
Machine Learning and Raspberry PI Cluster for Training and Detecting
Skin Cancer
Elias Rabelo Matos, Edward David Moreno
a
and Kalil Araujo Bispo
b
Programa de P
´
os-Graduac¸
˜
ao em Ci
ˆ
encia da Computac¸
˜
ao, Universidade Federal de Sergipe (UFS), Sergipe, Brazil
Keywords:
CNN Network, Machine Learning, Skin Cancer, Embedded Systems, High Performance Computing.
Abstract:
Context: Melanoma is the most popular and aggressive type of skin cancer with thousands of cases and deaths
worldwide each year. But melanoma isn’t the only type of skin lesion. Since 2016 the ISIC (International Skin
Cancer Challenge) has been launching challenges toward skin lesion detection. In this paper, we use the
HAM10000 dataset which is part of the ISIC archive and contains seven classes of skin lesions to train a
DenseNet network aiming to achieve state-of-the-art accuracy. Objective: We evaluate the use of a low-cost
cluster with four Raspberry PI to check the viability as a machine learning cluster for detecting one type of
skin cancer. Method: We trained a deep convolutional neural network using the pre-trained model of four
networks and we got 89% of accuracy which is a top state-of-art value. After we perform two experiments:
(i) we use the knowledge transfer technique to run an MLP model using four Raspberry and (ii) we train
the pre-trained DenseNet with a Raspberry PI cluster aiming to verify if a low-cost cluster is viable for this
approach. Results: We found that is not possible to train our network using only four Raspberry PI since it
has low computational resources but we show what more resources are needed to perform this task. Despite
this situation, we achieve 80% accuracy using the knowledge transfer technique and only four Raspberry Pi.
1 INTRODUCTION
Skin cancer is the most common cancer in Brazil.
Data from INCA (Brazilian National Institute of Can-
cer) shows that 180 thousand new cases are de-
tected every year. There are various skin cancer
types, among them the most common is melanoma.
Melanoma has the deadliest form of skin cancer
(American Cancer Society, 2014). Melanoma is a
worldwide danger, in the United States for example,
in 2017, 87 thousand cases were detected with 9730
deaths.
Skin cancer is provoked by abnormal growth cells
of the skin. These cells make layers and the way they
are organized determines the cancer type (sbd, ).
Melanoma is harder and deadliest but it is curable
since detected in its early stages. To detect skin can-
cer in early-stage and make the disease highly curable
dermatologists must be usually consulted for medi-
cal exams. However, in underdeveloped countries like
Brazil or countries without public health care like the
United States, visits to dermatologists not are acces-
a
https://orcid.org/0000-0002-4786-9243
b
https://orcid.org/0000-0001-8878-9293
sible to the poorest part of the population.
Dermoscopy is an important exam made by der-
matologists, however, due to the subjectivity of hu-
man diagnosis, then the medical result may differ
among different dermatologist. But, in general, any
dermatologists could examine a set of characteristics
in a skin lesion. These characteristics are called the
ABCDE rule, where: A is asymmetry, B is Border, C
is Colour, D is diameter and E is Elevation/Evolving
(Illig, 1987).
The intrinsic image analysis and pattern recog-
nition involved in skin cancer detection, naturally
makes it an interesting topic for image processing
and artificial intelligence. The first work published in
1994 by Binder et. al(BINDER et al., 1994) already
used dermoscopy images to train a neural network.
In 2016, the ISIC (International Skin Cancer Chal-
lenge) launch an annual competition for melanoma
detection in image analysis. The ISIC archive has
more than twenty thousand images with skin lesion
which are classified as benign or having a type of skin
cancer, and most part of them is related to melanoma.
In this paper, we use the HAM10000 dataset,
which is a dataset collected from different popula-
tions and is public to use in scientific experiments.
Matos, E., Moreno, E. and Bispo, K.
Machine Learning and Raspberry PI Cluster for Training and Detecting Skin Cancer.
DOI: 10.5220/0011575500003318
In Proceedings of the 18th International Conference on Web Information Systems and Technologies (WEBIST 2022), pages 75-82
ISBN: 978-989-758-613-2; ISSN: 2184-3252
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
75

It was described by (Tschandl et al., 2018) to train
a CNN (Convolutional Neural Network) with ResNet
pre-trained model in Google Colaboratory, and after
achieving the state-of-the-art we try to train the same
algorithm running in two platforms: (i) Google Colab,
and (ii) using an embedded and low-cost cluster, par-
ticularly, composed by Raspberry PI boards. We want
to evaluate the power of low-cost computers toward
skin lesion detection to make dermoscopy accessible
to the poorest people.
