
SANO: Score-based Anomaly Localization for Dermatology
Alvaro Gonzalez-Jimenez
1 a
, Simone Lionetti
2 b
, Ludovic Amruthalingam
1 c
,
Philippe Gottfrois
1 d
, Marc Pouly
2 e
and Alexander Navarini
3 f
1
University of Basel, Basel, Switzerland
2
Lucerne University of Applied Sciences and Arts, Rotkreuz, Switzerland
3
University Hospital of Basel, Basel, Switzerland
{alvaro.gonzalezjimenez, firstname.lastname}@{unibas.ch, hslu.ch, usb.ch}
Keywords:
Unsupervised Anomaly Localization, Score-based Diffusion Models, Dermatology, Jewelry.
Abstract:
Supervised learning for dermatology requires a large volume of annotated images, but collecting clinical data
is costly and it is virtually impossible to cover all situations. Unsupervised anomaly localization circumvents
this problem by learning the distribution of healthy skin. However, algorithms which use a generative model
and localize pathologic regions based on a reconstruction error are not robust to domain shift, which is a
problem due to the low level of standardization expected in many dermatologic applications. Our method,
SANO, uses score-based diffusion models to produce a log-likelihood gradient map that highlights potentially
abnormal areas. A segmentation mask can then be calculated based on deviations from typical values observed
during training. We train SANO on a public non-clinical dataset of healthy hand images without ornaments
and evaluate it on the task of detecting jewelry within images from the same dataset. We demonstrate that
SANO outperforms competing approaches from the literature without introducing the additional complexity
of solving a Stochastic Differential Equation (SDE) at inference time”.
1 INTRODUCTION
Skin diseases are among the leading non-fatal dis-
eases globally, accounting for a significant fraction of
visits to clinics. The scarce availability of experts to
treat these conditions is a serious issue in develop-
ing countries, where the ratio of dermatologists to the
general population is as low as 1 to 216,000 (Dlova
et al., 2017). Therefore, it is not surprising that devel-
oping a system capable of identifying and diagnosing
the most common dermatologic pathologies attracts
considerable interest, including from large organiza-
tions (Liu et al., 2020). Most efforts in this direction
are based on supervised Deep Learning (DL) algo-
rithms that achieve remarkable performance but cru-
cially depend on the availability of large amounts of
annotated data.
The problems with such a requirement in the field
a
https://orcid.org/0000-0002-1337-9430
b
https://orcid.org/0000-0001-7305-8957
c
https://orcid.org/0000-0001-5980-5469
d
https://orcid.org/0000-0001-8023-3207
e
https://orcid.org/0000-0002-9520-4799
f
https://orcid.org/0000-0001-7059-632X
of dermatology are manifold. First, although data col-
lection is straightforward compared to medical imag-
ing with specialized equipment, acquisition condi-
tions (such as camera model, lighting, view distance,
and angle) are often even less constrained than usual.
There is no established process to standardize im-
ages collected under such varied conditions. Sec-
ond, most current training data consists of white skin
samples, which results in a serious bias as perfor-
mance considerably deteriorates with different skin
tones (Kamulegeya et al., 2019; Adamson and Smith,
2018; Groh et al., 2021). This is a major obstacle
for the deployment of teledermatology in many de-
veloping countries, and highlights a potentially even
more complex problem in achieving fairness for eth-
nic minorities. Third, obtaining sufficient data for rare
pathologies is challenging, especially if the data dis-
tribution has a strong geographical dependence. For
instance, insect bites are common in Africa but rare in
Europe (World Health Organization, 2005; Kiprono
et al., 2015). Fourth, annotation is a time-consuming
task that requires clinical experience, which makes
it very costly to obtain detailed segmentation masks.
Even when effort is not an issue, the gold standard
for dermatologic diagnosis is histopathology, which
Gonzalez-Jimenez, A., Lionetti, S., Amruthalingam, L., Gottfrois, P., Pouly, M. and Navarini, A.
SANO: Score-based Anomaly Localization for Dermatology.
