
How Textual Datasets Enhance the PADI-Web Tool?
Mathieu Roche
1,3 a
, Elena Arsevska
2,3 b
, Sarah Valentin
1,2,3,4,5 c
, Sylvain Falala
2,6
,
Julien Rabatel
7 d
and Renaud Lancelot
2,3 e
1
UMR TETIS (Land, Environment, Remote Sensing and Spatial Information), University of Montpellier, AgroParisTech,
CIRAD, CNRS, INRAE, Montpellier, France
2
UMR ASTRE (Unit for Animals, Health, Territories, Risks and Ecosystems), University of Montpellier, CIRAD, INRAE,
Montpellier, France
3
French Agricultural Research for Development (CIRAD), France
4
Department of Biology, University of Sherbrooke, Sherbrooke, Canada
5
Quebec Centre for Biodiversity Science, McGill University, Montreal, Canada
6
National Research Institute for Agriculture, Food and the Environment (INRAE), France
7
Freelance Data Scientist, Montpellier, France
Keywords:
Text Mining, Information Retrieval, Named Entity Recognition, Event-based Surveillance, Epidemic Intelli-
gence.
Abstract:
The ability to rapidly detect outbreaks of emerging infectious diseases is a health priority of global health
agencies. In this context, event-based surveillance (EBS) systems gather outbreak-related information from
heterogeneous data sources, including online news articles. EBS systems, thus, increasingly marshal text-
mining methods to alleviate the amount of manual curation of the freely available text. This paper documents
the use of datasets obtained through an EBS system, PADI-Web (Platform for Automated extraction of Disease
Information from the web), dedicated to digital outbreak detection in animal health. This paper describes the
datasets used for improving 3 important tasks related to PADI-Web, i.e., news classification, information
extraction and dissemination.
1 INTRODUCTION
The ability to identify emerging and re-emerging dis-
eases is challenging for practitioners in the health
domain (WHO, 2014). In this context, event-based
surveillance (EBS) gathers information from hetero-
geneous data sources, including online news articles
(Barboza et al., 2013). EBS systems integrate text-
mining methods to deal with huge amounts of textual
data (Mutuvi et al., 2021). Text mining aims at dis-
covering new information from textual datasets (i.e.,
corpus) (Meng, 2021). Different text-mining meth-
ods associated with labeled textual datasets (i.e., news
data) are integrated into the main steps of EBS sys-
tems, these methods include data acquisition, infor-
mation retrieval (i.e., identification of relevant texts),
a
https://orcid.org/0000-0003-3272-8568
b
https://orcid.org/0000-0002-6693-2316
c
https://orcid.org/0000-0002-9028-681X
d
https://orcid.org/0000-0002-4742-923X
e
https://orcid.org/0000-0002-5826-5242
epidemiological information extraction and locating
information to communicate to end-users.
This paper focuses on the use of datasets to mine
news data pertaining to the health domain. These data
science approaches are integrated into an EBS system
called PADI-Web (Platform for Automated extraction
of Disease Information from the web). PADI-Web is
dedicated to animal health surveillance and addresses
disease-based and symptom-based surveillance.
PADI-web collects news articles based on Google
News due to its international coverage and flexible
RSS feeds. To detect news dealing with dedicated dis-
eases (e.g., avian influenza, West Nile virus, African
swine fever) or unknown diseases (i.e., Disease X),
the RSS feeds use specific keywords (e.g., disease
names, association of terms on hosts and symptoms).
The collected news items are automatically classified
as ”relevant” or ”irrelevant” using machine learning
techniques (Kowsari et al., 2019). The relevant news
corresponds to recent or current infectious animal
health events. Moreover, a more fine-grained classi-
Roche, M., Arsevska, E., Valentin, S., Falala, S., Rabatel, J. and Lancelot, R.
How Textual Datasets Enhance the PADI-Web Tool?.
