
Whole-slide Classification of H&E-stained Cervix Uteri Tissue using
Deep Neural Networks
Ferdaous Idlahcen
1 a
, Pierjos Francis Colere Mboukou
1
, Hasnae Zerouaoui
1 b
and Ali Idri
1,2 c
1
Modeling, Simulation, & Data Analysis -MSDA, Mohammed VI Polytechnic University -UM6P,
Ben Guerir 43150, Morocco
2
Software Project Management Research Team, ENSIAS, Mohammed V University -UM5, Rabat 10000, Morocco
Keywords:
Uterine Cervical Neoplasms, Whole-Slide Imaging (WSI), Digital Pathology (DP), Transfer Learning (TL),
Computer-aided Detection (CADe) and Diagnosis (CADx).
Abstract:
Cervical cancer (CxCa) is heavily swerved toward low- and middle- income countries (LMICs). Without
prompt actions, the burden is anticipated to worsen by 50% from 2020 to 2040 - nearly 90% of deaths to
occur in sub-Saharan Africa (SSA). Yet, uterine cervix neoplasms are readily avoidable due to a protracted
latent cancer period. As it stands, deep learning (DL) is a potent solution for enhancing the early detection of
cervical cancer. This work assesses and compares the performance of seven end-to-end learning architectures
to automatically recognize cervical lesions and carcinoma histotypes upon hematoxylin and eosin (H&E)-
stained pathology images. Pre-trained VGG16, VGG19, InceptionV3, ResNet50, MobileNetV2, Inception-
ResNetV2, and DenseNet201 were the implemented deep convolutional neural networks (dCNNs) throughout
the present empirical analysis. Experiments are conducted on two datasets: (i) Mendeley liquid-based cytol-
ogy (LBC) and (ii) The Cancer Genome Atlas (TCGA) Cervical Squamous Cell Carcinoma and Endocervical
Adenocarcinoma diagnostic slides. All tests were validated under a 5-fold cross-validation, with four key met-
rics, Scott-Knott (SK), and Borda count schemes. Both pathology data appear to promote InceptionV3 and
DenseNet201. Yet, while VGG16 is a weak-performing approach for liquid-based cytology, it evinces promise
in histopathology yielding 99.33% accuracy, 98.85% precision, 99.83% recall, and 99.34% F-measure.
1 INTRODUCTION
Cervical cancer (CxCa) remains a heavy cause of
malignancy-related morbimortality amongst women.
As per GLOBOCAN 2020, an estimated 342,000
deaths and 604,000 incident cases occurred overall
(Sung et al., 2021). Such statistics conceal a world-
wide inequity as 87-91% overall mortality rates are
recorded in low- human development index (HDI)
settings (Gravitt et al., 2021). The burden is expected
to worsen roughly if no further actions are applica-
ble. To that end, in May 2018, the Director-General
of the World Health Organization (WHO) promul-
gated a global call-to-action for cervical cancer elim-
ination over the next 100 years - in August 2020, the
World Health Assembly (WHA) adopted it (Wilailak
et al., 2021). As high-income countries (HICs) are
mostly fulfilling elimination goals, a call-to-action
a
https://orcid.org/0000-0001-5888-6404
b
https://orcid.org/0000-0001-7268-8404
c
https://orcid.org/0000-0002-4586-4158
paramount is the immediate adoption of sustainable
screening and treatment measures in LMICs.
The slow process of cervical carcinogenesis
(Laengsri et al., 2018) provides opportunities for pre-
vention, screening, and early-stage treatment. Typi-
cal CxCa screening methods for precancerous lesions
within cervix uteri involve conventional Papanicolaou
(Pap) smears, liquid-based cytology (LBC), and cer-
vicography (Eun and Perkins, 2020). Such lesions are
known as cervical intraepithelial neoplasia (CIN) and
are categorized as either low-grade intraepithelial le-
sions (LSIL), i.e. CIN1, or high-grade SIL (HSIL),
i.e. CIN2/CIN3. (Tainio et al., 2018). While low-
grades often revert to normal, high-grades need fur-
ther testing to establish a proper diagnosis and treat-
ment regimens (Tainio et al., 2018). As it stands,
biopsy is vital to conduct a thorough analysis of tumor
samples; yet pathological specimen interpretations
vary depending on subjective perspectives and mate-
rial resources, prompting the adoption of computer-
aided diagnostic/detection tools (Taqi et al., 2018).
322
Idlahcen, F., Mboukou, P., Zerouaoui, H. and Idri, A.
Whole-slide Classification of HE-stained Cervix Uteri Tissue using Deep Neural Networks.
DOI: 10.5220/0011578700003335
In Proceedings of the 14th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2022) - Volume 1: KDIR, pages 322-329
ISBN: 978-989-758-614-9; ISSN: 2184-3228
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

The contribution of medical imaging and intel-
ligent decision-making systems to pathology detec-
tion is soaring (Debelee et al., 2020). With the ex-
pansion of novel approaches, implementation is be-
coming cost-efficient, less labor-intensive, and partic-
ularly popular over cervical lesion screenings (Singh
and Sharma, 2019). Deep learning (DL) algorithms
are reportedly more accurate and have surpassed clas-
sical machine learning (ML) in medical image analy-
sis (Debelee et al., 2020). In previous work, we car-
ried out a systematic map (SMS) (Idlahcen and Idri,
2022) on the use of deep and machine learning in gy-
necologic (GYN) oncology. The present empirical
study is prompted by the aforesaid main findings.