DOI: 10.5220/0011539200003523
In Proceedings of the 1st Workshop on Scarce Data in Artificial Intelligence for Healthcare (SDAIH 2022), pages 53-58
ISBN: 978-989-758-629-3
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
53

raises ethical concerns for collecting data when a
biopsy is not clinically required. Finally, the an-
notation process is characterized by marked human
bias, as demonstrated by several reports of low inter-
annotator agreement in the field (Ribeiro et al., 2019).
Learning the appearance of healthy skin to find
unhealthy regions is a strategy that mitigates many
of the above problems. We call this approach unsu-
pervised anomaly localization, even if it is sometimes
termed semi-supervised when the training data is fil-
tered to be free of unhealthy examples.
1
Such a strat-
egy holds great potential in dermatology, where im-
ages of healthy skin are relatively effortless to obtain.
However, it does not produce a specific diagnosis and
typically results in less accurate segmentation masks.
In practice, one often learns to reconstruct healthy
images with a generative model and uses the differ-
ence between an image and its reconstructed version
to identify lesions. A variety of papers explore this
principle in combination either with Variational Au-
toencoders (VAEs) (Baur et al., 2020a; Baur et al.,
2019; Bergmann et al., 2019; Chen and Konukoglu,
2018; Chen et al., 2020) or with Generative Adver-
sarial Networks (GANs) (Schlegl et al., 2019; Ander-
matt et al., 2018; Baur et al., 2020b). These methods
demonstrated a high degree of success in clinical set-
tings where the image acquisition process is very stan-
dardized, but suffer significantly under less controlled
conditions (Heer et al., 2021). Recent works investi-
gated alternative strategies for unsupervised anomaly
localization (Cohen and Hoshen, 2020; Defard et al.,
2021; Yi and Yoon, 2020). In particular, it was noted
that the gradients of the log likelihood with respect to
inputs generate a normalcy score heatmap. Energy-
Based Models (EBMs) are particularly well-suited for
this purpose, as the energy itself is the log-likelihood
function up to an additive constant (Genc et al., 2021).
In this paper, we propose using score-based dif-
fusion models (Song et al., 2021b) for unsuper-
vised anomaly localisation, which we name Score-
based ANOmaly localization (SANO). These mod-
els directly approximate gradients of the log like-
lihood, achieving state-of-the-art likelihood values
even when this is not their explicit training objec-
tive (Song et al., 2021a). They are thus optimally
suited for unsupervised anomaly localization using
log-likelihood gradient distributions. In contrast with
(Wolleb et al., 2022), this does not require recon-
struction, whose computational complexity is one of
the main drawbacks of score-based diffusion models.
Moreover, we outline a procedure to determine abnor-
1
One often also speaks of “novelty” rather than
“anomaly” detection, a lexical distinction which would be
counterintuitive here.
mal regions that is distinct from the one proposed in
(Genc et al., 2021) for EBMs. We evaluate SANO on
the 11k Hands dataset (Afifi, 2019), which we aug-
mented with more than 3’000 ground-truth segmen-
tation masks for jewelry. These annotations are pub-
licly released for reproducibility and further research
(Gonzalez-Jimenez et al., 2022). We compare SANO
with a handful of competing approaches, and demon-
strate that it shows the best performance in this con-
text. These observations make SANO a promising
candidate for disease-agnostic segmentation of skin
pathologies in digital dermatology.
2 METHODS
2.1 Score-based Diffusion Models
Several generative modeling approaches were re-
cently unified under a single framework and grouped
under the common name of score-based diffusion
models (Song et al., 2021b). Models which belong to
this class are associated with a stochastic process x(t)
indexed by a time variable t ∈ [0, 1] which progres-
sively transforms a data point x(0) into a sample x(1)
from a prior distribution p
1
(x) representing random
noise. The transformation of data into noise admits
a reverse process which enables mapping a sample
x(1) from the prior to a data point x(0) following the
data distribution p
0
(x), i.e. it constitutes a generative
model. The reversible transformation process from
x(0) to x(1) is defined by a SDE and induces a one-
parameter family of probability distributions p
t
(x),
which smoothly interpolates between p
1
(x) and p
0
(x)
and may be factorized into the product of the prior
with a transition kernel p
t
′
(x
′
) = p
tt
′
(x
′
|x)p
t
(x).
