
PT-MESS: A Problem-transformation Approach for Multi-event
Survival Analysis
Michela Venturini
1,2 a
, Felipe Kenji Nakano
1,2 b
and Celine Vens
1,2 c
1
KU Leuven, Campus KULAK, Department of Public Health and Primary Care,
Etienne Sabbelaan 53, 8500 Kortrijk, Belgium
2
Itec, imec research group at KU Leuven, Etienne Sabbelaan 53, 8500 Kortrijk, Belgium
Keywords:
Data Scarcity, Right-censoring, Survival Analysis, Multi-target Regression.
Abstract:
Multi-event survival analysis is an under-explored field in literature, typically addressed by modeling each
event independently or implying specific event settings. In this context, problem transformations approaches
offer a promising alternative to rephrase the setting into standard multi-target regression. Nevertheless, they
also suffer from the intrinsic presence of partial information in time-to-event data, since their application often
requires the exclusion of censored observations, thus potentially discarding valuable information. In this work,
we propose a novel Problem Transformation Approach for Multi-event Survival analySis (PT-MESS), which
is capable of exploiting partial information, by encoding the survival outcome in a risk score based on the time-
to-event distribution estimation. This approach allows the use of any multi-target machine learning model to
address the original survival task. Using random forest as the underlying model, we conducted experiments
using real-data multiple benchmarks from the medical domain and synthetic datasets. Our results revealed
that PT-MESS provides superior or competitive results compared to competitors from the literature, especially
when the events considered had a similar survival distribution.
1 INTRODUCTION
Survival analysis (SA) refers to a field of statistics
that deals with time-to-event data, which often con-
cerns medical applications where the outcome of in-
terest is the time until occurrence of one (or more)
adverse outcomes (e.g., death or cancer recurrence).
SA is characterized by the presence of partial infor-
mation, referred to as censoring, mainly due to indi-
viduals that are either lost to follow-up or do not ex-
perience the event during the follow-up, thus leaving
the true time-to-event unknown. This phenomenon
can be considered as a type of data scarcity as it leads
to lack of labels and possibly poor prediction perfor-
mance in case of high censoring rate. SA has been
tackled either with algorithm-adaptation or problem-
transformation machine learning approaches. While
the first scenario implies adapting existing algorithms
to handle survival data (Ping Wang, 2019), the latter
one focuses on transforming time-to-event data to a
well-studied problem, such as classification and re-
a
https://orcid.org/0000-0002-9947-0218
b
https://orcid.org/0000-0002-4884-9420
c
https://orcid.org/0000-0003-0983-256X
gression, enabling off-the-shelf models to be straight-
forwardly applied (Vock et al., 2016). For instance,
Vock et.al. (Vock et al., 2016) introduced an ap-
proach that employs weights to address SA as a bi-
nary classification task, however it excludes censored
observations, potentially overlooking useful informa-
tion. Despite the existence of numerous machine
learning approaches for single-event SA, the literature
presents few studies on the multi-event setting (Tjan-
dra et al., 2021; Ishwaran et al., 2014). Furthermore,
these studies have proposed algorithm-adaptation ap-
proaches which are solely tailored for competing and
semi-competing risk analysis. That is, events are nec-
essarily mutually exclusive (e.g., death from heart
attack or breast cancer) or chronologically ordered
(e.g., Alzheimer’s disease onset and death), thus be-
ing inadequate for more general settings. Although
multiple outputs have been extensively studied in the
machine learning literature (Xu et al., 2020), to date
no problem-transformation approach exists to cast
multi-event survival analysis to a multi-output predic-
tion problem. In this work, we propose a problem-
transformation approach, namely Problem Transfor-
mation Multi-Event Survival AnalySis (PT-MESS),
Venturini, M., Nakano, F. and Vens, C.
PT-MESS: A Problem-transformation Approach for Multi-event Survival Analysis.
DOI: 10.5220/0011531600003523
In Proceedings of the 1st Workshop on Scarce Data in Artificial Intelligence for Healthcare (SDAIH 2022), pages 29-34
ISBN: 978-989-758-629-3
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
29

which can be employed in any multi-event survival
setting. More specifically, our approach relies on the
survival distribution relative to each event to encode
outcomes, allowing the inclusion of censored data in
the model. Specifically, PT-MESS encodes time and
censoring information into a score, enabling us to
treat any multi-event SA task as a multi-target regres-
sion problem. By performing experiments on pub-
licly available medical datasets, employing a standard
multi-target regression model, we showcase that PT-
MESS leads to superior or competitive results against
methods from the literature in the majority of the
cases.