The current study carries out an empirical eval-
uation that develops and evaluates the performances
of seven deep learning techniques over two datasets:
(i) liquid-based cytology whole-slide images (WSI)
and (ii) The Cancer Genome Atlas (TCGA) Cer-
vical Squamous Cell Carcinoma and Endocervical
Adenocarcinoma histopathology WSIs. To the best
of authors knowledge, this is the first attempt to
(i) employ both cytology and histopathology-related
whole-slide imaging and (ii) empirically assess seven
DL techniques, i.e. VGG16, VGG19, InceptionV3,
ResNet50, MobileNetV2, InceptionResNetV2, and
DenseNet201, under Scott-Knott (SK) and Borda
count voting schemes for cervical pathology classifi-
cation. Different fields like software engineering (Idri
et al., 2016; Ottoni et al., 2019), have utilized the SK
test to contrast clusters while ranking several ML ap-
proaches for parameter tuning. Ergo, we adopt the SK
test because (i) it picks the optimal non-overlapping
sets and (ii) it outperforms prior statistical schemes.
Similar, the Borda count serves to rate optimally SK-
elected approaches (Mart
´
ınez-M
´
as et al., 2019).
In this study, three key research questions (RQs)
are addressed:
• RQ1: How effective are DL networks at classify-
ing cervical H&E-stained WSIs?
• RQ2: Which DL networks perform foremost?
• RQ3: Are DL networks performed analogously
on cyto- and histo- pathology whole-slides?
The following are the key contributions of the
present empirical study:
• Designing seven DL networks for CxCa clas-
sification: VGG16, VGG19, InceptionV3,
ResNet50, MobileNetV2, InceptionResNetV2,
and DenseNet201.
• Avoiding overfitting through the use of weight de-
cay and L2 regularizers.
• Implementing the DL nets over two datasets per-
taining to cytology and histopathology materials.
• Assessing the performances through the use of
both SK clustering and Borda count.
This document is organized as follows. Some
prior related works relevant to CxCa are briefed in
Section 2. The proposed DL techniques details are re-
ported in Section 3. Data acquisition and processing
are described in Section 4. Section 5 reports the fol-
lowed empirical scheme. Experimental findings and
discussion are provided in Section 6. Section 7 con-
cludes this study.
2 RELATED WORK
(Idlahcen and Idri, 2022) carried a systematic map on
the use of ML in GYN oncology from 2011 to mid of
2021. Of the 2,807 potential records retrieved from
PubMed, IEEE Xplore, ScienceDirect, Springer Link,
and Google Scholar, 169 studies were in-depth ana-
lyzed according to four criteria: the year, the chan-
nel/source, the female genital tract (FGT) site, and the
medical discipline. The main findings were:
• The use of ML/DL in GYN cancers surged signif-
icantly in 2019 - most notably cervical. Most of
the papers (93.5%) were published in journals.
• Most of the articles dealt with cervical cancer
(63.3%) as it is a paradigm of global health in-
equity, with higher morbimortality rates than both
uterine and ovarian malignancies combined.
• The most investigated task was diagnosis
(52.07%) followed by screening (31.95%). The
”gold standard” diagnosis relies on visual as-
sessment of biopsied tissues. Yet it is inherently
subjective to biases requiring, then, CADx.
Table 1 summarizes some ML/DL-based classifi-
cation studies dealing with cervical cancer cytology.
3 DL CLASSIFIERS
ARCHITECTURES
This section outlines the pretrained DL networks pa-
rameters tuning. As per length, further details are
available upon request from the corresponding author.
3.1 Configuration
We build DL networks through multiple parameters
tuning experiments for binary classification of two
datasets, i.e. LBS (Hussain et al., 2020) and TCGA-
CESC (Idlahcen et al., 2020). Except for InceptionV3
Whole-slide Classification of HE-stained Cervix Uteri Tissue using Deep Neural Networks
323

Table 1: Prior research on Herlev-based Pap smear classification.
Authors Techniques Metrics Findings
(Kurnianingsih et al., 2019) VGG-Like Acc, Sen, The classifier yielded sensitivity scores
Net Spe, AUC exceeding 96% and 95% for 2-class and
7-class problems respectively.
(Lin et al., 2019) AlexNet, Acc The pre-trained models achieved accuracy
GoogLeNet, scores of 94.5%, 71.3%, and 64.5% for all
ResNet, 2-class, 4-class, and 7-class problems
DenseNet respectively.
(Promworn et al., 2019) Resnet101, Acc, Sen, The Densenet161 network was top-performer
AlexNet, Spe for both binary and multiclass classifications
Densenet161, with 94.38% and 68.54% respectively.
Vgg19-bn, AlexNet and ResNet attained a sensitivity of
Squeeznet1-1 100% for binary classification.
(Dong et al., 2020) Inception-V3 Acc, Sen, The proposed model attained an overall accuracy,
Spe sensitivity, and specificity of 98.2%, 99.4%, and
96.73% respectively for 2-class classification.
and InceptionResNetV2 models of which 299x299
is the default input size, all the images were down-
sized to 224x224. We then applied transfer-learning
using seven DL nets pre-trained on ImageNet (Fei-
Fei et al., 2009). The modified last dense layer nar-
rowed the output classes from 1,000 to normal and ab-
normal conditions. A ReLU-trained fully-connected
(FC) layer was succeeded by a dropout layer with
a probability of 0.5. For some models, L2 regular-
ization was used to avoid overfitting. Adaptive mo-
ment estimation (ADAM) algorithm has been applied
to optimize the models parameters. Parameters for
training were set to batch size of 32, an initial learn-
ing rate of 0. 000001, and epoch size of 200.