The training process for score-based diffusion
models consists in finding an approximation s
θ
(x,t)
for the gradient of the log likelihood with respect
to the inputs, ∇
x
log p
t
(x), which is also called the
(Stein) score function (Stein, 1972; Liu et al., 2016)
of p
t
(x). This can be achieved by minimizing the loss
J (θ) =
1
2
Z
1
0
E
p
0t
(x
′
|x)p
0
(x)
∥∇
x
′
log p
0t
(x
′
|x)
− s
θ
(x
′
, t)∥
2
2
dt. (1)
Note that, in this formulation, the analytic or numeric
tractability of the normalization factor for the time-
dependent probability distribution p
t
(x) is irrelevant.
Remarkably, it has been shown that although score-
based diffusion models do not directly optimize the
likelihood of the data, there is a way of weighting
the integrand in eq. (1) which turns J (θ) into a lower
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
54

bound for the likelihood (Song et al., 2021b; Song
et al., 2021a). Empirical results in the same refer-
ences demonstrate that score-based diffusion models
obtain very competitive likelihood values on a range
of practical tasks.
The cited works on score-based diffusion models
studied three types of SDE: Variance Exploding (VE),
Variance Preserving (VP), and sub-VP. Here we con-
sider the VP SDE, the simplest apart from VE which
empirically delivers worse likelihoods. The equation
reads
dx(t) = −
1
2
β(t)x(t)dt +
p
β(t)dw(t), (2)
where w denotes the standard Wiener process and
β(t) is a positive function. Following (Ho et al., 2020;
Song et al., 2021b), we set
β(t) =
¯
β
min
+t(
¯
β
max
−
¯
β
min
). (3)
In particular, we note that these definitions yield a
gaussian transition kernel, which significantly simpli-
fies calculations and indicates that the stochastic evo-
lution from t = 0 to t = 1 corresponds to gradual ad-
dition of gaussian noise.
2.2 Anomaly Localization with Scores
The principle we use to localize anomalies is the same
as in (Genc et al., 2021), i.e. gradients of the log like-
lihood with respect to input values are typically larger
for inputs that are unlike any training examples. Since
score-based diffusion models are directly trained to
predict ∇
x
log p
t
(x) from x, including the special case
t = 0, a single forward pass of s
θ
(x,t) is sufficient to
obtain these gradients. This is in contrast to EBMs,
where backpropagation is required at this stage.
The basic idea is to classify pixels as anomalies
when the probability distribution observed in the nor-
mal training data for the score is lower than a certain
threshold. Any density estimation technique can, in
principle, be used for this task.
2
We note that the
threshold parameter is associated with the expected
false positive rate, and reasonable values can even be
guessed without a validation set containing anoma-
lous data. In practice, provided that the score distri-
bution is centered around zero, a heuristic recipe is
to identify anomalies as those pixels whose gradients
deviate from zero by more than a certain number N of
training-data standard deviations. Indeed, even when
the distribution of gradients in the training data is non-
gaussian, its standard deviation will be dominated by
the longest tails and the Mahalanobis squared norm
2
A similar probabilistic approach can be adopted even
for reconstruction errors.
will provide a reasonably conservative anomaly crite-
rion.
In contrast to (Genc et al., 2021), we do not en-
counter evidence that considering a different distribu-
tion at each pixel position improves our results. We
simply combine color channels by averaging the ab-
solute score values for a given pixel, and compare the
result to the standard deviation of the distribution for
all color channels and pixels. Finally, to increase the
scale which defines an anomaly while retaining pixel-
level resolution, we apply a gaussian filter with σ = 2
to the anomaly score image.
3 EXPERIMENTS
3.1 11k Hands
It is a public dataset that contains 11,076 hand im-
ages with a resolution of 1600 × 1200 pixels (Afifi,
2019). Each hand was photographed from the dor-
sal and palmar sides with uniform white background
and the same indoor lighting, approximately at the
same distance from the camera. We manually anno-
tated a total of 3179 pixel-wise masks of jewels in
order to consider the task of segmenting jewelry as an
anomaly. We release these masks labels (Gonzalez-
Jimenez et al., 2022) for reproducing our results and
any other use.