2 PROPOSED METHOD
Survival outcome typically consists of two elements,
namely time to event (or censoring) and binary sta-
tus, indicating whether the individual experiences the
event or is censored at the given time point. We pro-
pose to encode multi-event SA as a multi-target re-
gression task, by transforming the survival outcome,
for each event, into a single score. Specifically, the
encoding process is based on the Kaplan-Meier esti-
mate (Kaplan and Meier, 1958) of the survival curve
relative to each event. Kaplan-Meier curves are used
to describe the event distribution in the population,
taking into account time and censoring information.
Typically, multi-event survival problems are encoded
as follows. Given N individuals and K events, each
individual i = 1, 2, ..., N is associated with three vari-
ables: the covariates vector x
(i)
, the censoring statuses
for each event c
(i)
and the time information o
(i)
. In
this context, the Kaplan-Meier curve estimate of the
survival curve for each event k,
ˆ
s
k
(t), is defined as
follows:
ˆ
s
k
(t) =
∏
j:t
j
≤t
(1 −
d
( j)
k
r
( j)
k
) (1)
with t
j
a time when at least one event k happened,
d
( j)
k
the number of events k that happened at time t
j
,
and r
( j)
k
the individuals still at risk for event k at time
t
j
, estimated from c
k
and o
k
. PT-MESS incorporates
time and censoring information into the new outcome
m
(i)
k
as follows:
m
(i)
k
= 1 −
R
o
(i)
k
t=0
ˆs
k
R
inf
t=0
ˆs
k
(2)
where
R
o
(i)
k
t=0
ˆs
k
is the restricted mean survival time, up
to o
(i)
k
, the time at which patient i experiences the
event or is censored, and
R
inf
t=0
ˆs
k
is the expected sur-
vival time of the population, for event k. Thus, Equa-
tion 2 can be seen as an indication of how early pa-
tient i is expected to experience event k w.r.t. the con-
sidered population. The higher the score, the lower
the risk of experiencing the event. The new outcome
for each individual i thus becomes m
(i)
, a vector con-
taining a risk score for each event, that can be the
outcome to any multi-target regression model. Given
their efficiency and flexibility in handling high dimen-
sional datasets, we employed multi-target random for-
est (Kocev et al., 2013) as the underlying regression
model in our approach. Such model intrinsically ex-
ploits correlation among events in the splitting rule of
the individual trees, by computing the average impu-
rity reduction across them.
3 EXPERIMENTAL SETUP
3.1 Datasets
We employed 5 publicly available datasets, 3 of
which are real world datasets from the medical do-
main (ADNI
1
, MIMIC (Johnson et al., 2016) and
CIBMTR
2
) and 2 were synthetic (scrData
2
and Syn-
thetic). All of the real ones contain semi-competing
risks, while the synthetic datasets contain both semi-
competing risk and multi-event (events are not mu-
tually exclusive and can happen in any order). Pre-
processing of ADNI and MIMIC, and creation of Syn-
thetic dataset were performed according to (Tjandra
et al., 2021). Further details are reported at Table 1.
It can be seen that these datasets present different
characteristics. Namely, MIMIC presents a consider-
able high number of features in comparison to ADNI.
Similarly, CIBMTR contains a rather limited number
of instances which are described by a considerably
higher number of features. It can also be noticed that
most of the datasets present scarce data where the cen-
soring rate is frequently above 80%.
1
Data used in the preparation of this article were ob-
tained from the Alzheimer’s Disease Neuroimaging Initia-
tive (ADNI) database (adni.loni.usc.edu). The ADNI was
launched in 2003 as a public-private partnership, led by
Principal Investigator Michael W. Weiner, MD. The pri-
mary goal of ADNI has been to test whether serial mag-
netic resonance imaging (MRI), positron emission tomog-
raphy (PET), other biological markers, and clinical and neu-
ropsychological assessment can be combined to measure
the progression of mild cognitive impairment (MCI) and
early Alzheimer’s disease (AD).
2
https://cran.r-project.org/web/packages/
SemiCompRisks/index.html
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
30

Table 1: Characteristics of the datasets employed in this work where semi-competing datasets are represented using (S) and
independent as (I). The terminal event in each dataset highlighted in bold.