3.2 Baseline CNN Architecture
The proposed baseline parameters involve a 244x244
RGB-three channel input layer; a convolutional layer;
a max-pooling layer set at 2x2 and 2 strides; a fully-
connected layer, i.e. Dense; and a last fully-connected
layer, i.e. output layer, using sigmoid activation func-
tion and two output filters for binary classification.
Table 2 reports the fine-tuned CNN layers. To per-
mit the variation at runtimes, each output shape has
”None” rather than the batch size.
4 DATA PREPARATION
This section describes the used datasets respective
preparation, consisting of (i) data acquisition and (ii)
data pre-processing as shown in Figure 1.
Table 2: Baseline CNN architecture.
Layer Type Output Shape #Param.
Conv2D (None, 222, 222, 64) 1792
MaxPooling2 (None, 111, 111, 64) 0
Flatten (None, 788544) 0
Dense (None, 128) 100933760
Dropout (None, 128) 0
Dense (None, 2) 258
Total Param. Trainable Param. Non Train.
100,935,810 100,935,810 0
Figure 1: Data preparation scheme.
4.1 Data Acquisition
We used two pathology-based datasets, i.e. LBC and
TCGA-CESC, to build DL nets binary classifications.
4.1.1 LBC Dataset
LBC data was collected by (Hussain et al., 2020) from
the Obstetrics and Gynecology department of Gauhati
Medical College and Hospital. The set comprises
963 cytological WSIs at 400x magnification, of which
613, 163, 113, and 74 images belonging to NILM,
LSIL, HSIL, and SCC respectively. For binary clas-
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
324

sification matter, we regard NILM to be a ”normal”
class and all LSIL, HSIL, and SCC as a single ”ab-
normal”.
4.1.2 TCGA-CESC Dataset
(Idlahcen et al., 2020) collected CESC data from the
Cancer Genome Atlas (TCGA) and pre-processed it.
The subset comprises 600 histopathological WSIs at
20x magnification, of which 300 images belonging to
ACC and SCC each. All the images are 1024x1024
px with at least 90% tissue and a histologic criterion.
4.2 Data Pre-processing
Turning data from one form to another in a relevant,
desirable, and user-friendly manner is known as data
processing.
The overfitting prospect and imbalanced classifi-
cations are mitigated with data augmentation. As per
LBC, 63% of images refer to NILM, indicating an im-
balance proportion. Else, TCGA-CESC comprises in-
sufficient images to train the DL models. As it stands,
both datasets underwent data augmentation for resam-
pling to conquer such limitation by the use of a ran-
dom 90-degree rotation or flip. In the rotation, coun-
terclockwise or clockwise rotation is selected, else,
horizontal or vertical flip is used. The results obtained
from this process are recapped in Table 3 and Table 4.
Once we got augmented sets, data for training was
set up. Indeed, the images range was rescaled by con-
verting 0-255 integers to 0-1 float values. The fixed-
size images were allowed by most of the DL nets.
Hence, we scaled the input images to related archi-
tecture as 299x299 and 224x244 for InceptionRes-
NetV2 and InceptionV3 models respectively. Note
that pickle files were used to store the resized images
to avoid repetition throughout the process.
Table 3: TCGA-CESC description after augmentation.
Class Size
Adenocarcinoma, ACC 600
Squamous cell carcinoma, SCC 600
Total 1200
Table 4: LBC description after augmentation.
Class Size
Normal (NILM) 613
Abnormal (LSIL, HSIL, SCC) 613
Total 1226
5 EMPIRICAL DESIGNS
The followed empirical design is given in this section:
• Models were evaluated using cross-validation.
• Total of seven DL architectures were evaluated
through the performance criteria.
• Accuracy values were used to cluster the DL tech-
niques as per the Scott-Knott statistical test.
• F-measure (F1), recall (Re), precision (Pr), and
accuracy (Acc) were used to rank the DL nets as
per Borda count.
• Empirical evaluations were performed using ex-
perimental process.
5.1 Cross-validation Technique
The stratified (k=5)-fold cross validation was adopted
to evaluate the models. Each fold has the same tar-
get class as per the entire set. Out of five folds, four
sets were allocated for training and one last for test-
ing purposes. We trained a new model in every iter-
ation on the training set while validating and storing
results from the test set. This process is repeated for
five times with validation on each and every fold. At
last, the final score was the average of the results ob-
tained.
5.2 Scott-Knott Test
It is a hierarchical clustering algorithm proposed by
Scott and Knott in 1974 (Ottoni et al., 2019). Variance
analysis (ANOVA) is its primary use, but also widely
included to obtain multiple comparisons of treatments
means for distinct homogeneous overlapping groups
distinction due to its simplicity yet robustness.
5.3 Borda Count Voting System
This method has various applications in decision
making situations such as elections. The candidate
received the points through the ranking. For instance,
the last choice got one point, similarly, second-to-
last gain two points, and so on till the candidate
reaches the top. The winner is decided the last based
on the best points option (Emerson and Emerson,
2011). In this study, the Borda count is used to deter-
mine the optimal DL net from the four metrics with
equal weight. Even different candidates and options
could be chosen instead of majorly preferred option
- the majority system is the opposite of it as per the
consensus-based voting mechanism. We performed
this to ensure the biases in choosing any particular
metric.
Whole-slide Classification of HE-stained Cervix Uteri Tissue using Deep Neural Networks
325
5.4 Performance Measures
The four evaluation measures covered in the previous
sections are: F1-score, recall, precision, and accuracy.