3.2 Training
We divided 7682 images of the 11K Hands without
jewels into two sets with no patient overlap to train the
score-based diffusion model. From the 7862 images
without jewelry, we used 6146 images for training and
the remaining 1536 images for validation. There is no
overlapping of patient images between the two sets
and we used the validation images to select models
based on the value of the loss function, which mea-
sured the accuracy of the estimated score for healthy
images not used during training. We resize the im-
ages to 256 × 256 pixels and do not employ any
data augmentation. We approximate the score using
a U-Net for s
θ
(x,t) as suggested by (Dhariwal and
Nichol, 2021). We set the training objective as in
eq. (1) with the choice in eq. (3), 1000 diffusion time
steps, β
min
= 10
−4
, and β
max
= 0.02, using a public
codebase
3
adapted for the purpose. We set the batch
size to 64 and perform 400 iterations using the Adam
(Kingma and Ba, 2017) optimizer with a learning rate
of 2 × 10
−4
.
3
https://github.com/yang-song/score sde pytorch
SANO: Score-based Anomaly Localization for Dermatology
55
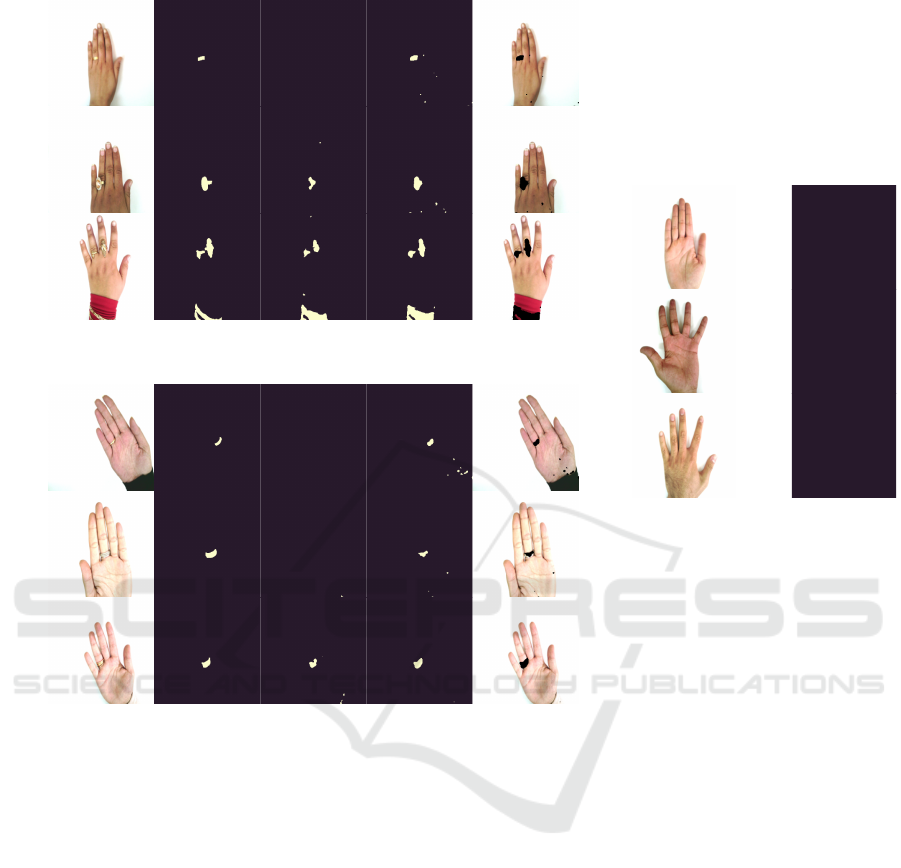
Image Ground truth mask VQ-VAE SANO SANO overlay
(a) Segmentation masks on the dorsal side.