To achieve this evaluation we try two experiments:
In the first experiment, we performed a ”knowledge
transfer” from a densenet to an MLP running in Rasp-
berry PI. In the second experiment, we try to train a
full dense network on an embedded cluster composed
of only four Raspberry nodes. We found that is not
possible to train our network with only four Raspberry
PI but despite this, we achieve 80% of accuracy using
knowledge transfer.
This paper is organized into six sections as fol-
lows: section 2 presents the related works, section
3 shows the methodology used and the experiment´s
planning, and section 4 shows the experiments made.
Section 5 shows and discusses the results. Finally,
section 6 presents the main conclusions of this re-
search.
2 RELATED WORK
Some papers from the skin cancer detection literature
were useful for our research. In this section, we pre-
sented them with their main contribution and results.
Initially, Abedini et al. (2016) (Abedini et al.,
2016) presented a great contribution to the pre-
processing aspects of skin lesion images. They used
two combined datasets and achieved an accuracy of
94%.
Tajeddin and Asl (2017) (Tajeddin and Asl, 2017)
presented a general algorithm to process image seg-
mentation in skin lesions They used the algorithm
for skin lesion images but claimed that it serves a
general-purpose with different properties and defi-
ciencies. They used a multi-step pre-processing phase
with hair removal, illumination correction, etc. The
proposed algorithm was trained with the ISIC dataset
of 900 images and archived Dice and Jaccard coeffi-
cient values of 0.89 and 0.79, respectively.
The work by Majtner et al. (2017) (Majtner et al.,
2017) was among pioneer´s works that used deep
learning for classifying skin lesion images. Their
combined deep learning methods with hand-crafted
Rsuf Features, which is an idea where the feature
set is based on dividing the image into parallel se-
quences of intensity values from the upper-left cor-
ner to the bottom-right corner, and Local Binary pat-
terns achieved 0.826 of accuracy with the sensitivity
of 0.533 and specificity of 0.898. Their dataset was
compounded by 900 images of benign and malignant
melanoma provided by the ISIC dataset.
The paper by Romero Lopez et al (2017) (Romero
Lopez et al., 2017) presented transfers of knowledge
from a pre-trained CNN method, the VGGNet, to a
dermoscopy approach to a two-class melanoma clas-
sifier between malign and benign melanoma. They
used the ISIC dataset and had encouraging results
with 81.33% of accuracy on the test dataset.
Sousa and Moraes (2017) (Sousa and de Moraes,
2017) presented an approach that considers not only
melanoma but Seborrheic Keratosis lesions as well as
their trained different algorithms with GoogleNet and
AlexNet models. Their resized images from dataset to
256x256 in pre-processing phase and user 72 epochs
in GoogleNet 256, 50 epochs in GoogleNet 224, and
30 epochs in AlexNet. Their result of the arithmetic
mean of these three networks was 84.7%.
The main related work is the paper by Sahu et
al. (2018) (Sahu et al., 2018). It used a Rasp-
berry Pi to evaluate the skin lesions analysis in a
hand-held computer, like Raspberry itself, a smart-
phone, or SoCs (System-on-a-chip). The authors pro-
posed and implemented a hybrid approach based on
the use of deep learning models and specific knowl-
edge domains bases on features that dermatologists
use to detect skin cancer. In the process of obtain-
ing the deep learning features, the authors used a pre-
trained Google MobileNet model. In addition, they
also trained the Raspberry PI and detected that the
training in Movidius Neural Compute Stick is five
times faster than Raspberry PI. Their final task was
to transfer the knowledge to an SVM and run it in a
Raspberry. They used the ISIC 2017 challenge dataset
with 2700 images and achieved a result among the top
10 challengers.
3 METHODOLOGY AND
PLANNING
3.1 Methodology
The main objective of this work is to evaluate the pos-
sibility to build a low-cost cluster of Raspberry PI to
train a network capable of detecting skin cancer. The
first step of work is to search the literature in this re-
search area and know the state-of-art in skin lesion
detection. After we plan and describe our experiment,
firstly training a network on Google Colab. With this
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
76

trained network, we train an MLP algorithm using the
teacher-student method. Immediately after we try to
train our fully dense network in a raspberry PI cluster.
The results are shown in section 5.