Dataset (Setting) Features Instances Events Event (Censoring rate)
MIMIC (S) 13801 3822 3 1 (0.87), 2 (0.91) and 3 (0.92)
ADNI (S) 1917 1024 2 1 (0.82) and 2 (0.99)
CIBMTR (S) 9651 30 2 1 (0.82) and 2 (0.62)
ScrData (S) 2000 4 2 1 (0.34) and 2 (0.50)
Synthetic (I) 5000 15 2 1 (0.70) and 2 (0.71)
3.2 Comparison Methods
For a fair comparison, we focused on competitive
methods that also employ ensembles of decision
trees. To the best of our knowledge, the literature
only presents a single study on random forests for
multi-event SA (Ishwaran et al., 2014), nonetheless
it is specifically tailored for competing risks, assum-
ing that each patient can only experience one event.
Hence, applying this method would require adap-
tations which are not straightforward. Further, the
method proposed by Tjandra et.al. was excluded
(Tjandra et al., 2021). Although prominent, the au-
thors have proposed a deep learning approach which
requires time-consuming parameters tuning and lacks
in interpretability. Thus, we compared the following
methods:
• Random survival forest (RSFi): this approach
learns a separate RSF (Ishwaran et al., 2008) per
event and assumes that the time-to-events among
all the events are independent.
• Inverse probability of censoring weighting
(IPCWi): this problem transformation approach
translates the survival task into a binary classifi-
cation (Dong et al., 2020). Similarly to RSFi, this
approach learns a separate model (random forest
classifier) for each event.
• PT-MESS independent (PT-MESSi): A variant
of our problem transformation approach, which
learns a separate single-target random forest re-
gressor for each event;
• (PT-MESS): A variant of our approach that builds
a multi-target random forest regressor that consid-
ers all events at once;
Each ensemble model was trained with 200 trees
and all other parameters were left to their default
values
34
. RSF is probably the most prominent ap-
proach using ensemble of trees, thus its comparison is
mandatory. Similarly, IPCW is employed as compar-
ison because it is a problem-transformation approach
3
https://scikit-learn.org/stable
4
https://scikit-survival.readthedocs.io/en/stable
comparable to ours, nonetheless it is worth mention-
ing that this method discards censored data. More-
over, both RSF and IPCW are originally designed for
single-event applications. Thus, to make them com-
parable, it is necessary to build an independent model
per event.
3.3 Evaluation
To estimate predictive performances of the models,
we employed Harrell’s concordance index(Harrell
et al., 1982) (C-index), one of the most used metrics
in survival analysis. C-index is given by
C =
∑
i,l
I(T
i
> T
l
) × I(r
l
> r
i
) × ∆
l
∑
i,l
I(T
i
> T
l
) × ∆
l
(3)
where i and l refer to pairs of observations in the sam-
ple i, l = 1, ..., N with i ̸= l, r
i
∈ R is the outcome risk
score and T
i
is the observed time-to event. ∆
l
discards
pairs of observations that are not comparable because
the smaller survival time is censored. The C-index
estimates how well a predicted risk score ranks obser-
vations according to their true time-to-event, taking
into account censored data. Moreover, it is easy to in-
terpret: C = 0.5 indicates a non-informative model
prediction, while C = 1 indicates that the model is
perfectly capable of separating patients with differ-
ent outcomes. We evaluated our results based on
per-event C-index as well as averaged C-index across
events.