DOI: 10.5220/0011590400003318
In Proceedings of the 18th International Conference on Web Information Systems and Technologies (WEBIST 2022), pages 327-331
ISBN: 978-989-758-613-2; ISSN: 2184-3252
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
327
fication using machine learning approaches has been
implemented to highlight more accurate topics. To
identify key pieces of epidemiological information in
the news (i.e., location and date of outbreaks, affected
hosts, their numbers and encountered clinical signs,
and so forth), PADI-web integrates information ex-
traction (IE) methods (Schmitt et al., 2019). Finally,
our system proposes automatic notifications to end-
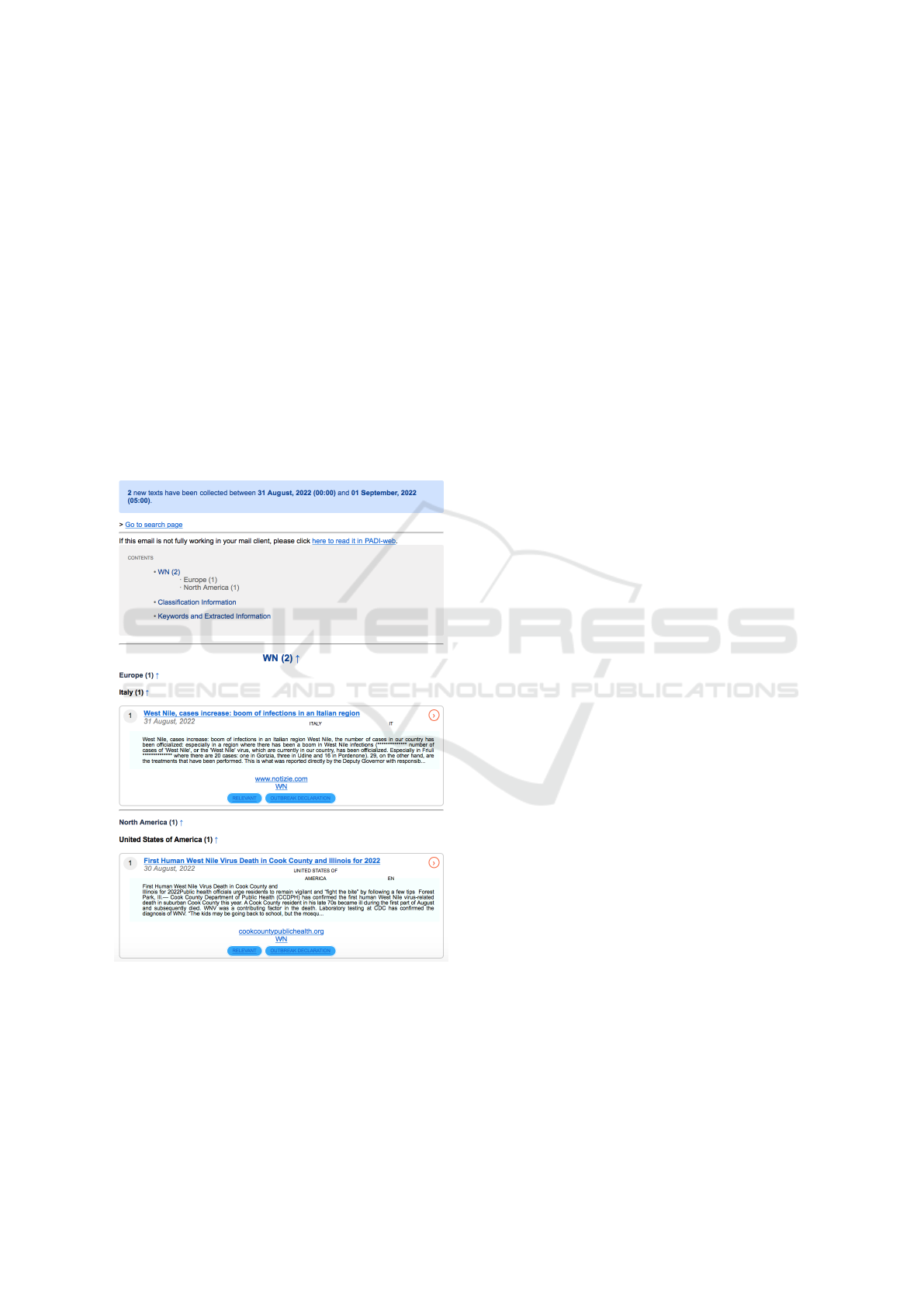
users (see Figure 1). The outputs of the PADI-Web
system are accessible at https://padi-web.cirad.fr/en/
This paper is an overview of the datasets used and
produced to improve the modules of PADI-Web and
for its dissemination. The paper is structured as fol-
lows: In Section 2 we describe the datasets used for
improving PADI-Web (i.e., the classification, extrac-
tion and dissemination tasks), while Section 3 pro-
poses a discussion of the future directions of this
work.