3.2 Planning
Context Selection: We select the HAM1000 dataset
in the first step while we research the literature. We
have selected this dataset because it is a simple and
already validated data set for skin lesion tasks. Af-
ter, we trained our model to validate the results of our
network using this dataset.
3.3 Hypothesis Formulation
We formulate two hypotheses, one for each experi-
ment that we will perform.
3.3.1 Hypothesis 1
Hypothesis: When we perform a knowledge transfer
from our pre-trained network we lose some accuracy.
Independent Variable: None. Dependent Variable:
Accuracy.
3.3.2 Hypothesis 2
Hypothesis: We think that with the same network
trained in two different environments, the accuracy
will stay the same, but the time spent in training will
be different. Independent Variable: Accuracy. De-
pendent Variable: Time of training.
Instrumentation: The instrumentation process is
to use the Google Colab and the Raspberry PI cluster
configuration. We used the same instrumentation for
both experiments.
4 ROADMAP OF THE
EXPERIMENTS
In this section, we present how we defined, imple-
mented, and ran experiments for this work. We use
the HAM1000 dataset to train a pre-trained DenseNet
using the Pytorch library.
4.1 The Dataset
The dataset chosen to run our experiment was the
HAM1000 dataset described by (Tschandl et al.,
2018). The dataset is called Human ‘Against Machine
with 10000 Images‘, but in fact, this dataset contains
10015 images. These images were collected from dif-
ferent populations and nowadays they are public and
available via the ISIC archive. This dataset is used for
comparisons among machines and human experts and
more than 50% of the lesions are confirmed by pathol-
ogy. This dataset includes seven unbalanced classes
of different types of skin cancer, in Figure 1 one of
each type is shown. They are:
Figure 1: Skin lesion types.
1. Actinic Keratoses
2. Basal cell carcinoma
3. Benign keratosis
4. Dermatofibrom
5. Melanocytic neviare
6. Melanoma
7. Vascular skin lesions
4.2 The Densenet
The Densenet was presented by (Huang et al., 2017)
and is a convolutional neural network (CNN) deeper,
more accurate, and efficient if the connection be-
tween layers of input and outputs is closer. The au-
thors of Densenet say that it has the following ad-
vantages: they alleviate the vanishing-gradient prob-
lem, strengthen feature propagation, encourage fea-
ture reuse, and substantially reduce the number of pa-
rameters. The efficiency of Densenet is measured by
achieving the state-of-art in CIFAR-10, CIFAR-100,
SVHN, and ImageNet datasets. In this paper, we used
the DenseNet model provided by the Pytorch library.
4.3 The Resnet
The ResNet network is the winner of the ILSVRC
2015 image classification, detection, and location, in
addition to having also won the MS COCO 2015 in
detection and replacement (). As ResNet networks
can be very deep, reaching up to 152 layers learning
Machine Learning and Raspberry PI Cluster for Training and Detecting Skin Cancer
77

residual representation functions instead of direct sig-
nal representation. The ResNet network introduces
the skip connection concept to circumvent the Vanish-
ing / Exploding Gradient problems Common convo-
lutional networks usually have completely connected
convolutional layers without any skipoushotcurt, thus
being called flat networks. When these networks get
deep the problem of gradient degradation/explosion.
That is: during the backpropagation phase when a
partial derivative has an error in the current weight
this effect is propagated n times, where n is the num-
ber of layers. When a network is deep a small error
multiplied by n will become close to zero (disappear),
while a large error will cause this error to grow (ex-
ploded). problem adding the input of a layer x directly
to the output of a layer ahead. In this way even if the
wandering / exploding gradient is mitigated by x com-
ing directly from some previous layers
4.4 The VGG
The VGG network is an evolution of AlexNet, the
winning network of ILSVRC 2012. It is focused on
smaller entries and larger steps in the first convolu-
tional layer and has a greater focus on network depth
than previous convolutional networks (). The VGG
input uses a 224x224 pixel RGB channel, each con-
volutional layer has a very short 3x3 filter and also
a 1x1 filter that works like a linear transform. VGG
has a fully connected layer train where the first two
have 4096 channels and the last one has 1000 chan-
nels. All of its hidden layers use a ReLU and do
not use local response normalization. The main in-
novations regarding AlexNet are the smaller 3x3 en-
tries compared to 11x11 entries regarding the use of
3 ReLU units instead of just one, the function of de-
cision manages to be more discriminative. With this,
the VGG needs fewer parameters to work. The use
of 1x1 filter on the layers makes the decision function
more non-linear. With the convolution size, less VGG
has a greater number of layers.
