuge the cells (at 300 rpm) for 3 min,
discard 600μL supernatant, resuspend the rest of the
thalli and transfer 200μL liquid smear to the
ampicillin-resistance LB solid tablet. Cultivate at
37℃ for about 20-22h.
Collect the transformant and patch on the
ampicillin- resistance LB solid tablet and recultivate
at 37℃.
5) Colony PCR
Dissolve some thalli in 20μL ddH
2
O, at 95℃ for
10 min. Use the supernatant as the template of PCR.
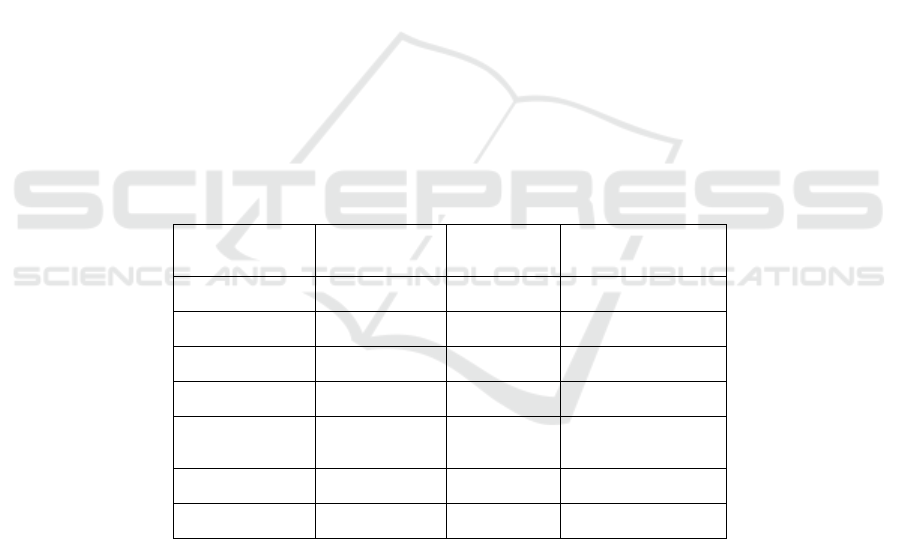
Table 1: Colony PCR.
Reagents
Dosages
(μL)
Steps Conditions
Hief 5 1 94℃, 5 min
F 0.5 2 94℃, 30 s
R 0.5 3 57℃, 30 s
Template 2 4 72℃, 30 s/kb
ddH
2
O 2 5
Go to step2 for 33
times more
Up to 10 6 72℃, 10 min
7 16℃, forever
Detect the PCR product by agarose gel, choose the
suspected positive clone strain, inoculate to LB/
Amp+ fluid nutrient medium, at 37℃, and cultivate
overnight. Extract the recombinant plasmids to
sequence the gene.
2.2.2 Acquisition of the Host Bacteria
Activate the S. cerevisiae BJ5464/ p414-TEF1p-
Cas9- CYC1t culture to make competent cells that
will be used as host bacteria.
Select a single colony and place it into 5ml SD
(without tryptophan) liquid culture medium, at 30℃,
and cultivate overnight, until the bacteria solution OD
increases to 3.0-5.0.
After the bacteria solution grows to the expected
OD600, transfer a proper amount of solution into
another 5ml SD (without tryptophan) liquid culture
medium. Set the starting OD600 of the solution
between 0.2 and 0.4, cultivate at 30℃, 200 rpm for
about 4-6 hours. The OD600 should be kept between
0.8-1.0.
Gene Editing of Saccharomyces Cerevisiae Using CRISPR
405

Transfer 1 mL solution to a 1.5 mL centrifuge
tube, centrifuge for 3 min, at normal temperature, at
7000 rpm, and discard all the supernatant.
Wash away the residual medium, prepare 1 mL
wash solution to suspend the bacteria, centrifuge for
3 min, at normal temperature, at 3000 rpm, and
discard all the supernatant.