Image Ground truth mask VQ-VAE SANO SANO overlay
(b) Segmentation masks on the palmar side.
Image SANO
(c) Segmentation masks on hands
with no rings.
Figure 1: Some examples of the segmentations obtained by SANO and VQ-VAE. SANO obtained a more detailed segmenta-
tion mask under a variety of jewelry and skin tones.
Alongside the score-based diffusion model we
consider several other DL models for compari-
son. More specifically, we train an Autoencoder
(AE), a Variational Autoencoder (VAE) (Kingma and
Welling, 2014; Rezende et al., 2014), a context-
encoding Variational Autoencoder (ceVAE) (Zim-
merer et al., 2018), VQ-VAE (Razavi et al., 2019),
and AnoVAEGAN (Baur et al., 2019) optimizing the
L
2
reconstruction loss. In these cases we relied on
public implementations with manual hyperparameter
tuning on a validation set consisting of healthy images
from 11k Hands, without changing their architecture.
3.3 Evaluation
We evaluate all approaches using traditional metrics
for unsupervised anomaly localization. Although the
jewelry detection problem could be formulated as an
instance segmentation task, we label, train, and eval-
uate for binary semantic segmentation. We report the
Area Under the Precision-Recall Curve (AUPRC) and
the Area Under the Receiver Operating Characteristic
curve (AUROC) at the level of pixels over the entirety
of images in the datasets. From the AUPRC we ob-
tain the Operating Point (OP) as threshold to generate
the final segmentation mask, and we compute Dice
coefficient and Intersection over Union (IoU) over the
obtained pixel-wise predictions.
4 RESULTS
All considered algorithms achieve reasonably good
performance, as shown in table 1. The reported uncer-
tainties are estimates of expected variations due to the
finite size of the evaluation set, computed as the stan-
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
56

Table 1: Results for the localization of jewelry in 11k Hands dataset. (⋆) indicates that the threshold locate the anomaly is set
without accessing to abnormal images.
Model AUROC AUPRC Dice IoU
AE 0.944 ± 0.002 0.123 ± 0.014 0.200 ± 0.013 0.111 ± 0.008
VAE 0.945 ± 0.002 0.123 ± 0.011 0.199 ± 0.011 0.110 ± 0.007
ceVAE 0.941 ± 0.002 0.120 ± 0.014 0.196 ± 0.012 0.108 ± 0.007
VQ-VAE 0.967 ± 0.002 0.446 ± 0.022 0.467 ± 0.015 0.305 ± 0.013
AnoVAEGAN 0.945 ± 0.002 0.121 ± 0.013 0.196 ± 0.015 0.109 ± 0.009
SANO
⋆
N=3
0.966 ± 0.003 0.488 ± 0.020
0.559 ± 0.024 0.388 ± 0.023
SANO 0.618 ± 0.027 0.448 ± 0.027
dard deviations of 50 bootstrap runs where random
selection with replacement was stratified over individ-
uals in the dataset. Although the AUROC of SANO
is lower than of VQ-VAE, the table shows that even
without access to anomalous pictures, SANO obtains
better Dice and IoU scores.
Some example masks obtained with SANO are il-
lustrated in fig. 1. The model is able to correctly
segment jewels on both the dorsal (fig. 1a) and pal-
mar (fig. 1b) sides of hands in a variety of skin tones.
Note that sleeves were present in the training set, and
that SANO also gets qualitatively reasonable results
on wrist jewelry. Finally, in fig. 1c we observe that
SANO does not yield any false positives on a few
hand images without jewelry
4
.
5 CONCLUSIONS
In this paper we proposed SANO, an approach
that uses the log-likelihood gradient magnitude
from score-based diffusion models for unsupervised
anomaly localization. Unlike many other techniques
which leverage generative modeling, it does not re-
quire reconstruction to determine abnormal regions.
We publicly released manually labeled masks for jew-
elry in 11k Hands and showed that SANO performs
competitively to previous unsupervised approaches
for various jewelry objects and skin tones. As our next
step, we plan to evaluate the performance of SANO
on the localisation of pathologic regions in a clinical
dermatology dataset.