4.5 Inception
The inception network is a milestone in the develop-
ment of convolutional networks because before it the
most popular networks just got deeper in hopes of get-
ting more performance. Inception on the other hand
used several tricks to gain performance, both in speed
and accuracy. It was developed sequentially, thus hav-
ing four famous versions (). In the first version the
authors found that choosing the ideal size of the con-
volution kernel is a very complicated task because
while a larger kernel is better for identifying infor-
mation distributed globally, a smaller kernel is used
for local information. Another problem is that very
deep networks can cause overfitting and just making
the network larger can be computationally very costly.
For this, the authors decided to use multiple fil-
ters in the same layer, so three convolutional opera-
tions are performed on each layer with three different
filters, 1x1, 3x3, and 5x5. After the filters, a max
pooling operation is performed and the concatenated
result is sent to the next layer. The 1x1 filter is used
to decrease the number of channels, thus making con-
volutions less costly. Versions 2 and 3 were published
in the same work and propose some updates that im-
prove inception performance (). In the second ver-
sion, the 5x5 filter was replaced by two 3x3 convo-
lutions, as it is still less costly than performing a 5x5
convolution. Version 3 introduced and optimized the
(), factorized 7x7 convolutions, the use of BatchNorm
in sorting aids, and the Label Smoothing technique
used to prevent overfitting. For version 4 (SZEGEDY
et al., 2017) the authors identified that some network
modules were much more complex than necessary, to
mitigate this problem and gain performance, the set
of operations performed before each inception block
was altered.
4.6 Pre-processing, Dataset Split and
Run
We download the dataset from Kaggle
1
and before
running into the Densenet model provided by Pytorch
we run a function to compute the mean and standard
deviation between image sizes of the dataset and use
these values to Pytorch input normalization.
After that, we use the ”CSV” archive that contains
the class of each image and create a [image, label] tu-
ple for each image and its respective class. We divide
the dataset into three splits: (I) 75% to train and 25%
to test, (II) 80% to train and 20% to test, and (III) fi-
nally, 90% to train and 25% to test. On each split, we
divide 20% of the train set for validation.
We ran the Densenet training in Google Colabora-
tory which provides a 15 GB GPU.
4.7 The Knowledge Transfer
As one of our goals is to use a low-cost embedded
platform and therefore with less computational ca-
pacity, we need to have lighter algorithms from a
computational point of view. For this reason, we
need lightweight algorithms. To run a lightweight
1
https://www.kaggle.com/kmader/skin-cancer-mnist-
ham10000
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
78

algorithm with good accuracy on the Raspberry PI
we need to translate the knowledge achieved by the
pre-trained networks to an MLP model using the
method teacher-student which is based on the distill-
ing knowledge purpose by (Hinton et al., 2015) that
is a very simple way to improve the performance of
almost any machine learning algorithm. Thus, in this
paper we used knowledge transfer concepts, specifi-
cally the teacher-student technique.
This method uses the previous knowledge that
makes predictions using a whole ensemble of models
which is cumbersome and may be too computation-
ally expensive. Then compress the knowledge in an
ensemble into a single model which is much easier to
deploy. This process makes a small model computa-
tionally cheaper than the previous network.
The called teacher-student technique uses a fully
trained DenseNet network as a teacher and the knowl-
edge is passed to an MLP algorithm using the knowl-
edge and making the MLP algorithm achieve more
accuracy in fewer epochs. The technique works as
follows: the highly complex teacher network is first
trained separately using the complete dataset. This
step requires high computational performance. Then
a correspondence between the teacher and student
network must be created. In the third step, the data
is passed through the teacher network to get all in-
termediate outputs. Now the outputs from the teacher
network are used to make the correspondence relation
to the student network and backpropagate error in the
network.
4.8 PyTorch
PyTorch is an open framework for machine learning
that is suitable for use in research until use in pro-
duction. Pytorch works on top of Python, making it
possible to run on all operating systems and architec-
tures with Python interpreters, including ARM archi-
tectures. In this paper, we use Pytorch’s predefined
functions to write our experiment code.
4.9 Apache Hadoop
Apache Hadoop is a framework written mainly in Java
that allows the development of distributed process-
ing of a large amount of data in clusters using sim-
ple programming models. The framework is designed
to be scalable from single servers to hundreds of ma-
chines in a cluster, offering local Hadoop processing
and storage.