Herein, the measure of correctly identified cases
is known as accuracy. The right malignant prediction
quantification is called precision. Further, a recall is
a number of correct malignant predictions count mea-
sure, it minimizes the total benign cases considered
under malignant. In last, the weighted average (har-
monic mean) of precision and recall named as F1-
score. As a result, both false negatives and false posi-
tive were considered in it.
5.5 Experiment Scheme
The used methodology is based on prior research (Idri
et al., 2016; Idri and Abnane, 2017; Idri et al., 2018).
The five steps of this process are defined as follows:
1. Each variant of DL architecture, i.e. VGG16,
VGG19, InceptionV3, ResNet50, MobileNetV2,
InceptionResNetV2, and DenseNet201, was eval-
uated on the basis of accuracy using LBC and
TCGA-CESC dataset.
2. We considered the accuracy higher than 5% to se-
lect the DL outperforming baseline CNN.
3. DL model accuracy needs to be transformed us-
ing Box-cox method as Scott-Knott test takes the
normally distributed inputs.
4. Scott-Knott test was applied to cluster the elected
DL networks and to choose the SK top-cluster
based on accuracy and statistical indifference.
5. The four performance metrics were used to rank
the DL techniques using Borda count for the best
SK cluster and find the top DL architecture.
6 RESULTS & ANALYSIS
The empirical findings of the implemented DL net-
works are depicted and analyzed in this section. As
stated, four metrics were used for DL techniques as-
sessment. First, the CNN baseline model is compared
with each DL technique based on accuracy. If such
is more than 5% as compared to the baseline CNN
model, we kept those DL techniques. Further, SK sta-
tistical test is used to cluster the elected techniques for
Borda count ranking into the SK top-cluster.
6.1 DL Networks Accuracy Assessment
The accuracy of the CNN baseline model was com-
pared with the seven DL techniques. Intel® CoreTM
i5-7200U CPU @ 2.50GHz × 4 and 4 Go in RAM
with Ubuntu 18.04.5 LTS operating system was used
for the implementation. As backend, Keras and Ten-
sorflow frameworks were used in Python 3. SK clus-
tering was performed in Scott-Knott R-package.
6.1.1 LBC Dataset
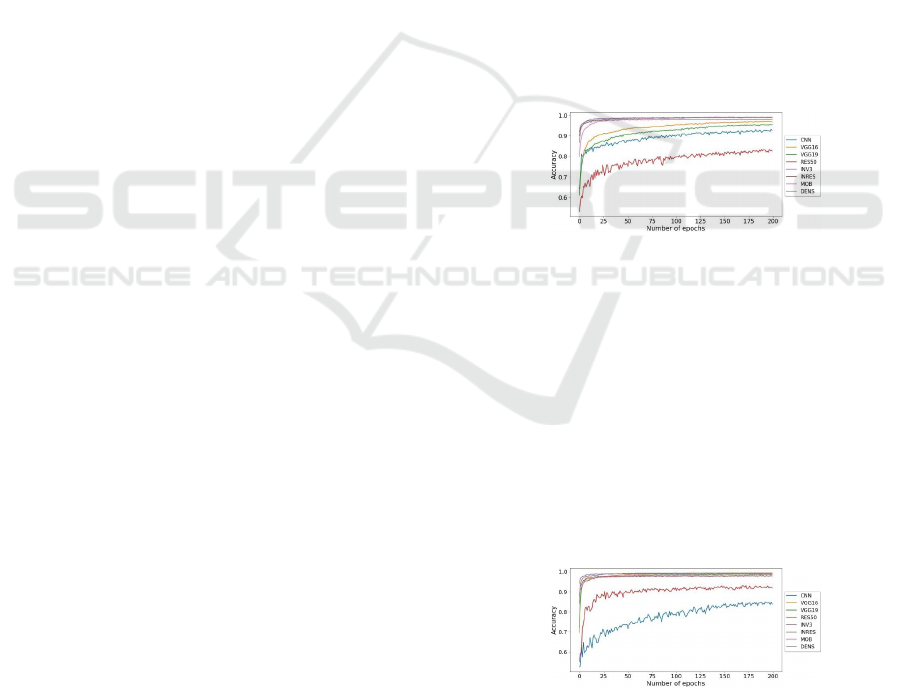
Table 5 and Figure 2 display the DL nets accuracy
over epochs for the LBC. The accuracy given is then
compared with the baseline CNN. We observe that In-
ception V3, InceptionResNetV2, DenseNet201, and
MobileNetV2 models are performing better than
baseline CNN. Yet, the accuracy of ResNet50, Incep-
tionV3, VGG19, and VGG16 were not able to con-
quer our 5% limit. Besides, ResNet50 accuracy is
only 82.51%, which is even less than of the baseline.
But, InceptionV3 gives 99.02% accuracy which is the
best among all DL nets, followed by MobileNetV2
with 98.94%, and DenseNet201 with 98.94% accu-
racy. Thus, MobileNetV2, DenseNet201, Inception-
ResNetV2, and InceptionV3 were chosen for evalua-
tion in the forthcoming process.
Figure 2: Accuracies of the used DL networks and baseline
as per LBC dataset.
6.1.2 TCGA-CESC Dataset
The accuracy values of MobileNetV2, ResNet50,
InceptionV3, InceptionResNetV2, DenseNet201,
VGG19, VGG16, and baseline CNN over TCGA-
CESC are shown in Figure 3 and Table 5. It should
be noted that all models were able to cross our 5%
accuracy slab. A reason why we selected all the net-
works for SK. Out of these, 99.33% accuracy is given
by VGG16. Overall, cervical cells were correctly
classified by all the models under consideration.
Figure 3: Accuracies of the used DL networks and baseline
as per TCGA-CESC dataset.
