
Technology Transfer of Convolutional Neural Networks: An Example
Thanakij Wanavit
1 a
, Samuel Sallee
1
, Chedtha Puncreobutr
2,3
, Leslie Klieb
1 b
and Pin Pin Tea-Makorn
1,4 c
1
Tenxor Inc, 401 Harrison St. Unit 41C, San Francisco, 94105 U.S.A.
2
Department of Metallurgical Engineering, Faculty of Engineering, Chulalongkorn University, Bangkok, Thailand
3
Biomedical Engineering Biomechanics Research Center, Meticuly Co., Ltd., Chulalongkorn University, Bangkok, Thailand
4
Sasin Graduate Institute of Business Administration, Chulalongkorn University, Bangkok, Thailand
Keywords:
Convolutional Neural Networks, Bone Segmentation, Technology Transfer to Industry, U-net, LSTM, Dice
Similarity Coefficient, Hausdorff Distance.
Abstract:
A number of university groups have shown that neural networks, especially U-nets, can satisfactorily segment
CT-scans of bones. Segmentation, labelling the scans where bone and enamel are and where not, can be used
to make a 3D model of the skull. This paper gives an overview of efforts to transfer university-based research
work for use to a company that manufactures titanium meshes for brain surgery. It discusses issues and pitfalls
in such a transition. A working prototype is discussed.
1 INTRODUCTION
In this position paper, we argue that there are many
industrial niches where Machine Learning and Arti-
ficial Intelligence can have substantial benefits. As
an example, we present a project by start-ups Tenxor
Inc and Meticuly Co.,Ltd. that aims to benefit brain
surgery.
It is not always straightforward to recognize those
niches and potential applications. Research on neu-
ral networks and artificial intelligence is still mainly
done in academic environments. These often miss the
detailed insight into industry to which some of their
work may be useful and applicable. Moreover, there
is often a big gap between a low-budget academic trial
project and a robust implementation in industry, even
if the opportunity for implementation is recognized
and the core idea is clear. This paper hopes to con-
tribute to making such implementations of academic
research more feasible than one might expect by pro-
viding an explicit example.
In this paper, we do our best to explain the medical
side in a simplified way.
Tenxor Inc is a virtual company whose employees
are scattered over three continents. It is led by the
a
https://orcid.org/0000-0001-7291-394X
b
https://orcid.org/0000-0002-0881-5330
c
https://orcid.org/0000-0003-3219-9264
main author of this paper, Thanakij Wanavit. Tenxor
Inc received sufficient venture capital funding that it
is less constrained by concerns about costs of training
neural network models than academic researchers. It
is involved in several projects; this paper focuses on
its joint development experiences with Meticuly Co.,
Ltd.
Meticuly Co., Ltd. is a relatively young medical
company based in Thailand. In its production process
for titanium meshes used as implants, CT scans need
to be analysed. After a conversion by the software in
the CT scanner, the CT-scan consists as layers of par-
allel images, usually with a slice thickness of around
1 mm. The titanium meshes are used as implants by
neurosurgeons when there are openings in the skull
(from accidents, stroke, brain surgery, maxillofacial
surgery — surgery related to face, jaw, mouth or neck
—, bone tumors, and other reasons) to cover up those
openings. The mesh is fixed to the edges of the open-
ing. Therefore, an exact 3D model of the outer surface
of the area around the hole is necessary in order to de-
sign the best possible mesh. Software with manual in-
put generates a wireframe for the mesh that operates a
metal 3D printer operated via Selective Laser Melting
(SLM) and produces a mesh.
CT scans have superior hard tissue contrast and
spatial resolution (van Eijnatten et al., 2018). Bones
and the enamel of teeth (from here on not men-
tioned separately anymore) are the densest parts in the
Wanavit, T., Sallee, S., Puncreobutr, C., Klieb, L. and Tea-Makorn, P.
Technology Transfer of Convolutional Neural Networks: An Example.
DOI: 10.5220/0011540200003332
In Proceedings of the 14th International Joint Conference on Computational Intelligence (IJCCI 2022), pages 375-380
ISBN: 978-989-758-611-8; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
375

head, besides possibly metal from previous implants,
crowns, and other human interventions. To make a
personalized mesh tailored to the curves of the skull,
every pixel in the scan has to be labelled with a 1 for
bone and or a 0 for not-bone. The process of labelling
“bone” or “not bone” is called segmentation. The cus-
tomary way to accomplish this labelling is manually
by a radiologist. The work is tedious, not particularly
rewarding because there is no involvement in clini-
cal decision making, and prone to inconsistencies be-
tween practitioners when it is not clear if pixels rep-
resent bones or other tissues or scattering from metal
objects.