Although most of the framework is written in Java
there are also parts written in C and Shell Script. In
this way it is possible through Hadoop Streaming, to
write code in several programming languages, includ-
ing Python. The core of Hadoop is mainly made of
four modules, all Hadoop modules were developed
with the fundamental assumption that the hardware
can fail, so Hadoop, via software, can automatically
deal with these failures (Bappalige, 2014).
4.10 The Experiments Environment
The experiments were executed in the google Colab
and a Raspberry PI Cluster.
4.10.1 Google Colab
Google Collaboratory is a free-to-use tool provided
by Google as part of encouraging the use and learning
of both data science and machine learning. The only
requirement for using the tool is a Google account.
Colab allows the user to create a notebook for the
Python language, in that notebook the codes can be
divided into sections called cells, where the result of
the cells can be stored, making it not need to be ex-
ecuted every time it is necessary. This makes it easy
to store parts of machine learning´s process, such as
pre-processing.
In addition to a Python development environment
with all the language tools, it includes the possibility
of installing new libraries. Colab also allows the user
to execute their codes using CPU, GPU, or TPU.
4.10.2 The Raspberry Cluster
The cluster used in this work is formed by a Rasp-
berry PI cluster with four Raspberry PI 2 Model B
with 1GB of RAM memory connected by a gigabit
switch running an Apache Hadoop Cluster.
4.11 The Experiment Architecture
Figure 2 represents, in the diagram, how experiments
were performed. On top, we have two sets of raw data
that represents the HAM1000 dataset.
In the first step to be able of using correctly the
dataset, we make a mapping to attach each image pro-
vided by the dataset to metadata that represents the
lesion using the image ”id”. With this, we can work
effectively on the lesions in the next steps. The pre-
processing phase is performed as we described in 4.6.
We execute the training phase in both ways: (1)
using the Google Colab and (2) using our raspberry
Cluster to make the DenseNet. With our DenseNet,
we apply for the Knowledge Transfer and built our
MLP.
Machine Learning and Raspberry PI Cluster for Training and Detecting Skin Cancer
79
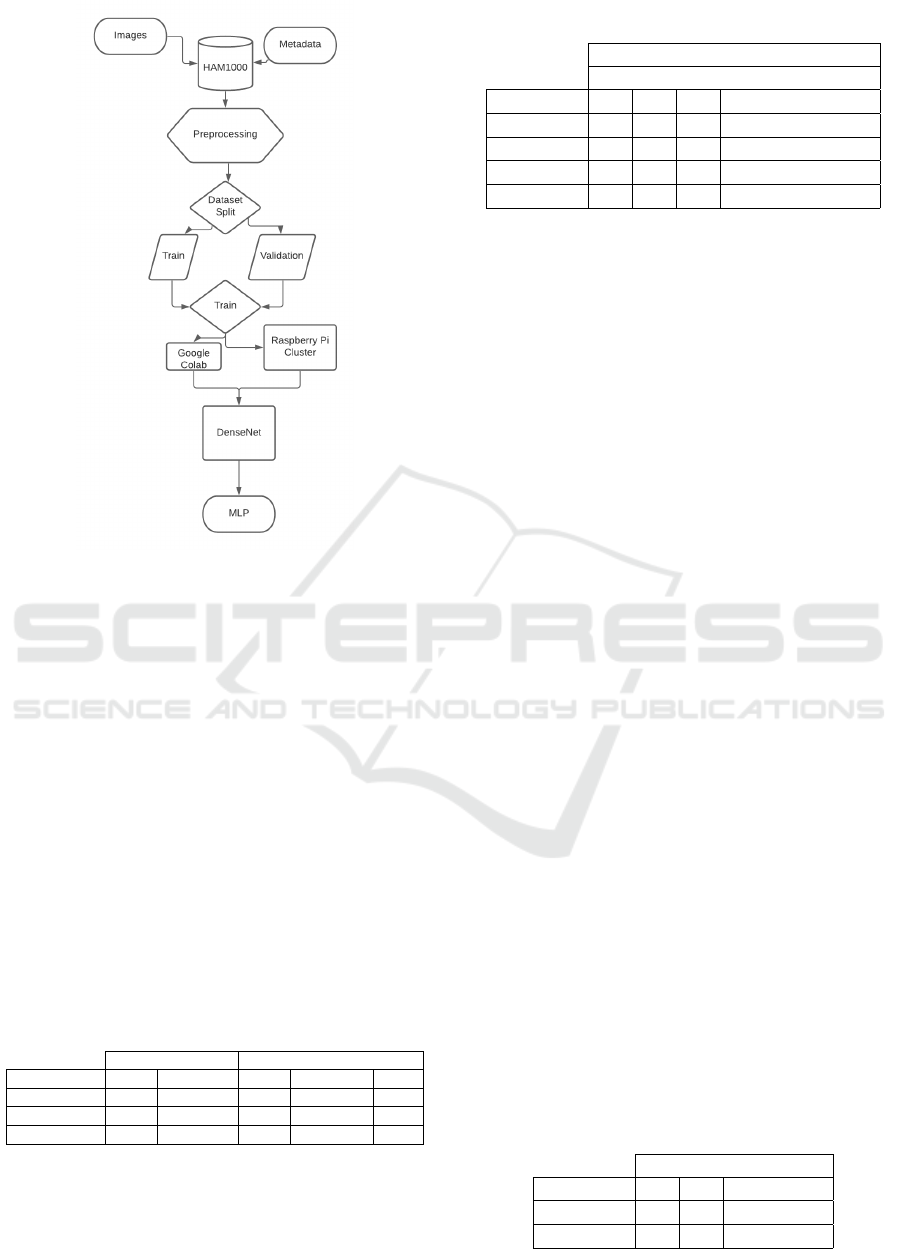
Figure 2: Experiment Architecture.
5 RESULTS AND DISCUSSION
As we said in Section 4 we split the dataset in
three ways to execute our experiments because the
HAM10000 does not have a traditional standard di-
vision between trainset and testset.
On Table 1 we show each split and how many im-
ages were used for training and testing. For each ex-
periment, we split the trainset into 80% for the train-
ing phase and 20% for the validation phase.
For each experiment, we ran the training 10 times
with ten epochs, and in each run, due to the pre-