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
326
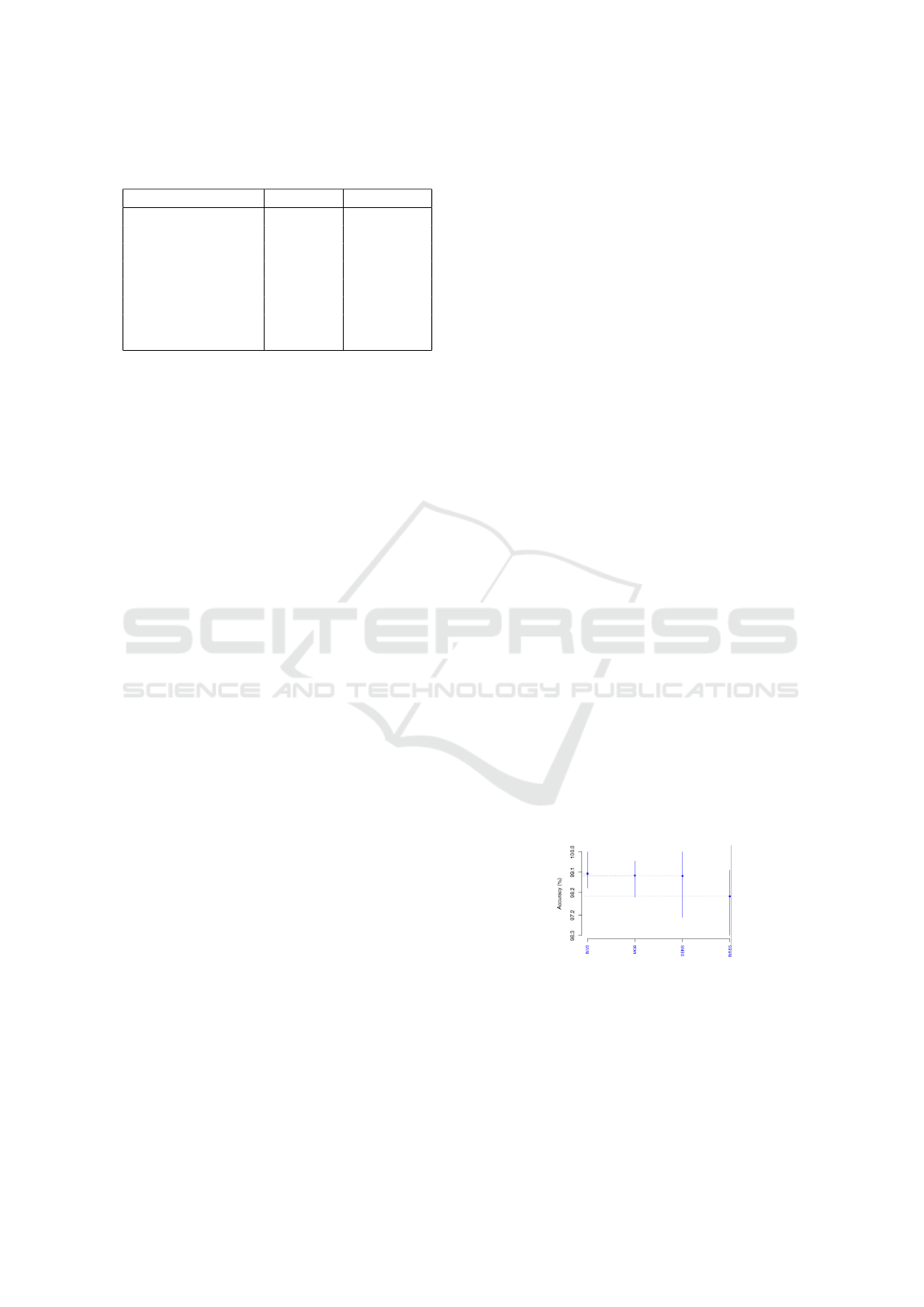
Table 5: Accuracy values[%] obtained per LBC and TCGA-
CESC dataset.
DL Networks LBC Acc CESC Acc
InceptionV3 99.02 99.08
DenseNet201 98.94 99.17
MobileNetV2 98.94 98.50
InceptionResNetV2 98.04 97.83
VGG16 96.81 99.33
VGG19 95.43 98.67
CNN 92.65 83.75
ResNet50 82.51 91.75
6.1.3 Analysis
Both pathology data appear to promote InceptionV3
and DenseNet201. But while VGG16 is a weak per-
forming approach for cytology, it appears to profit
more from histopathology. Here, we could demon-
strate a slight difference between cervix cytologi-
cal and histopathological findings although the con-
formity, as well as the certainty that deeper neural
networks do not automatically outperform shallower
counterparts in whole-slide imaging analysis.
In part, cervical cytology slides present deftly dif-
ferentiated nuclei with no complex tissue structures,
whereas nuclei in histopathology are very heteroge-
nous with an inter-/intra-instance pluralism in mor-
phology (e.g., size and shape), chromatin patterns,
etc., even within a single tissue specimen. Yet, quite
like erratic features, distinguishing nuclear aspects is
more challenging given the variation of hematoxylin
& eosin (H&E) stain intensity, artifacts/batch effects
(e.g., tissue-folds, air bubbles, etc.), and the exis-
tence of healthy tissue, denoting that “not” all sin-
gle WSI regions are representative. However, despite
cytology WSIs exhibiting minor nuclear mimics and
a pronounced contrast, the histopathology data pre-
processing and classification tasks have boosted the
performance of neural (esp. shallower) networks. If
we compare both datasets, only free-artifacts TCGA-
CESC WSIs were included, each one was split into
multiple region-of-interest (ROIs) at 20x magnifica-
tion, with at least one histologic criterion and >90%
tissue, then stain normalized; while LBC slides were
fed wholly into the neural networks asserting that
such a training set comprises noise. To achieve a
normal/abnormal prediction task regarding cytology,
we regarded NILM to be a ”normal” category and all
LSIL, HSIL, and SCC as a single ”abnormal” cat-
egory. This task may not provide performance ad-
vantages over a histotype classification task as there
is an apparent difference in the aspect of SCC and
ACC cells but difficult to tell the difference between
LSIL/NILM or LSIL/HSIL. Such lesions have several
common features, most notably LSIL with immature
metaplastic cytoplasm.