4 RESULTS
We present our results in Figure 1. All experiments
were repeated using 5-fold cross-validation, strati-
fied according to the rarest event(considering the cen-
soring distribution) in the dataset. Average C-index
(per single event, and overall) is reported, together
with standard deviation. As can be seen, our pro-
posed method often has the upper-hand. More specif-
ically, PT-MESS achieves the overall superior perfor-
mance in 3 out of 5 datasets (CMBTR, scrData and
Synthetic). Furthermore, when analyzing the perfor-
PT-MESS: A Problem-transformation Approach for Multi-event Survival Analysis
31
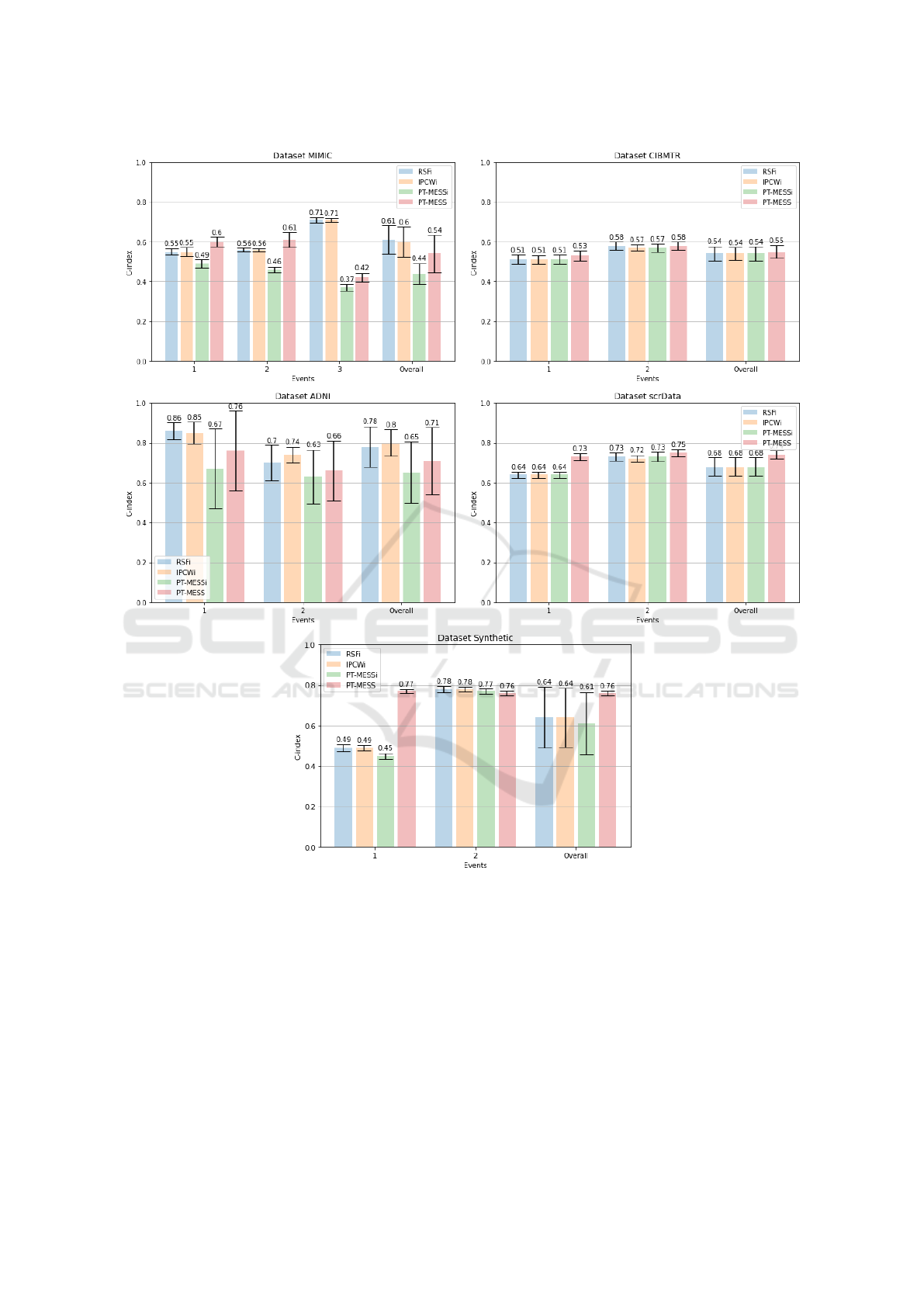
Figure 1: Results obtained on each dataset. We report the average performance obtained using the C-index and its standard
deviation obtained using 5-fold cross validation.
mance per single event, our proposed method sur-
passed the competitors in 7 out of 11 cases. As
opposed to that, PT-MESSi managed, at its best, to
be competitive with RSF and IPCW. We interpret
this as an indication that PT-MESS is preferable over
its counterpart and the competitors, considering both
its performance and its computational complexity, as
only one single model is required.
Additionally, we observed that PT-MESS out-
performed its problem-transformation approach com-
petitor, IPCWi, in several cases, as seen in events 1
and 2 of MIMIC, in event 1 of Synthetic and in the
entire scrData dataset. As reported in Table 1, these
datasets present a very high censoring rate. Hence,
we may assume that PT-MESS is capable of correctly
encoding the information of data which did not expe-
rience the event, whereas overlooking this data might
lead to sub-optimal results.