Figure 1: PADI-web notification for West Nile virus (WN).
2 PADI-Web DATASETS
After a web-crawling approach is used to collect on-
line news texts, PADI-Web uses a classification mod-
ule to identify the relevant texts and sentences that
describe outbreak-related events. The system extracts
epidemiological information, such as hosts, locations,
dates and symptoms, from the collected news text
data. The PADI-Web pipeline summarized in Fig-
ure 3 and detailed in (Arsevska et al., 2018; Valentin
et al., 2020a; Valentin et al., 2021a) collected more
than 400,000 news items dealing with animal health
to extract the epidemiological events. Several datasets
have been built to test and validate the usefulness of
PADI-Web as an EBS system, as well as to create
new features (e.g., modules) in successive versions
of the tool. This paper summarizes the multiuse of
the PADI-Web datasets freely available on Dataverse
(Arsevska et al., 2017; Valentin et al., 2019; Rabatel
et al., 2017; Valentin et al., 2020b; Roche and Arse-
vska, 2018) for the different tasks summarized in the
following subsections.
2.1 Corpora for Improving
Classification
In the second version of PADI-Web (Valentin et al.,
2020a), a dedicated module uses a supervised ma-
chine learning approach to predict if a news item is
relevant. Relevant news is text dealing with a new,
suspected or unknown outbreak. For this classifica-
tion task, specific labeled data must be used. For
instance, a dataset of news can be used to learn a
model for classification (Rabatel et al., 2017). Cur-
rently, the trained classification models integrated into
PADI-web reach a mean accuracy score of 0.94 for
the article-level relevance task using a random forest
classifier. Moreover, this corpus is used for the infor-
mation extraction process (see the following subsec-
tion).
In the third version of PADI-Web, a fine-
grained classification on a sentence level is proposed
(Valentin et al., 2021a). A dataset of sentences has
been manually labeled to train the model for sentence
classification (Valentin et al., 2019). Each sentence
has two labels, an event label (e.g., current event, old
event, general information) and an information label
(e.g., descriptive epidemiology and distribution, pre-
ventive and control measures, economic and political
consequences).
Epidemiological information can be extracted
from relevant news and sentences as detailed in the
following subsection.
2.2 Corpora for Improving Extraction
To build a model for extracting information, epidemi-
ological entities (e.g., locations, diseases, hosts, dates,
number of cases) are also manually labeled in the
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
328

news (Rabatel et al., 2017). In the first version of
PADI-Web (Arsevska et al., 2018), the information
extraction module consisted of two stages: (1) can-
didate identification and (2) candidate verification.
Candidate identification aims to detect all the possi-
ble candidates for each type of epidemiological entity
extracted from the news data. Each candidate in the
labeled dataset is associated with elements that de-
scribe its context, e.g., its surrounding words. Rules
to identify a candidate based on its context are auto-
matically discovered using a data mining technique.
These rules represent the features of supervised ma-
chine learning approaches. A support vector machine
(SVM) model is trained on the labeled dataset to pre-
dict whether the candidates are relevant. To learn this
named entity recognition model, the labeled data (Ra-
batel et al., 2017) have been used (Arsevska et al.,
2018).