REFERENCES
Adamson, A. S. and Smith, A. (2018). Machine Learning
and Health Care Disparities in Dermatology. JAMA
4
We re-trained and ran SANO using a split where the
evaluation set also contains 1953 images without jewelry,
obtaining AUROC=0.945, AUPRC=0.400, Dice=0.508,
and IoU=0.340.
Dermatology, 154(11):1247.
Afifi, M. (2019). 11k hands: gender recognition and bio-
metric identification using a large dataset of hand im-
ages. Multimedia Tools and Applications.
Andermatt, S., Huck, A., Pezold, S., and Cattin, P. (2018).
Pathology Segmentation Using Distributional Differ-
ences to Images of Healthy Origin.
Baur, C., Denner, S., Wiestler, B., Albarqouni, S., and
Navab, N. (2020a). Autoencoders for Unsupervised
Anomaly Segmentation in Brain MR Images: A Com-
parative Study.
Baur, C., Graf, R., Wiestler, B., Albarqouni, S., and Navab,
N. (2020b). SteGANomaly: Inhibiting CycleGAN
Steganography for Unsupervised Anomaly Detection
in Brain MRI. In Medical Image Computing and Com-
puter Assisted Intervention – MICCAI 2020, pages
718–727. Springer International Publishing.
Baur, C., Wiestler, B., Albarqouni, S., and Navab, N.
(2019). Deep Autoencoding Models for Unsuper-
vised Anomaly Segmentation in Brain MR Images. In
Brainlesion: Glioma, Multiple Sclerosis, Stroke and
Traumatic Brain Injuries, pages 161–169. Springer
International Publishing.
Bergmann, P., L
¨
owe, S., Fauser, M., Sattlegger, D., and Ste-
ger, C. (2019). Improving Unsupervised Defect Seg-
mentation by Applying Structural Similarity to Au-
toencoders:. In Proceedings of the 14th International
Joint Conference on Computer Vision, Imaging and
Computer Graphics Theory and Applications, pages
372–380.
Chen, X. and Konukoglu, E. (2018). Unsupervised Detec-
tion of Lesions in Brain MRI using constrained adver-
sarial auto-encoders.
Chen, X., You, S., Tezcan, K. C., and Konukoglu, E. (2020).
Unsupervised Lesion Detection via Image Restoration
with a Normative Prior.
Cohen, N. and Hoshen, Y. (2020). Sub-image anomaly
detection with deep pyramid correspondences. arXiv
preprint arXiv:2005.02357.
Defard, T., Setkov, A., Loesch, A., and Audigier, R. (2021).
Padim: a patch distribution modeling framework for
anomaly detection and localization. In International
Conference on Pattern Recognition, pages 475–489.
Springer.
Dhariwal, P. and Nichol, A. (2021). Diffusion models beat
gans on image synthesis. Advances in Neural Infor-
mation Processing Systems, 34.
SANO: Score-based Anomaly Localization for Dermatology
57

Dlova, N., Chateau, A., Khoza, N., Skenjane, A., Mkhize,
M., Katibi, O., Grobler, A., Tsoka-Gwegweni, J., and
Mosam, A. (2017). Prevalence of skin diseases treated
at public referral hospitals in KwaZulu-Natal, South
Africa. The British journal of dermatology, 178.
Genc, E. U., Ahuja, N., Ndiour, I. J., and Tickoo, O.
(2021). Energy-based anomaly detection and local-
ization. arXiv preprint arXiv:2105.03270.
Gonzalez-Jimenez, A., Lionetti, S., Amruthalingamnd, L.,
Gottfrois, P., Pouly, M., and Navarini, A. (2022). Jew-
elry segmentation masks for the 11k Hands dataset.
Groh, M., Harris, C., Soenksen, L., Lau, F., Han, R., Kim,
A., Koochek, A., and Badri, O. (2021). Evaluating
Deep Neural Networks Trained on Clinical Images in
Dermatology with the Fitzpatrick 17k Dataset.