Computer scientists will readily recognize this as
a binary classification problem for images, which can
be solved by convolutional neural networks (Rawat
and Wang, 2017).
A number of academic groups have worked on
bone segmentation via convolutional neural networks
(CNNs). Here we first describe the workflow for the
production of such titanium meshes, showing how
segmentation is a necessary preliminary step. Then
we will discuss briefly the academic research that has
led to a reasonable understanding and resolution of
the segmentation problem for skulls. After that we
discuss our planned path for technology transfer. We
present briefly our progress, and conclude with issues
that still have to be resolved.
2 WORK FLOW TO PRODUCE
MESHES
Brain surgery is older than one would expect. The In-
cas carried out already trepanation (drilling or scrap-
ing a hole in the human skull) (Kushner et al., 2018).
This is remarkable, because the skull is thick, around
7-10 mm, depending also on the location on the skull
(Mahinda and Murty, 2009). In Peru during the Inca
era, the patients survived in around 80% of the cases,
compared with only 50% during the USA civil war.
With current brain surgery techniques, a drill is
used to scrape the bone away to make a hole. It stops
automatically as soon as the tip is no longer in the
bone. The head of the patient is fixated and a grid is
used that guides the probe of the surgeon through the
bored hole (stereo-tactic surgery). The filling of the
opening at the end of the surgery can be done with a
graft, either autologous (using the scraped bone from
the same individual, which carries a risk of infection),
by using a synthetic bone substitute in liquid form, by
using prefabricated solid biomaterial, or by covering
the opening with a titanium mesh. The surgeon de-
cides which technique to use. With a mesh the bone
will grow under the mesh and use the mesh as a scaf-
fold.The bone will grow and will get so solid that it
affords enough protection.
This paper focuses on the processes needed to
manufacture the titanium meshes. Meticuly receives
the CT scans that are performed after a portion of
skull is removed and/or opened. Sometimes there are
also bone fragments on those scans, for instance from
accidents, and existing crowns and other metal im-
plants may make the scan not as clear as desirable. CT
scans are done with low-energy X-rays to minimize
radiation risks. Images are typically received in a di-
mension of 500x530 or 560x560 pixels, and cropped
and resized for this research to a more convenient size
for a CNN of 512x512 pixels. Image quality is suffi-
cient for segmentation. The company tries to limit
the variation in CT-scan parameters, especially slice
thickness and interval, as those can influence accuracy
(van Eijnatten et al., 2018). CT-Scans are received in
DICOM format. Internally also NIfTI is used. Af-
ter an anonymizing process (to remove any patient’s
information), the image is loaded in software to per-
form manual labelling and 3D rendering of the skull.
Subsequently, knobs to fix the mesh to the bone, the
outside edge of the mesh, the grid of the mesh and
other details are added. Figure 1 gives an impression.
After that, an STL file is generated for the mesh that
is used to manufacture the titanium implant by a 3D
SLM printer.
It is seen that the quality of the initial segmenta-
tion is essential for the rest of the process and that
good segmentation is labour-intensive. Academic
groups accomplished segmentation with CNNs. It is
therefore tempting to automate that part. The next
section will give an overview what those academic
groups accomplished and show how their results are
already close to enabling industrial applications.
Figure 1: Titanium Mesh for Mid-Face reconstruction.
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
376
3 A LIMITED LITERATURE
OVERVIEW OF SKULL
SEGMENTATION
3.1 Overview
A quick sketch of some relevant literature is given
here as background for the decisions that were made
in trying to use available academic research. Be-
fore AI was used to do segmentation, researchers at-
tempted a number of other methods. Thresholding for
edge detection was attempted from the difference in
grey scale; however, it was very difficult to decide ac-
curately which voxel belongs to the bone region or
which one does not. Manual postprocessing was usu-
ally required. It was the most often used method for
the skull [van Eijnatten 2018]. Data clustering like K-
means does not always reach an optimal solution. Re-
searchers more and more gravitated to the use of neu-
ral networks. Convolutional Neural Networks are es-
pecially suitable for image processing. Several types
have been researched for segmentation. Most groups
seem to have used the U-net architecture, but other
choices have been made. Most interesting for the tran-
sition to industry is that the difference in quality is
often quite small between various approaches. It is
actually very difficult to assess differences in quality
of the results between research groups because of in-
compatibility in metrics and test sets. Often there are
not even enough details in the reports in the scientific
literature to be able to faithfully replicate the work.