Prepare 200μL lithium cation solution to suspend
the bacteria, which is the BJ5464- p414- Cas9
competent cells. Sub-package 25μL in each tube, then
either start the transformation of the plasmid or store
it in an ultra-low temperature refrigerator at -80℃.
2.2.3 Transformation of the S. cerevisiae
The donor DNA has been pre-designed. After the
PCR proliferation, use SDS-PAGE to detect if the
plasmids contain the donor sequences. Then purify
the plasmids to get the donor DNA. Mix the plasmids
transformed by gRNA and the donor DNA to
transform the S. cerevisiae. Perform coated-plate
culture and colony PCR, then obtain the transformed
colony.
Donor Primers:
LEU-Donor- F:
TCATCTCCGATGAAGCCTCCGTTATCCCA
GGTTCCTTGGGCATAGCTTCAAAATGTTTC
LEU-Donor-R:
AGTAAGCTACTATGAAAGACTTGTCTGGC
AAAGAGGCCAAGGACGCAGATGGCAACAAA
.
Table 2. PCR (Amplify the donor fragment)
Reagents
Dosages
(μL)
Steps Conditions
PrimeStar Max
Premix(2x)
25 1 94℃, 2 min
F (20 μM) 0.5 2 98℃, 10 s
R (20 μM) 0.5 3 52℃, 30 s
Template 0.2 4 72℃, 2 min
ddH
2
O 23.8 5
Go to step2 for 29
times more
Up to 50 6 72℃, 10 min
7 16℃, forever
2.2.4 Acquisition and Detection of the
Products
Cultivate the positive transformant in a tube with
YPD culture medium for 12 h, transfer to a 50 mL
conical flask to cultivate. Control the starting OD600
to be 0.05. Cultivate in 10 mL YPD, at 30℃, at 200
rpm, for 12 h. Place 200μL solution in a black,
transparent 96-pore plate to perform the fluorescence
test.
Excitation wavelength: 485nm.
Emission wavelength: 528nm.
3 RESULTS
Figure 2 represents the p426-pSNR52-gRNA (LEU)-
tSUP4 colony PCR results. It shows the suspected
positive strain.
Figure 1: The plate of p426-pSNR52-gRNA (LEU)-tSUP4.
ICBB 2022 - International Conference on Biotechnology and Biomedicine
406

Figure 2: Results of p426-pSNR52-gRNA (LEU)-tSUP4
colony PCR.
Figure 3: Results of concentration detection of p426-
pSNR52-gRNA (LEU)-tSUP4 plasmid extraction.
Figure 4: Results of BJ5464-p414-Cas9 streaking of the
strain.
This above figure shows the results of the
activated S. cerevisiae.
Figure 5: Results of electrophoresis detection of donor
fragments on the LEU site.
From Figure 5, we can see that the donor sequence
is less than 2000bp.
Figure 6: Results of BJ5464::LEU-pTEF1-YPet-tFBA1
transformant coating.
Figure 6 represents that the colonies have been
transformed to S. cerevisiae.
Figure 7: Results of BJ5464::LEU-pTEF1-YPet-tFBA1
colony PCR.
Gene Editing of Saccharomyces Cerevisiae Using CRISPR
407
From Figure 7, we know the target sequence is
3000bp, so the result numbered 1 in the figure is a
positive stripe.