In another part, the use of smaller layers’ net-
works require shorter training times and fewer com-
putational resources, enabling the assessment of high-
resolution and multi-scale image training such as
pathology WSIs, where complex and irregular visual
elements (attempted to detect abnormalities) could be
wasted through downscaling. Since relevant ROIs in
TCGA-CESC dataset are prioritized, we postulate it
is the reason behind the great performance of smaller
VGG16 against state-of-the-art DCNNs. Otherwise,
the relevant information may be lost. Instead, the ef-
fectiveness of DenseNet201 is assumed to be due to
its structure adapted to avoid feature redundancy us-
ing fewer parameters. ResNet50 performs the worst
in both datasets indicating it is not a suitable approach
for this task. This is because ResNet50 exhibits heavy
pooling and little details are further missed.
6.2 Scott-Knott & Borda Count
The selected DL techniques in step 6.1 were clus-
tered using SK, then ranked through the Borda count
method.
6.3 LBC Dataset
The LBC retained InceptionV3, IncResNetV2,
DenseNet201, and MobileNetV2. Figure 4 depicts
the SK test by means of accuracies. Manifestly, the
SK test generated just one cluster with all four DL
networks, indicating a statistical similarity. Hence,
such were selected to be ranked through Borda count.
Table 7 presents the LBC Borda count ranking per ac-
curacy, precision, recall, and F1-score. As in Table 6,
InceptionV3 is rated top, then DenseNet201, which
scored akin to MobileNetV2 and IncResNetV2.
Figure 4: Accuracies of the four SK top-cluster DL net-
works as per LBC dataset.
6.4 TCGA-CESC Dataset
Figure 5 displays TCGA-CESC SK results as per ac-
curacy. Two clusters were obtained, and the best com-
prises all the DL techniques except ResNet50. Table 9
shows the Borda count ranking for the TCGA-CESC
Whole-slide Classification of HE-stained Cervix Uteri Tissue using Deep Neural Networks
327
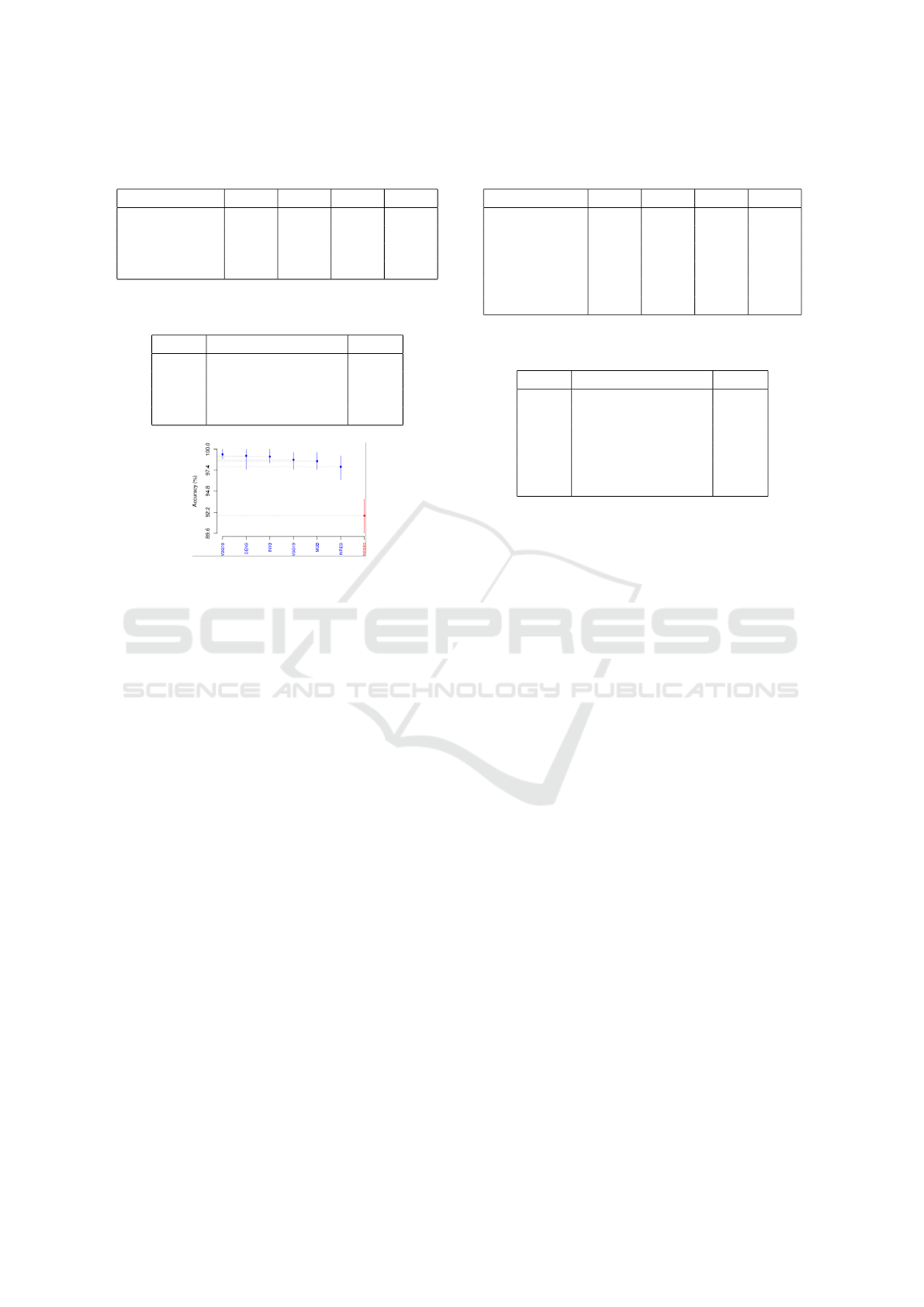
Table 6: Performance[%] of the SK top-cluster DL net-
works as per LBC dataset.