Luckily, most approaches are more than good enough
for the purpose of automatic segmentation (possibly
helped by manual postprocessing). This is even true
for approaches that are very different in a theoretical
way, like building the model from segmented slices
(2D approach) or segmenting the model in one swoop
(3D approach).
3.2 Other Networks than U-net
As an early example, (Minnema et al., 2018) used an
adaptation from Aldenborgh for MRI. The quality of
their results (The Dice similarity coefficient DSC —
see section 4— around 0.94) is not very different from
what other groups later obtained in different ways.
This group put in an enormous effort in establish-
ing ground truth for training. They used STL wire-
frames from 20 clinical patients who had previously
undergone craniotomy (the surgical removal of part
of the bone from the skull to expose the brain) and
cranioplasty (repair of a skull bone defect) for which
3D manufactured skull implants were used served as
“gold standard” models during CNN training. The
ground truth was determined using global threshold-
ing with manual corrections. Our group used an op-
posite approach with respect of setting ground truth.
The anonymized DICOM files containing skull de-
fects (e.g. voids and holes) were used instead of nor-
mal full skull bones to represent a typical CT con-
dition for cranioplasty. In the discussion section, we
will discuss what kind of effect (if any) retaining skull
openings has on the quality of the results. One can see
how difficult it is to compare results: their work cal-
culates the Dice similarity coefficient using the voxels
labelled for the full segmentation and comparing that
with the 3D ground truth, so it calculates the 3D Dice
similarity coefficient (compare section 4 on metrics).
That is correct. But then they report for statistical pur-
poses the arithmetic average of the 20 Dice similarity
coefficients, instead of the harmonic mean (see again
section 4 on metrics). The difference may be so small
that it does not significantly influences their results.
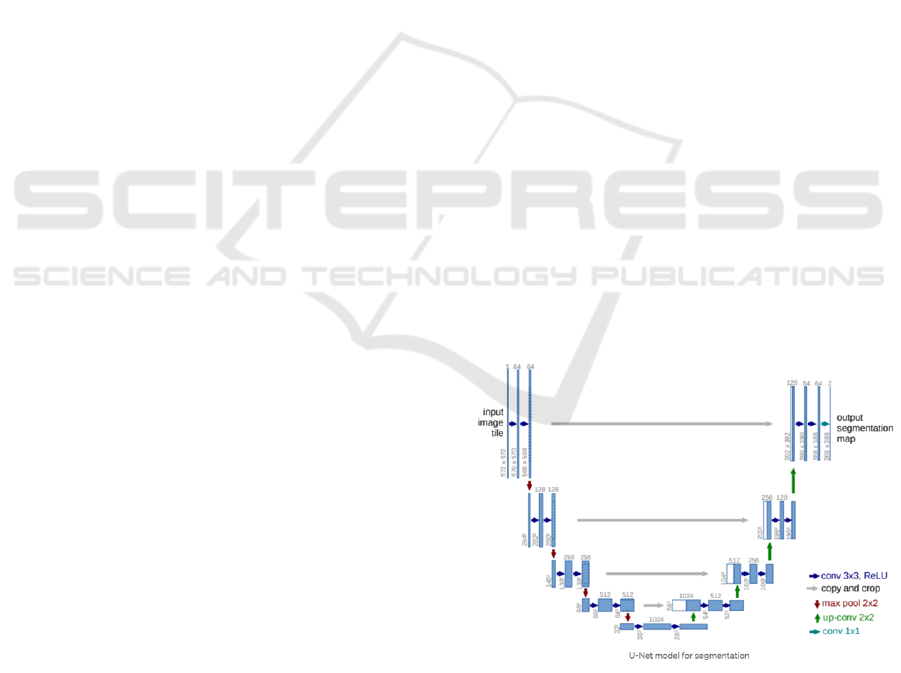
3.3 U-net
U-net is the most popular CNN for skull segmenta-
tion (and probably image segmentation in general).