Heer, M., Postels, J., Chen, X., Konukoglu, E., and Albar-
qouni, S. (2021). The OOD Blind Spot of Unsuper-
vised Anomaly Detection.
Ho, J., Jain, A., and Abbeel, P. (2020). Denoising Diffusion
Probabilistic Models.
Kamulegeya, L. H., Okello, M., Bwanika, J. M., Musin-
guzi, D., Lubega, W., Rusoke, D., Nassiwa, F., and
B
¨
orve, A. (2019). Using artificial intelligence on der-
matology conditions in Uganda: A case for diversity
in training data sets for machine learning.
Kingma, D. P. and Ba, J. (2017). Adam: A method for
stochastic optimization.
Kingma, D. P. and Welling, M. (2014). Auto-Encoding
Variational Bayes.
Kiprono, S. K., Muchunu, J. W., and Masenga, J. E. (2015).
Skin diseases in pediatric patients attending a tertiary
dermatology hospital in Northern Tanzania: A cross-
sectional study. BMC Dermatology, 15(1):16.
Liu, Q., Lee, J., and Jordan, M. (2016). A Kernelized Stein
Discrepancy for Goodness-of-fit Tests. In Proceed-
ings of The 33rd International Conference on Machine
Learning, pages 276–284. PMLR.
Liu, Y., Jain, A., Eng, C., Way, D. H., Lee, K., Bui,
P., Kanada, K., de Oliveira Marinho, G., Gallegos,
J., Gabriele, S., Gupta, V., Singh, N., Natarajan, V.,
Hofmann-Wellenhof, R., Corrado, G. S., Peng, L. H.,
Webster, D. R., Ai, D., Huang, S. J., Liu, Y., Dunn,
R. C., and Coz, D. (2020). A deep learning sys-
tem for differential diagnosis of skin diseases. Nature
Medicine, 26(6):900–908.
Razavi, A., van den Oord, A., and Vinyals, O. (2019). Gen-
erating Diverse High-Fidelity Images with VQ-VAE-
2.
Rezende, D. J., Mohamed, S., and Wierstra, D. (2014).
Stochastic backpropagation and approximate infer-
ence in deep generative models. In International
conference on machine learning, pages 1278–1286.
PMLR.
Ribeiro, V., Avila, S., and Valle, E. (2019). Handling Inter-
Annotator Agreement for Automated Skin Lesion Seg-
mentation.
Schlegl, T., Seeb
¨
ock, P., Waldstein, S., Langs, G., and
Schmidt-Erfurth, U. (2019). F-AnoGAN: Fast Unsu-
pervised Anomaly Detection withGenerative Adver-
sarial Networks. Medical Image Analysis, 54.
Song, Y., Durkan, C., Murray, I., and Ermon, S. (2021a).
Maximum Likelihood Training of Score-Based Diffu-
sion Models.
Song, Y., Sohl-Dickstein, J., Kingma, D. P., Kumar, A., Er-
mon, S., and Poole, B. (2021b). Score-Based Gener-
ative Modeling through Stochastic Differential Equa-
tions. In 9th International Conference on Learning
Representations,ICLR.
Stein, C. (1972). A bound for the error in the normal ap-
proximation to the distribution of a sum of dependent
random variables. In The Sixth Berkeley Symposium
on Mathematical Statistics and Probability, Volume 2:
Probability Theory, volume 6.2, pages 583–603. Uni-
versity of California Press.
Wolleb, J., Bieder, F., Sandk
¨
uhler, R., and Cattin, P. C.
(2022). Diffusion Models for Medical Anomaly De-
tection.
World Health Organization (2005). Epidemiology and man-
agement of common skin diseases in children in devel-
oping countries. (WHO/FCH/CAH/05.12).
Yi, J. and Yoon, S. (2020). Patch svdd: Patch-level svdd for
anomaly detection and segmentation. In Proceedings
of the Asian Conference on Computer Vision.
Zimmerer, D., Kohl, S. A. A., Petersen, J., Isensee, F., and
Maier-Hein, K. H. (2018). Context-encoding Varia-
tional Autoencoder for Unsupervised Anomaly Detec-
tion.
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
58