In the last version of PADI-Web (Valentin et al.,
2021a), a well-known named entity recognition tool,
spaCy, was integrated into the version. Classic mod-
els associated with this tool recognize generic named
entities, such as locations and organizations. More-
over, specific models for entity recognition related
to the animal disease surveillance domain are imple-
mented using the spaCy tool. The labeled dataset de-
scribed above (Rabatel et al., 2017) is used to enrich
the current spaCy models.
The epidemiological data that can be downloaded
(see Figure 2) are used in different studies summa-
rized in the following subsection.
Figure 2: PADI-web data to download.
2.3 Datasets for Analysis and
Dissemination
The relevant signals detected with PADI-Web for
African swine fever, foot-and-mouth disease, blue-
tongue and avian influenza in 2016 are manually la-
beled in (Arsevska et al., 2017). Each row represents
the collected news’s extracted epidemiological infor-
mation about a potential outbreak (a signal). Each
signal is constructed with one location automatically
detected as correct in the news, combined with all
other epidemiological information types detected in
the same news article. This enables us to conduct a
retrospective analysis highlighting the sensitivity of
PADI-Web to detect primary outbreaks. From Jan-
uary to June 2016, PADI-Web detected signals for
64% of all the primary outbreaks of African swine
fever, 53% of the avian influenza outbreaks, 25% of
the bluetongue outbreaks and 19% of the foot-and-
mouth disease outbreaks.
Another retrospective study proposed to evaluate
the capacity of the three EBS systems (i.e., ProMED,
HealthMap and PADI-Web) to detect early COVID-
19 emergence signals (Valentin et al., 2021b; Valentin
et al., 2020b). We discussed how an EBS system fo-
cusing on animal health (i.e., PADI-web) could detect
a newly emerging pathogen. This aspect is crucial in
a One Health context and for syndromic surveillance.
Finally, specific corpora are built for training and
dissemination (e.g., courses
1
, tutorials
2
, etc.). In this
context, the corpus dealing with African swine fever
disease called ”ASF corpora” (Roche and Arsevska,
2018) is used (i) for classification tasks using ma-
chine learning algorithms with the Weka tool
3
and (ii)
for terminology extraction based on the BioTex soft-
ware
4
. This enables highlighting different directions
so the same corpus can be used as if dealing with an-
imal disease surveillance.
3 DISCUSSION
In this paper, we presented a summary of the datasets
used to improve some modules of PADI-Web. More
specifically, the datasets described in the previous
subsection are used to learn models to extract and
analyze epidemiological information in news articles.
These events are based on: (i) The classification of
documents and sentences and; (ii) information extrac-
tion.
Based on these PADI-Web modules, end-users
could apply specific methods to extract events (i.e.,
the detection or suspicion of a disease at a specific
location and date). First, a fine-grained classifica-
tion enables highlighting texts dealing with ”Out-
break declaration” (i.e., document classification). The
second step consists of extracting a group of epidemi-
ological information, such as locations, hosts and
1
https://agritrop.cirad.fr/600001/
2
https://agritrop.cirad.fr/597999/
3
http://old-www.cms.waikato.ac.nz/ml/weka
4
https://github.com/sifrproject/biotex
How Textual Datasets Enhance the PADI-Web Tool?