Our group also takes the U-net architecture as a base
model. The original work on U-net was reported in
(Ronneberger et al., 2015). U-net can start with any
type of feature extraction. For skull segmentation that
is usually not necessary. In the original design, there
is a contracting encoder part to analyse the whole im-
age and a successive expanding decoder part to pro-
duce a full-resolution segmentation. U-Net requires
much smaller sample sizes than many other methods.
Figure 2: U-Net architecture. Each blue box corresponds
to a multi-channel feature map. The number of channels is
denoted on top of the box. The x-y-size is provided at the
lower left edge of the box. White boxes represent copied
feature maps. The arrows denote the different operations.
(From (Ronneberger et al., 2015)).
Technology Transfer of Convolutional Neural Networks: An Example
377

Seamless segmentation of images of any size is
accomplished by an overlap-tile strategy. This limits
the GPU footprint of the network itself. The upsam-
pling path mirrors in some way the down path, from
there the name “U-net”. To compensate for small
sample sizes, deformation augmentation was used. U-
net was used in a number of very successful segmen-
tation research projects, like (Klein et al., 2019). That
research used a combination of a regularized form of
the Dice similarity coefficient and Cross Entropy Loss
to accomplish segmentation for full-body CT scans
for patients with myeloma. A number of parame-
ters were adjusted in that work by experimentation.
Aggressive augmentation was used. Dice similarity
coefficients around 0.92 were obtained for full-body
segmentation (not just skulls). Another version of U-
net, (Mader, nd), publicly available, of the program
(Klein et al., 2019) used, was used by us in this work.
It needed considerable upgrades, though. Our train-
ing was only done using skull CT scans with holes,
therefore a good comparison of results was not possi-
ble.
3.4 LSTM Networks
We are also experimenting with adding a few LSTM
(Long short-term memory) layers in front of U-net. A
theoretical advantage of this type of neural network
is that it can process the sequence of image CT-scan
layers instead of treating every layer separately. Very
preliminary results indicate a slight improvement in
accuracy.
4 METRICS
4.1 Dice Similarity Coefficient
A convolutional network used for bone Segmentation
needs a metric how similar ground truth (determined
maybe by a radiologist manually) and its correspond-
ing segmented image is in order to iterate toward the
best solution. The most often used metric to gauge the
similarity between two arbitrary samples is the Dice
similarity coefficient DSC (Jimenez et al., 2016):
DSC = 2
|X ∩Y |
|X| + |Y |
(1)
Numerator and denominator are measured in the
same units and therefore DSC is independent of the
measurement unit. Its value can lie between 0 and 1,
where 0 indicates no similarity at all (no overlap) and
1 indicates perfect similarity with complete overlap.
The | bars indicate a size or value. When X and Y are
sets, the original definition, one uses the cardinality of
the set (how many members it has) and DSC is quan-
tity/quantity like counting pixels in segmentation. For
two-dimensional areas, the measurement unit of the
full expression is m
2
/m
2
. For three-dimensional cal-
culations, the Dice similarity coefficient compares the
two volumes and is m
3
/m
3
. Because every voxel in
every slice has the same volume in a CT scan, this
is also the DSC between ground truth and segmented
image as measured in pixels. The calculation of sums
and overlap in pixels is a sum in a loop over the slices
in the 3D images.
Taking the average of the Dice similarity co-
efficients of each 2D slice usually gives a differ-
ent answer than calculating by volume. With two
slices, if one slice has a size of 100 for both ground
truth and segmented image, and 50 overlap, and
the other one is 200 for both images and also 50
overlap, the correct Dice similarity coefficient is
2*(50+50) /(100+100+200+200) = 1/3 (in whatever
units the size is calculated). However, the average of
2*50/(100+100) and 2*50/(200+200) is (0.5+0.25)/2
= 0.375. “The Dice metric measures volumetric over-
lap between segmentation results and annotations”
(Structseg2019, 2019). Theoretically, the correct av-
erage to calculate the volumetric DSC is the harmonic
mean
¯x = n(
n
∑
i=1
1
x
i
)
−1
(2)
The two separate Dice similarity coefficient in the
example were ¼ and ½. The Harmonic Mean is
2(4 + 2)
−1
= 1/3, identical to the calculation where
voxels were counted. Using the Arithmetic average
overestimates DSC, because for positive numbers the
Harmonic Mean is always lower than the Geometric
Mean, which is lower than the Arithmetic Mean, un-
less all numbers are the same (Xia et al., 1999). Av-
eraging the DSC of 2D layers overestimates slightly
the volume DSC. After regularization, 1 − DSC can
be used as a loss function for the CNN to optimize
the learning.