trained Densenet, we achieved good accuracy in fewer
epochs. We save the best result models for each ex-
periment and archived an accuracy between 82% and
89% for the Densenet train.
Table 1: Dataset split and image quantity for each experi-
ment.
Split Percentagem Images Quantity
Dataset Split Train Test Train Validation Test
I 75 25 6000 1500 2500
II 80 20 6400 1600 2000
III 90 10 7200 1800 1000
Among the pre-trained networks, the network that
did better in all dataset divisions was the DenseNet
network. With an accuracy of 89% in the III division
of the dataset making it the best result obtained by
Table 2: Accuracy on Four Different Pretrained Networks.
Acurracy
Dataset split
Network I II III Network average
DenseNet 83 82 89 84,6
Resnet 82 81 83 82
VGG 76 80 83 79,6
Inception 79 82 82 81
the model. It is possible to identify with the analysis
of the entire table, that for all networks the division
of number 3 of the dataset is always better. While
divisions I and II ended up being worse. However, it is
possible to identify that the average between the three
data divisions is very close for the four networks.
The analysis of this result indicates that there may
be overfitting when a training set is used. To solve
this doubt it is necessary to investigate the number of
entries of each class that was in the test set and if there
was greater learning in any of the classes. However,
this problem was not explored in this work, since its
main objective is to work with networks on the Rasp-
berry platform, as it would be an investigation closer
to the part of artificial intelligence, it was excluded
from this work. work is the use of the Raspberry
PI Platform, such doubt was little explored by this
work. For direct deployment on Raspberry, this work
addresses two solutions, direct training on Raspberry
PI using a Hadoop cluster and the knowledge transfer
model known as the teacher and student method.
After these tests, we can perform our two experi-
ments using the Raspberry PI.
5.1 Experiment 1: The Knowledge
Transfer
With the best model saved for each experiment, we
execute the transfer process using the four pre-trained
networks to MLP models and run each MLP network
in Raspberry. We measure the accuracy for each MLP
network using the same set of training.
5.1.1 Densenet
Table 3 summarises the accuracy for each experiment
in Densenet and MLP networks. We achieve accuracy
between 73 and 80%.
Table 3: Accuracy on Densenet and MLP Networks.
Experiment Accuracy
Network I II II
DenseNet 83 82 89
MPL 74 73 80
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
80
As expected while transferring the knowledge we
lost some accuracy and yet achieved 80% of accuracy
on experiment III which we considered to be a good
result.
5.1.2 Resnet
Table 4 summarises the accuracy for each experiment
in Densenet and MLP networks. We achieve accuracy
between 65 and 72%.