Surprisingly, RSF and IPCWi yielded consider-
ably better results in specific cases with very high cen-
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
32
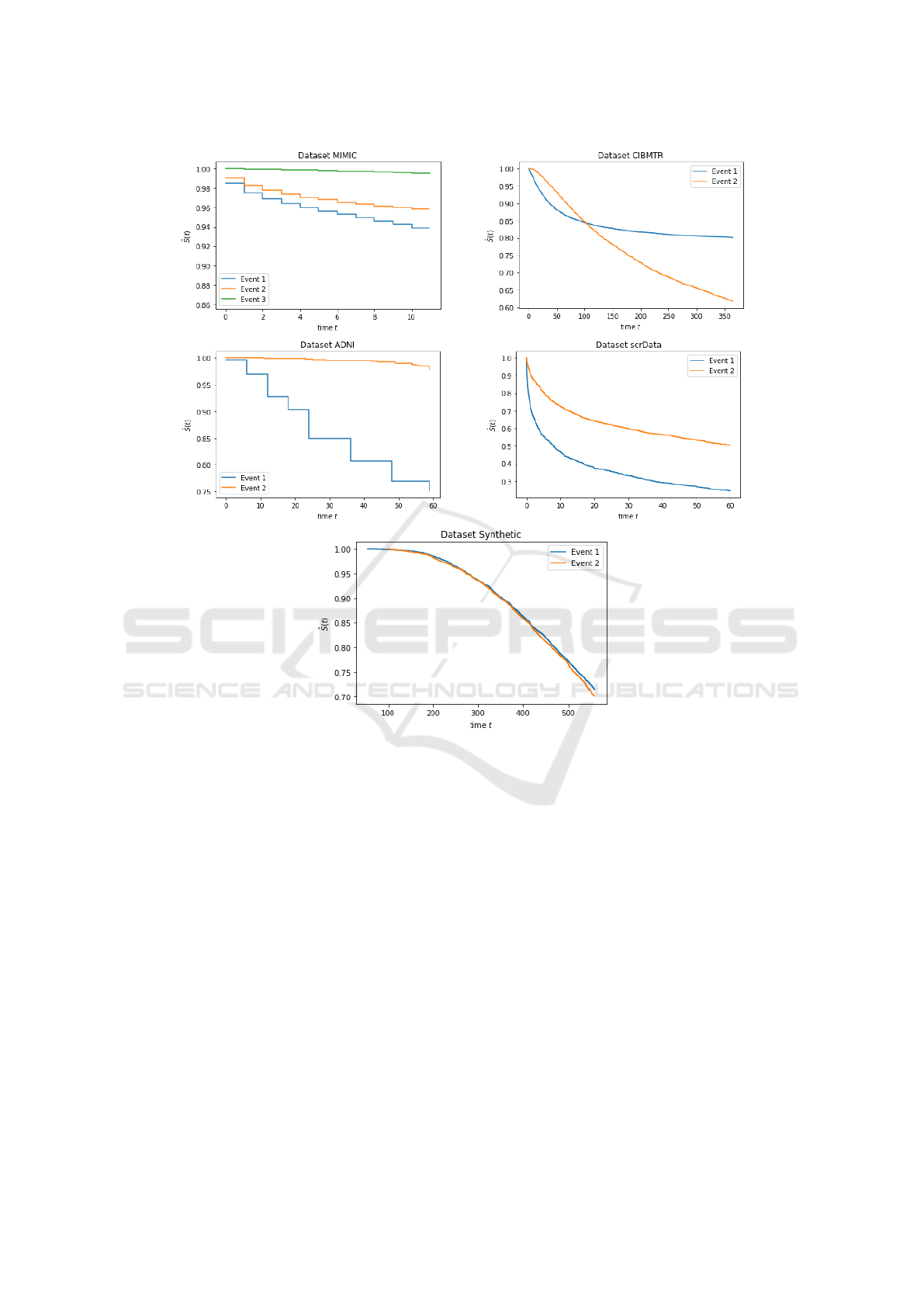
Figure 2: Kaplan-Meier estimate of the survival distribution for each event in the datasets: MIMIC, CIBMTR, ADNI and
scrData, Synthetic.
soring rate, as in MIMIC-event 3, and in the ADNI
dataset. We believe that this is related to the low cor-
relation of the events. As shown in Figure 2, their
Kaplan Meier curves reveal that events in ADNI have
very different survival curves (or time-to-event dis-
tributions). A similar behavior is observed in the
MIMIC dataset where events 1 and 2 behave iden-
tically, whereas the curve associated to event 3 fol-
lows a significantly different tendency. This finding is
further reinforced by the curve of scrData where the
events appear to be substantially correlated, leading
to superior results by our method.