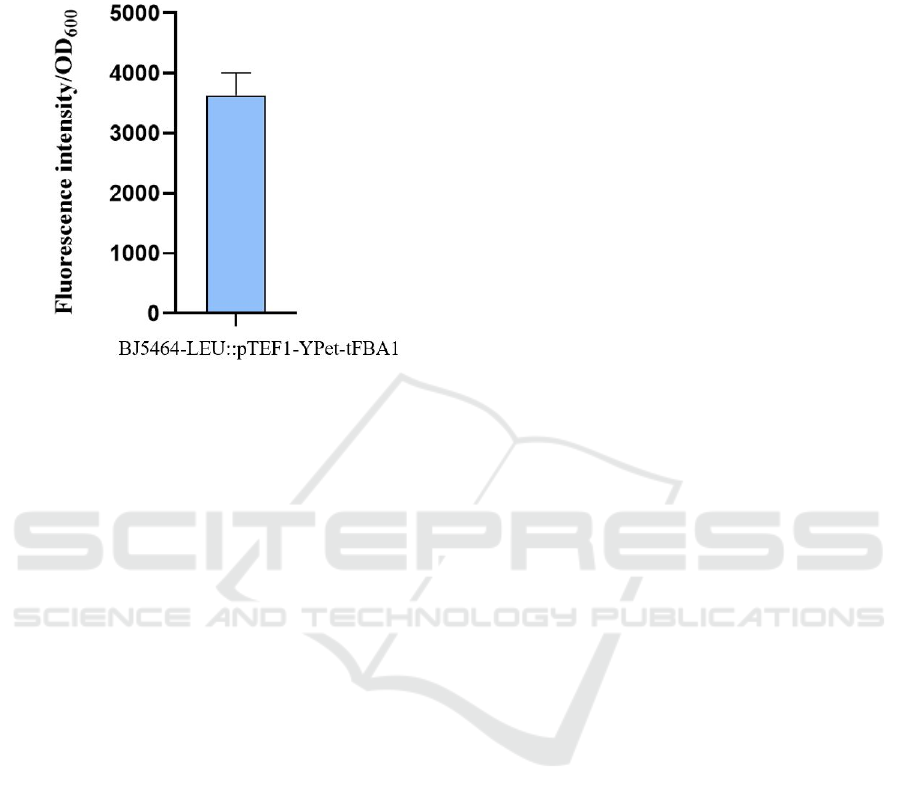
Figure 8: BJ5464::LEU-pTEF1-YPet-tFBA1.
Figure 8 shows the relative fluorescence value of
BJ5464::LEU-pTEF1-YPet-tFBA1. The result shows
the OD600 is between 3000 and 4000.
4 CONCLUSION
In this experiment, we use the technology of
CRISPR/Cas9 to transform the genetic material of S.
cerevisiae to produce specific fluorescent protein.
The CRISPR/Cas9 system was discovered in the
Streptococcus pyogenes CRISPR pathway, and it is
widely used in research (
Vasu, 2021)
. CRISPR is
also applicable in clinical treatments, with the
challenges and prospects to realize the clinical
potential widely studied. The transformation method
of CRISPR-Cas9 technology is the same as that of
most gene technologies, but due to the specificity of
Cas enzyme, it can successfully bind to the gene chain
at a specific site (Cheng, 2021; Lu, 2015).
CRISPR/Cas9 has the potential in many
application prospects, for example, cancer treatment
(Khajuria, 2021; Wang, 2021), AIDS (Acquired
Immune Deficiency Syndrome) therapies (Xiao,
2019) and cardiac diseases (Schreurs, 2021).
Cancer is one of the major causes of death, and
humans have worked hard to treat it for many years.
CRISPR/Cas9 optimizes the way people usually treat
tumours (Xing, 2020). Experiments have shown that
CRISPR/Cas9 technology can be used to methylate
DNA to alter genetic performance, which is of great
significance for the treatment of many genetic
diseases (Katayama, 2021). Another successful trial
is that researchers from the University of Sichuan,
China, had been able to inject genetically modified
lymphocytes for the first time to a patient with lung
cancer as a therapeutic approach to promote the
immune system's response for eliminating malignant
tumor cells (Castillo, 2016).
AIDS is a disease that seriously threatens human
health. The duplication of HIV (human
immunodeficiency virus) can only be restrained for
the time being. Using CRISPR/Cas9 as an effective
tool to edit the gene is a possible approach to cure
AIDS (Pelletier, 2015).
CRISPR/Cas9 is a technology with great promise.
There have also been successes in treating other
disease. Sickle cell disease is the name for a group of
inherited health conditions that affect the red blood
cells. The most serious type is called sickle cell
anaemia. Researchers successfully induced
hemoglobin for the treatment of sickle cell disease
(SCD) (Demirci, 2021; Philippidis, 2021).