4.2 Hausdorff Distance
Another metric that is often used in skull segmen-
tation is the Hausdorff distance. It has been de-
scribed informally as “the extent to which each point
of a ‘model’ set lies near some point of an ‘im-
age set’” (Huttenlocher et al., 1993). The image set
is the ground truth and the model set the segmen-
tation. The Hausdorff distance describes at which
point(s) the surface of the segmentation is not fol-
lowing well enough the surface of the ground truth.
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
378

(Structseg2019, 2019). In principle, this is a better
metric for the final goal of manufacturing mesh im-
plants, because it prioritizes shape over skull volume
and slice area overlap.
Figure 3: Hausdorff distance diagram. (from (Struct-
seg2019, 2019)).
The Hausdorff distance is the maximum of d
XY
and d
Y X
in Figure 3. Because meshes are fitted to the
surface of the skull, the largest deviation is more im-
portant for practical applications than if the skull vol-
ume is segmented correctly. The Hausdorff distance
has unpleasant characteristics as a loss function, but
can be easily regularized. Unfortunately, its calcula-
tion is expensive, even if algorithms to speed it up are
available. Its long training runtime might have ham-
pered its use in academic research.
5 PRELIMINARY RESULTS
The following has been accomplished at the time of
writing.
• While self-configuring implementations of U-net
have been developed, it was chosen to use a lean
version that could be optimized for this particular
segmentation issue (Mader, nd)
• Usually implants like those used in hip-
replacement need to fit in a 3D dimensional
space. Therefore the ubiquitous use of the Dice
similarity coefficient (which as discussed is a
volumetric gauge) in the academic literature is
understandable when segmentation of various
bones in the body is discussed. However, for the
best fit of the titanium meshes, it is important that
deviation in the surface is as small as possible.
It was found that a loss function combination
of (1 − DSC) and Hausdorff distance gave good
results in modeling the surface of the skull on
both the outside and the inside. An additional
benefit is that the thickness of the skull is well-
determined. This makes it easier to decide where
to put screws that hold the mesh to the skull at the
edges of the holes in the skull.
• A prototype of the segmentation module is work-
ing. It is still slow (several minutes computation
time), mainly because the calculation of the Haus-
dorff distance is time-consuming.
• A prototype of a web-based interface was devel-
oped that enables uploads of a DICOM file and
serverless cloud-based execution of segmentation.
The users can then download the segmented file to
their local workstations for further processing.
• A considerable amount of development and test-
ing will still be necessary. Data security has not
been addressed, and while unit testing was done,
integration testing and testing in practice have also
not been done yet.
6 CONCLUSIONS
We estimate that now this project is approximately
half way for potential use, a couple of conclusions and
areas of concerns are already getting into focus. Most
probably, similar issues will arrive with every project
that aims to incorporate academic AI research into an
industrial environment and to increase productivity.
• Academic papers rarely contain all the informa-
tion that is necessary to recreate the academic re-
search. It helps if an informant from the academic
group is available.
• Open source repositories quickly become obsolete
from upgrades in libraries, decisions where to run
the programs (cloud, local, etc.), version upgrades
in Python, deprecation of features, etc. This adds
to the time needed to recreate an academic project
outside academia.
• (Not typical for only this kind of projects): time
estimates are usually wildly optimistic.
• Available funding is important. This makes more
experimentation possible and ensures that over-
runs in estimates or in training time have less im-
pact.
• Academic papers try to impress with progress in
areas that sometimes are not relevant for indus-
trial applications. A concrete example: From the
literature it seems that improving the Dice similar-
ity coefficient is very important, but many groups
show marginal improvements that are not relevant
or actually meaningless.
Technology Transfer of Convolutional Neural Networks: An Example
379

• Rarely discussed is the quality of the ground truth,
whether the set of skulls to train is cherry-picked
in some way, and how the Dice similarity coef-
ficient is exactly calculated (average or volume-
metric). While one paper calculates a statistical
error, systematic errors are never discussed. For
application use, it is much more important that
different groups with different methods all obtain
good enough results. That points to the necessary
maturity of the field. This is actually a problem
with a lot of medical research.