329
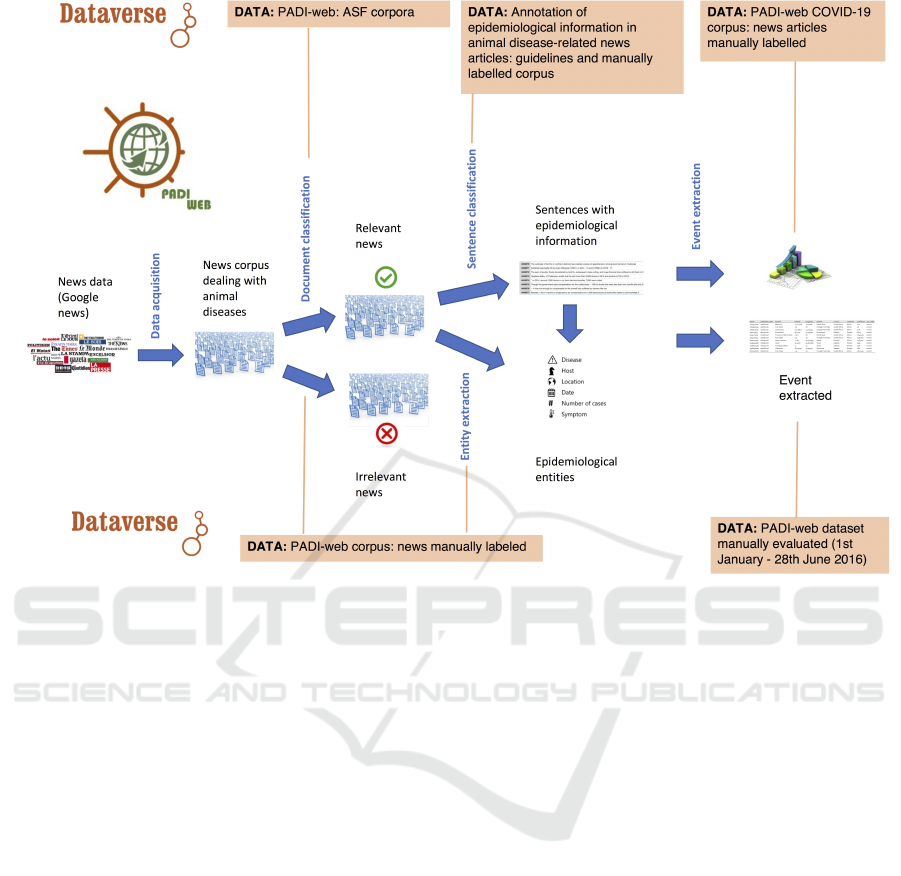
Figure 3: PADI-web process and datasets used.
symptoms, from these specific documents. The algo-
rithm implemented into PADI-Web consists of: (i) se-
lecting sentences with locations (pivot sentence); (ii)
extracting epidemiological information in a context of
3 sentences (i.e., pivot sentence + one sentence before
and after the pivot sentence). This type of extraction is
called ”location-based information” in the PADI-Web
system. An extension of this algorithm will be inte-
grated into our EBS system. In this extension, pivot
sentences will be sentences associated with the label
”Current event” (i.e., sentence classification).
In summary, in the next extensions of PADI-Web,
five types of extractions will be proposed to end users:
• All events extracted in relevant articles based on
locations extracted with spaCy;
• All events extracted in relevant articles based on
locations extracted with spaCy learnt with labeled
data;
• All events extracted at the beginning of the arti-
cles;
• All events extracted in Outbreak articles (i.e.,
document-based classification);
• All events extracted in Outbreak articles (i.e.,
document-based classification) and Current event
sentences (i.e., sentence-based classification).
End-users will be able to use and compare epi-
demiological information obtained with these five
strategies for surveillance.
ACKNOWLEDGEMENTS
This work was funded by the French General Di-
rectorate for Food (DGAL) and the French Agri-
cultural Research Centre for International Develop-
ment (CIRAD). This work was also supported by the
French National Research Agency under the Invest-
ments for the Future Program, referred to as ANR-
16-CONV-0004. This study was partially funded
by EU grant 874850 MOOD and is catalogued as
MOOD041. The contents of this publication are the
sole responsibility of the authors and do not necessar-
ily reflect the views of the European Commission.
REFERENCES
Arsevska, E., Valentin, S., Rabatel, J., de Go
¨
er de Herv
´
e,
J., Falala, S., Lancelot, R., and Roche, M. (2017).
PADI-web dataset manually evaluated (1st January -
28th June 2016) - CIRAD Dataverse.