5 CONCLUSIONS
We introduced a problem transformation approach
to address multi-event survival analysis, where data
scarcity is present as right-censored outcomes. Pre-
cisely, our approach encodes the typical survival out-
come in a single score per event, based on Kaplan-
Meier estimate of the survival curve and allows to in-
clude the censored observations in the model. As the
underlying predictive model, we chose multi-target
random survival forest. Our results revealed that PT-
MESS is capable of providing superior results, indi-
cating that predicting all events concurrently is bene-
ficial over addressing them separately in the majority
of the cases. In the challenging datasets, we could
identify that our method, in its current form, struggles
to predict events which are not correlated.
Hence, future work should aim to amend such de-
ficiency. In this direction, we instigate the investi-
gation of hybrid approaches, where global and local
methods, may be used in cooperation to achieve su-
perior results. That is, we will extend our method
PT-MESS: A Problem-transformation Approach for Multi-event Survival Analysis
33

to automatically detect and address correlated events
during its building time, similarly to the concept of
predictive bi-clustering trees used in multi-label clas-
sification (Zamith et al., 2020). Finally, we would
also like to further validate our method by performing
more experiments, specially regarding datasets with
competing risks, as seen in (Ishwaran et al., 2014).
AUTHORS CONTRIBUTIONS
Conceptualization, M.V.; methodology, M.V. and
F.K.N.; software, M.V.; writing—original draft prepa-
ration, M.V. and F.K.N.; writing—review and editing,
C.V.; supervision, funding acquisition, C.V. All au-
thors have read and agreed to the published version of
the manuscript
FUNDING
This research was funded by the Research Fund
Flanders (through research project G080118N and
G0A2120N). The authors also acknowledge the
Flemish Government (AI Research Program).
REFERENCES
Dong, G., Mao, L., Huang, B., Gamalo-Siebers, M.,
Wang, J., Yu, G., and Hoaglin, D. C. (2020). The
inverse-probability-of-censoring weighting (ipcw) ad-
justed win ratio statistic: an unbiased estimator in the
presence of independent censoring. Journal of bio-
pharmaceutical statistics, 30(5):882–899.
Harrell, F., Califf, R., Pryor, D., Lee, K., and Rosati, R.
(1982). Evaluating the yield of medical tests. JAMA,
247(18):2543—2546.
Ishwaran, H., Gerds, T. A., Kogalur, U. B., Moore, R. D.,
Gange, S. J., and Lau, B. M. (2014). Random survival
forests for competing risks. Biostatistics, 15(4):757–
773.
Ishwaran, H., Kogalur, U. B., Blackstone, E. H., and Lauer,
M. S. (2008). Random survival forests. The annals of
applied statistics, 2(3):841–860.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). Mimic-
iii, a freely accessible critical care database. Scientific
Data, 3(1):160035.
Kaplan, E. L. and Meier, P. (1958). Nonparametric esti-
mation from incomplete observations. Journal of the
American Statistical Association, 53(282):457–481.
Kocev, D., Vens, C., Struyf, J., and D
ˇ
zeroski, S. (2013).
Tree ensembles for predicting structured outputs. Pat-
tern Recognition, 46(3):817–833.
Ping Wang, Yan Li, C. K. R. (2019). Machine Learning for
Survival Analysis: A Survey. ACM Comput. Surv.,
51(6).
Tjandra, D., He, Y., and Wiens, J. (2021). A hierarchical
approach to multi-event survival analysis. Proceed-
ings of the AAAI Conference on Artificial Intelligence,
35(1):591–599.
Vock, D. M., Wolfson, J., Bandyopadhyay, S., Adomavi-
cius, G., Johnson, P. E., Vazquez-Benitez, G., and
O’Connor, P. J. (2016). Adapting machine learn-
ing techniques to censored time-to-event health record
data: A general-purpose approach using inverse prob-
ability of censoring weighting. J Biomed Inform,
61:119–131.
Xu, D., Shi, Y., Tsang, I. W., Ong, Y.-S., Gong, C., and
Shen, X. (2020). Survey on multi-output learning.
IEEE Transactions on Neural Networks and Learning
Systems, 31(7):2409–2429.
Zamith, B., Nakano, F. K., Cerri, R., and Vens, C. (2020).
Predictive bi-clustering trees for hierarchical multi-
label classification. ECML PKDD 2020.
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
34