DL Networks Acc Re Pr F1
InceptionV3 99.02 98.4 99.67 99.03
IncResNetV2 98.04 97.58 98.53 98.05
DenseNet201 98.94 98.43 99.51 98.95
MobileNetV2 98.94 99.18 98.70 98.94
Table 7: Borda count ranking of the SK top-cluster DL net-
works as per LBC dataset.
Rank DL Networks Score
1 InceptionV3 14
2 DenseNet201 12
3 MobileNetV2 11
4 InceptionResNetV2 4
Figure 5: Accuracies of the four SK top-cluster DL net-
works as per TCGA-CESC dataset.
according to accuracy, precision, recall, and F1-score.
Table 8 presents the Borda count elected DL nets from
SK-best cluster: VGG16 was ranked first, followed by
DenseNet201, then InceptionV3.
To sum up the findings of SK test and Borda count
voting system under both datasets, we note:
1. DenseNet201 yielded favourable results. It is the
second-best cluster for both datasets.
2. Inception yielded promising results. It is ranked
first for the LBC and third for the TCGA-CESC.
3. VGG16 is an optimal TCGA-CESC approach.
Yet not selected for the LBC SK test.
4. VGG16, VGG19, and ResNet50 accuracy over
LBC were subpar when compared to the baseline
CNN, thus omitted from the SK test. VGG19 and
ResNet50 were preserved for the TCGA-CESC
SK test, yet belong to the last cluster.
5. VGG16, MobileNetV2, and InceptionResNetV2
perform well. Such belongs to the top SK cluster
despite a low Borda count.
We infer that DenseNet201 outperformed in all the
metrics regardless of any dataset. This is the best op-
tion for classifying cervical tissue. As runner-up, In-
ceptionV3 also provides significant performance.
Table 8: Performance[%] of the SK top-cluster DL net-
works as per TCGA-CESC dataset.
DL Net Acc Re Pr F1
VGG16 99.33 98.85 99.83 99.34
VGG19 98.67 98.03 99.33 98.67
InceptionV3 99.08 98.38 99.83 99.09
IncResNetV2 97.83 96.74 99.00 97.86
DenseNet201 98.94 98.43 99.51 98.95
MobileNetV2 98.50 98.18 98.83 98.51
Table 9: Borda count ranking of the SK top-cluster DL net-
works as per TCGA-CESC dataset.
Rank DL Networks Score
1 VGG16 23
2 DenseNet201 20
3 InceptionV3 18
4 VGG19 12
5 MobileNetV2 8
6 InceptionResNetV2 5
7 CONCLUSION & FUTURE
WORKS
This empirical study assesses and compares seven
deep CNNs classifiers. The models were evaluated
under four key metrics, Scott-Knott, and Borda count
schemes over two cervix pathological datasets. The
main findings are as follows:
RQ1: How effective are DL networks at
classifying cervical H&E-stained WSIs? Incep-
tionV3, DenseNet201, MobileNetV2, and Inception-
ResNetV2 outperformed the CNN baseline regardless
the used dataset. Conversely, ResNet50 performs the
worst in both datasets.
RQ2: Which DL networks perform foremost?
DenseNet201 is the best option for classifying cervi-
cal tissue as it is positioned second for both datasets.
InceptionV3 is a viable alternative as it ranks first on
the LBC dataset and third on the TCGA-CESC.
RQ3: Are DL networks performed analogously
on cyto- and histo- pathology whole-slides? Both
pathology data appear to promote InceptionV3 and
DenseNet201. But while VGG16 is a weak perform-
ing approach for cytology, it appears to profit more
from histopathology. Herein, we could demonstrate
a slight difference between cervix cytological and
histopathological findings although the conformity.
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
328

ACKNOWLEDGEMENTS
This work was conducted under the research project
“Machine Learning based Breast Cancer Diagnosis
and Treatment”, 2020-2023. The authors would like
to thank the Moroccan Ministry of Higher Education
and Scientific Research, Digital Development Agency
(ADD), CNRST, and UM6P for their support.
REFERENCES
Debelee, T. G., Kebede, S. R., Schwenker, F., and She-
warega, Z. M. (2020). Deep Learning in Selected Can-
cers’ Image Analysis—A Survey. Journal of Imaging
2020, Vol. 6, Page 121, 6(11):121.
Dong, N., Zhao, L., Wu, C., and Chang, J. (2020). Inception
v3 based cervical cell classification combined with ar-
tificially extracted features. Applied Soft Computing,
93:106311.