Arsevska, E., Valentin, S., Rabatel, J., de Go
¨
er de Herv
´
e, J.,
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
330

Falala, S., Lancelot, R., and Roche, M. (2018). Web
monitoring of emerging animal infectious diseases in-
tegrated in the French Animal Health Epidemic Intel-
ligence System. PLOS ONE, 13(8):e0199960.
Barboza, P., Vaillant, L., Mawudeku, A., Nelson, N. P.,
Hartley, D. M., Madoff, L. C., Linge, J. P., Collier, N.,
Brownstein, J. S., Yangarber, R., Astagneau, P., and
on behalf of the Early Alerting, Reporting Project of
the Global Health Security Initiative (2013). Evalua-
tion of epidemic intelligence systems integrated in the
Early Alerting and Reporting project for the detection
of A/H5N1 influenza events. PLoS ONE, 8(3):e57252.
Kowsari, K., Jafari Meimandi, K., Heidarysafa, M., Mendu,
S., Barnes, L., and Brown, D. (2019). Text classifica-
tion algorithms: A survey. Information, 10(4).
Meng, X.-L. (2021). Enhancing (publications on) data
quality: Deeper data minding and fuller data confes-
sion. Journal of the Royal Statistical Society: Series
A (Statistics in Society), 184(4):1161–1175.
Mutuvi, S., Boros, E., Doucet, A., Lejeune, G., Jatowt, A.,
and Odeo, M. (2021). Multilingual epidemic event ex-
traction. In Ke, H., Lee, C. S., and Sugiyama, K., edi-
tors, Towards Open and Trustworthy Digital Societies
- 23rd International Conference on Asia-Pacific Dig-
ital Libraries, ICADL 2021, Virtual Event, Decem-
ber 1-3, 2021, Proceedings, volume 13133 of Lecture
Notes in Computer Science, pages 139–156. Springer.
Rabatel, J., Arsevska, E., de Go
¨
er de Herv
´
e, J., Falala, S.,
Lancelot, R., and Roche, M. (2017). PADI-web cor-
pus: news manually labeled - CIRAD Dataverse.
Roche, M. and Arsevska, E. (2018). PADI-web: ASF cor-
pora - CIRAD Dataverse.
Schmitt, X., Kubler, S., Robert, J., Papadakis, M., and Le-
Traon, Y. (2019). A replicable comparison study of
ner software: Stanfordnlp, nltk, opennlp, spacy, gate.
In 2019 Sixth International Conference on Social Net-
works Analysis, Management and Security (SNAMS),
pages 338–343.
Valentin, S., Arsevska, E., Falala, S., Go
¨
er, J. D., Lancelot,
R., Mercier, A., Rabatel, J., and Roche, M. (2020a).
Padi-web: A multilingual event-based surveillance
system for monitoring animal infectious diseases.
Comput. Electron. Agric., 169:105163.
Valentin, S., Arsevska, E., Rabatel, J., Falala, S., Mercier,
A., Lancelot, R., and Roche, M. (2021a). PADI-web
3.0: A new framework for extracting and disseminat-
ing fine-grained information from the news for animal
disease surveillance. One Health, 13:100357.
Valentin, S., De Waele, V., Vilain, A., Arsevska, E.,
Lancelot, R., and Roche, M. (2019). Annotation of
epidemiological information in animal disease-related
news articles: guidelines and manually labelled cor-
pus - CIRAD Dataverse.
Valentin, S., Mercier, A., Lancelot, R., Roche, M., and Ar-
sevska, E. (2020b). PADI-web COVID-19 corpus:
news articles manually labelled - CIRAD Dataverse.
Valentin, S., Mercier, A., Lancelot, R., Roche, M., and Ar-
sevska, E. (2021b). Monitoring online media reports
for early detection of unknown diseases: Insight from
a retrospective study of COVID-19 emergence. Trans-
boundary and emerging diseases, 68:981–986.
WHO (2014). Early detection, assessment and response to
acute public health events: implementation of early
warning and response with a focus on event-based
surveillance. WHO Press, Geneva: The Organization,
interim version edition.
How Textual Datasets Enhance the PADI-Web Tool?
331