REFERENCES
Castillo Andres. Gene editing using CRISPR-Cas9 for the
treatment of lung cancer. [J].Colombia medica (Cali,
Colombia), 2016,47(4),178-180.
Cheng Hao; Zhang Feng; Ding Yang.CRISPR/Cas9
Delivery System Engineering for Genome Editing in
Therapeutic Applications. [J]. Pharmaceutics, 2021,
13(10).
Demirci Selami; Leonard Alexis; Essawi Khaled; Tisdale
John F..CRISPR-5Cas9 to induce fetal hemoglobin for
the treatment of sickle cell disease [J]. Molecular
Therapy - Methods & Clinical Development, 2021, 23,
276-285.
Katayama Shota; Andou Masao. Editing of DNA
methylation using CRISPR/Cas9 and a ssDNA template
in human cells. [J]. Biochemical and biophysical
research communications,2021,581,20-24.
Khajuria Ocean; Sharma Neha. Epigenetic targeting for
lung cancer treatment via CRISPR/Cas9 technology [J].
Advances in Cancer Biology - Metastasis, 2021,3.
L.S.Q., M.H.L., L.A.G. and X.W. wrote the manuscript.
J. S.W. , W.A.L. an d L. S.Q. supervised the research;
CRISPR interference (CRISPRi) for sequence-specific
control of gene expression. Nat Protoc. 2013 Nov;
8(11): 2180–2196e.
Lu Xiao-Jie, Xue Hui-Ying, Ke Zun-Ping, Chen Jin-Lian,
Ji Li-Juan.CRISPR-Cas9: a new and promising player
in gene therapy[J].Journal of Medical Genetics, 2015,
52(5).
Luyao Wang, Yurong Chen, [...], and Xiangpeng Dai; The
Application of CRISPR/Cas9 Technology for Cancer
Immunotherapy: Current Status and Problems. Front
Oncol. 2021; 11: 704999.
ICBB 2022 - International Conference on Biotechnology and Biomedicine
408

Pelletier Stephane, Gingras Sebastien, Green Douglas
R.Mouse genome engineering via CRISPR-Cas9 for
study of immune function.[J].Immunity,2015,42(1),18-
27.
Philippidis Alex.CRISPR-Cas9 Gene-Edited Therapy
Shows Sustained Treatment Response. [J]. Human gene
therapy,2021,32(13-14),642-644.
Qiaoqiao Xiao, Deyin Guo, and Shuliang Chen;
Application of CRISPR/Cas9-Based Gene Editing in
HIV-1/AIDS Therapy. Front Cell Infect Microbiol.
2019; 9: 69.
Schreurs Juliët; Sacchetto Claudia; Colpaert Robin M. W.;
Vitiello Libero; Rampazzo Alessandra; Calore Martina.
Recent Advances in CRISPR/Cas9-Based Genome
Editing Tools for Cardiac Diseases [J]. International
Journal of Molecular Sciences, 2021, 22(20), 10985-
10985.
Vasu Kommireddy; Fox Paul L.Screening of CRISPR-
Cas9-generated point mutant mice using MiSeq and
locked nucleic acid probe PCR.[J].STAR protocols,
2021,2(4), 100785-100785.
Xuebing Wu; Andrea J. Kriz; Phillip A. Sharp. Target
specificity of the CRISPR-Cas9 system [J].
Quantitative Biology, 2014,2(2),59-70.
Xing Hui, Meng Ling hua.CRISPR-cas9: a powerful tool
towards precision medicine in cancer treatment[J].Acta
Pharmacologica Sinica,2020,41(5), 583-587.
Yan Rihui; Lin Xianwu. CRISPR/Cas9-Mediated Genome
Editing System in Insect Genomics and Pest
Management. [J]. Methods in molecular biology
(Clifton, N.J.), 2022,2360,347-366.
Gene Editing of Saccharomyces Cerevisiae Using CRISPR
409