• Quality control will be necessary, by manual in-
spection or otherwise. This aspect has not been
considered yet.
• We did not encounter any problems in using skulls
with openings in training, probably because the
holes were never in the same location and there-
fore each hole was influencing only a small part
of the sample.
In this paper we tried to emphasize a few salient
points for transfer of academic research to industry.
First, how particular academic research can be used
in industry is not always very clear. Mesh manufac-
turing seemed originally more a problem in metal-
lurgy than a medical problem. Second, the transition
from papers in academic journals to reusing the work
elsewhere is more painful than academic researchers
seem to realize.
As a more general conclusion, we want to present
a more positive view by this example, given a general
pessimism that research is having diminishing returns
in boosting productivity, as for instance defended in
(Bloom et al., 2017). Bloom et. al. explicitly dismiss
a possible role of AI in growth of productivity. It is
true, as stated in that article, that most of IT efforts
have been spent on increasing choices (more choice
in streams instead of more time to listen, more fonts
instead of easier to understand documents, etc.), and
that AI has played a minor role in that. However,
the current example in this paper of technology trans-
fer provides a counter-example to that pessimism. It
shows that relatively low investments still can lead to
meaningful productivity improvements. AI can play
a significant role in that.
ACKNOWLEDGEMENTS
We thank our colleagues at Meticuly for their help and
contributions.
REFERENCES
Bloom, N., Jones, C., Van Reenen, J., and Webb, M. (2017).
Ideas aren’t running out, but they are getting more ex-
pensive to find. https://voxeu.org/article/ideas-aren-t-
running-out-theyare-getting-more-expensive-find.
Huttenlocher, P., Klanderman, G., and Rucklidge, W.
(1993). Comparing images using the Hausdorff dis-
tance. In IEEE Transactions on Pattern Analysis and
Machine Intelligence, volume 15(9), page 850. IEEE.
Jimenez, S., Gonzalez, F. A., and Gelbukh, A. (2016).
Mathematical properties of soft cardinality: Enhanc-
ing Jaccard, Dice and cosine similarity measures with
element-wise distance. Information Sciences, 367-
368:373–389.
Klein, A., Warszawski, J., Hillengaß, J., and Maier-Hein,
K. (2019). Automatic bone segmentation in whole-
body CT images. International journal of com-
puter assisted radiology and surgery, 14(1):21–29.
https://doi.org/10.1007/s11548-018-1883-7.
Kushner, D. S., Verano, J. W., and Titelbaum, A. R.
(2018). Trepanation procedures/outcomes:
Comparison of prehistoric Peru with other an-
cient, medieval, and American civil war cranial
surgery. World Neurochirurgy, 114:245–251.
doi:10.1016/j.wneu.2018.03.143.
Mader, K. (n.d.). 4Quant/Bone-Segmenter.
https://github.com/4Quant/Bone-Segmenter.
Mahinda, H. and Murty, O. (July-December 2009). Vari-
ability in thickness of human skull bones and sternum
– an autopsy experience. Journal of Forensic Medicine
& Toxicology, 26(2):26–31.
Minnema, J., van Eijnatten, M., Kouw, W., Diblen, F., Men-
drik, A., and Wolff, J. (2018). CT image segmentation
of bone for medical additive manufacturing using a
convolutional neural network. Computers in Biology
and Medicine, 103:130–139.
Rawat, W. and Wang, Z. (2017). Deep convolutional neural
networks for image classification: A comprehensive
review. Neural Computation, 29(9):2352–2449.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net:convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention – MICCAI 2015. Lec-
ture Notes in Computer Science, Volume 9351,
Springer. https://doi.org/10.1007/978-3-319-24574-
4
28.
Structseg2019 (2019). Structseg 2019 auto-
matic structure segmentation for radio ther-
apy challenge. https://structseg2019.grand-
challenge.org/Evaluation/.
van Eijnatten, M., van Dijk, R., Dobbe, J., Streekstra, G.,
Koivisto, J., and Wolff, J. (2018). CT image segmen-
tation methods for bone used in medical additive man-
ufacturing. Medical Engineering & Physics, 51:6–16.
Xia, D.-F., Xu, S.-L., and Qi, F. (1999). A proof of the arith-
metic mean – geometric mean – harmonic mean in-
equalities. RGMIA Research Report Collection, 2(1).
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
380