Table 4: Accuracy on Resnet and MLP Networks.
Experiment Accuracy
Network I II II
Resnet 82 81 83
MLP 65 72 71
In this network, we achieved 72% of accuracy but
curiously split II has more accuracy than split III.
5.1.3 VGG
Table 6 summarises the accuracy of each experiment
in VGG and MLP networks. We achieve accuracy
between 71 and 77%.
Table 5: Accuracy on VG and MLP Networks.
Experiment Accuracy
Network I II II
VGG 75 80 83
MLP 71 74 77
5.1.4 Inception
Table 6 summarises the accuracy of each experiment
in Inception and MLP networks. We achieve accuracy
between 71 and 74%.
Table 6: Accuracy on Inception and MLP Networks.
Experiment Accuracy
Network I II II
Inception 79 82 82
MLP 71 72 74
In this network, we achieved 74% of accuracy in
the best case.
5.2 Experiment 2: The Training in Our
Raspberry Cluster
We select the split number III which contains 90%
for the train set and 10% of the test set to perform
the training phase in our Raspberry PI Hadoop cluster.
We know that accuracy stays the same as the split and
the code are the same. We chose the time of train as
a variable to investigate the viability of the low-cost
cluster with four Raspberry PIs.
To have a good comparison we ran the algorithm
12 times in Google Colab, then we exclude the higher
and lower times to calculate the mean of 10 times left.
And we detect that our network spent a mean of 154.5
minutes to achieve 89% of accuracy in ten epochs.
After this phase, we pass to try to train the same
network in our Raspberry PI Hadoop cluster and we
expected to achieve the same accuracy with a slower
time of training. But this reveals not to be the truth,
the Raspberry PI that we build proved to not be capa-
ble of training our network.
And because of this limitation, we shift our focus
and try to understand why our cluster cannot run the
training phase of our network. We ran again the train-
ing phase of the network in Google Colab to measure
the amount of Memory RAM needed to perform this
training processing, and we detected that it need be-
tween 3.5GB and 5GB of RAM.
Since our cluster has 4GB of RAM in each plat-
form, and the Raspbian OS uses about 80MB of RAM
in the better scenario, and to build the Hadoop cluster
in each node using a java virtual machine uses about
254 MB, then we lose 334MB of RAM available in
each node. So, a minimum of 1.33 GB of RAM is
required to build the cluster.
Therefore, we detected that is unviable to use a
machine learning cluster with just four nodes of Rasp-
berry PI, for the training phase in this cluster. Despite
this result, we can use the single embedded cluster
(using only four Raspberries) for detecting skin can-
cer lesions with 80% of accuracy after using knowl-
edge transfer and making the training phase out of this
cluster, and running the MLP models into the embed-
ded cluster.
The initial objectives of this work were: Train a
network on the Raspberry platform using only Py-
torch and/or do training using an embedded clus-
ter using Apache Hadoop. Experiment number two
shows results that meet this objective as it was not
possible to carry out the training with a cluster of only
four nodes. However, this result is not disheartening
for Low-Cost High-Performance Computing, as there
are still chances of these goals being achieved us-
ing horizontal scaling, adding more nodes, or choos-
ing another platform that provides greater computing
power. For example using other platforms, such as
Banana PI, and Odroid, among others.
Machine Learning and Raspberry PI Cluster for Training and Detecting Skin Cancer
81

6 CONCLUSION
In this paper, we used machine learning techniques
for evaluating and detecting skin lesions. We used the
HAM10000 dataset, which is a dataset collected from
different populations and is public for use in scientific
experiments. We have trained a CNN (Convolutional
Neural Network) with DenseNet pre-trained model in
Google Colab, and after achieving the state-of-the-art
we trained the same algorithm running on two plat-
forms: (i) Google Colab, and (ii) using an embedded
and low-cost cluster, particularly, composed by Rasp-
berry Pi boards.
Generally, traditional machine learning is expen-
sive in processing cost but it can achieve good results.
We have nowadays plenty of options of supercomput-
ers available and many of them are free like Google
Colab. Thus, our model was initially trained for run-
ning in Google Colab and we achieve 89% of accu-
racy, but we also used transfer knowledge for reduc-
ing the machine learning models and running them
in a low-cost computer as mentioned in (Sahu et al.,
2018).