Emerson, P. and Emerson, P. (2011). The original Borda
count and partial voting. Social Choice and Welfare
2011 40:2, 40(2):353–358.
Eun, T. J. and Perkins, R. B. (2020). Screening for Cer-
vical Cancer. The Medical clinics of North America,
104(6):1063.
Fei-Fei, L., Deng, J., and Li, K. (2009). ImageNet: Con-
structing a large-scale image database. Journal of Vi-
sion, 9(8):1037–1037.
Gravitt, P. E., Silver, M. I., Hussey, H. M., Arrossi, S.,
Huchko, M., Jeronimo, J., Kapambwe, S., Kumar, S.,
Meza, G., Nervi, L., Paz-Soldan, V. A., and Woo, Y. L.
(2021). Achieving equity in cervical cancer screen-
ing in low- and middle-income countries (LMICs):
Strengthening health systems using a systems think-
ing approach. Preventive Medicine, 144:106322.
Hussain, E., Mahanta, L. B., Borah, H., and Das, C. R.
(2020). Liquid based-cytology Pap smear dataset for
automated multi-class diagnosis of pre-cancerous and
cervical cancer lesions. Data in Brief, 30:105589.
Idlahcen, F., Himmi, M. M., and Mahmoudi, A. (2020).
CNN-based Approach for Cervical Cancer Classifica-
tion in Whole-Slide Histopathology Images.
Idlahcen, F. and Idri, A. (2022). Systematic Map of Data
Mining for Gynecologic Oncology. pages 466–475.
Idri, A. and Abnane, I. (2017). Fuzzy Analogy Based Effort
Estimation: An Empirical Comparative Study. IEEE
CIT 2017 - 17th IEEE International Conference on
Computer and Information Technology, pages 114–
121.
Idri, A., Abnane, I., and Abran, A. (2018). Evaluating
Pred(p) and standardized accuracy criteria in software
development effort estimation. Journal of Software:
Evolution and Process, 30(4):e1925.
Idri, A., Hosni, M., and Abran, A. (2016). Improved esti-
mation of software development effort using Classical
and Fuzzy Analogy ensembles. Applied Soft Comput-
ing, 49:990–1019.
Kurnianingsih, Allehaibi, K. H. S., Nugroho, L. E.,
Widyawan, Lazuardi, L., Prabuwono, A. S., and Man-
toro, T. (2019). Segmentation and classification of
cervical cells using deep learning. IEEE Access,
7:116925–116941.
Laengsri, V., Kerdpin, U., Plabplueng, C., Treeratanapi-
boon, L., and Nuchnoi, P. (2018). Cervical Cancer
Markers: Epigenetics and microRNAs. Lab Medicine,
49(2):97–111.
Lin, H., Hu, Y., Chen, S., Yao, J., and Zhang, L. (2019).
Fine-grained classification of cervical cells using mor-
phological and appearance based convolutional neural
networks. IEEE Access, 7:71541–71549.
Mart
´
ınez-M
´
as, J., Bueno-Crespo, A., Khazendar, S.,
Remezal-Solano, M., Mart
´
ınez-Cend
´
an, J. P., Jassim,
S., Du, H., Assam, H. A., Bourne, T., and Timmer-
man, D. (2019). Evaluation of machine learning meth-
ods with Fourier Transform features for classifying
ovarian tumors based on ultrasound images. PLOS
ONE, 14(7):e0219388.
Ottoni, A. L., Nepomuceno, E. G., de Oliveira, M. S.,
and de Oliveira, D. C. (2019). Tuning of reinforce-
ment learning parameters applied to SOP using the
Scott–Knott method. Soft Computing, 24(6):4441–
4453.
Promworn, Y., Pattanasak, S., Pintavirooj, C., and Piyawat-
tanametha, W. (2019). Comparisons of pap smear
classification with deep learning models. Proceed-
ings of the 14th Annual IEEE International Confer-
ence on Nano/Micro Engineered and Molecular Sys-
tems, NEMS 2019, pages 282–285.
Singh, J. and Sharma, S. (2019). Prediction of Cervical
Cancer Using Machine Learning Techniques. Inter-
national Journal of Applied Engineering Research,
14(11):2570–2577.
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soer-
jomataram, I., Jemal, A., and Bray, F. (2021). Global
Cancer Statistics 2020: GLOBOCAN Estimates of In-
cidence and Mortality Worldwide for 36 Cancers in
185 Countries. CA: a cancer journal for clinicians,
71(3):209–249.
Tainio, K., Athanasiou, A., Tikkinen, K. A., Aaltonen, R.,
C
´
ardenas, J., Hern
´
andes, Glazer-Livson, S., Jakobs-
son, M., Joronen, K., Kiviharju, M., Louvanto, K.,
Oksjoki, S., T
¨
ahtinen, R., Virtanen, S., Nieminen, P.,
Kyrgiou, M., and Kalliala, I. (2018). Clinical course
of untreated cervical intraepithelial neoplasia grade
2 under active surveillance: systematic review and
meta-analysis. The BMJ, 360.
Taqi, S. A., Sami, S. A., Sami, L. B., and Zaki, S. A.
(2018). A review of artifacts in histopathology. Jour-
nal of Oral and Maxillofacial Pathology : JOMFP,
22(2):279.
Wilailak, S., Kengsakul, M., and Kehoe, S. (2021). World-
wide initiatives to eliminate cervical cancer. Interna-
tional journal of gynaecology and obstetrics: the of-
ficial organ of the International Federation of Gynae-
cology and Obstetrics, 155 Suppl 1(S1):102–106.
Whole-slide Classification of HE-stained Cervix Uteri Tissue using Deep Neural Networks
329