So, in this paper, we try to evaluate what is pos-
sible and achieve good results with small comput-
ers like Raspberry Pi. We made the experiment
and achieved 80% accuracy using the teacher-student
method as knowledge transfer. This is a good result
for machine learning research, but it is not adequate
as medical precision. Despite this result, we believe
that is possible low-cost clusters or smartphones can
help dermatologists in their diagnoses.
Attempting to run the training using the Apache
Hadoop cluster proved to be meaningless due to tech-
nical limitations presented in a cluster with only
four nodes. On the other hand, the 80% accuracy
achieved with the teacher-student technique is con-
sidered promising, as this rate is state-of-the-art for
an MLP network. high accuracy artificial intelligence
at low-cost, as it is possible to overcome this limita-
tion with other platforms or with the addition of more
nodes to the cluster.
Therefore, from this work it is possible to propose
future works divided into two fronts: In the first of
them, talking about artificial intelligence, it is possi-
ble to carry out a work using eight, sixteen, or thirty-
two nodes of Raspberry to build a cluster with higher
computational performance, as well as running tests
on other platforms such as Banana PI or Odroid. For
the artificial intelligence part, it is possible to propose
studies with different pre-trained networks, with dif-
ferent weights and dataset configurations, it is also
possible to carry out works isolating the accuracy for
each of the existing lesions in the dataset HAM10000.
REFERENCES
C
ˆ
ancer da pele - sociedade brasileira de dermatologia.
Abedini, M., Codella, N., Chakravorty, R., Garnavi, R.,
Gutman, D., Helba, B., and Smith, J. R. (2016). Multi-
scale classification based lesion segmentation for der-
moscopic images. Proc. of the Annual Intl. Confer-
ence of the IEEE Engineering in Medicine and Biol-
ogy Society, EMBS, 2016-Octob:1361–1364.
American Cancer Society (2014). American Cancer Soci-
ety: Cancer Facts & Figures 2014. Cancer Facts and
Figures.
Bappalige, S. P. (2014). An introduction to apache hadoop
for big data.
BINDER, M., STEINER, A., SCHWARZ, M., KNOLL-
MAYER, S., WOLFF, K., and PEHAMBERGER, H.
(1994). Application of an artificial neural network in
epiluminebdcence microscopy pattern analysis of pig-
mented skin lesions: a pilot study. British Journal of
Dermatology.
Hinton, G., Vinyals, O., and Dean, J. (2015). Distilling
the Knowledge in a Neural Network. arXiv e-prints,
pages 1–9.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. Proceedings - 30th IEEE Conference on Com-
puter Vision and Pattern Recognition, CVPR 2017,
2017-Janua:2261–2269.
Illig, L. (1987). Epidemiologic aspects of malignant
melanoma. (Review).
Majtner, T., Yildirim-Yayilgan, S., and Hardeberg, J. Y.
(2017). Combining deep learning and hand-crafted
features for skin lesion classification. 2016 6th In-
ternational Conference on Image Processing Theory,
Tools and Applications, IPTA 2016.
Romero Lopez, A., Giro-I-Nieto, X., Burdick, J., and
Marques, O. (2017). Skin lesion classification from
dermoscopic images using deep learning techniques.
Proceedings of the 13th IASTED International Con-
ference on Biomedical Engineering, BioMed 2017,
pages 49–54.
Sahu, P., Yu, D., and Qin, H. (2018). Apply lightweight
deep learning on internet of things for low-cost and
easy-to-access skin cancer detection. In Zhang, J. and
Chen, P.-H., editors, Medical Imaging 2018: Imag-
ing Informatics for Healthcare, Research, and Appli-
cations, volume 10579, pages 254 – 262. Intl. Society
for Optics and Photonics, SPIE.
Sousa, R. T. and de Moraes, L. V. (2017). Araguaia Medical
Vision Lab at ISIC 2017 Skin Lesion Classification
Challenge. arXiv e-prints.
Tajeddin, N. Z. and Asl, B. M. (2017). A general algorithm
for automatic lesion segmentation in dermoscopy im-
ages. 23rd Iranian Conference on Biomedical Engi-
neering and 1st Intl. Iranian Conference on Biomedi-
cal Engineering, ICBME 2016, (Nov):134–139.
Tschandl, P., Rosendahl, C., and Kittler, H. (2018). Data de-
scriptor: The HAM10000 dataset, a large collection of
multi-source dermatoscopic images of common pig-
mented skin lesions. Scientific Data, 5:1–9.
WEBIST 2022 - 18th International Conference on Web Information Systems and Technologies
82
