
Towards Reducing Segmentation Labeling Costs for CMR Imaging using
Explainable AI
Alessa Stria
a
and Asan Agibetov*
b
Medical University of Vienna, Center for Medical Statistics, Informatics and Intelligent Systems (CeMSIIS),
Institute of Artificial Intelligence, Vienna, Austria
∗
corresponding author
Keywords:
Deep Learning, Segmentation, Classification, Explainable AI, Class Activation Map, Labeling Costs, Scarce
Data, Sample Size Dependence, MRI, Cardiology
Abstract:
Provided with a sufficient amount of annotated data, deep learning models have been successfully applied
to automatically segment cardiac multi-structures from MR images. However, manual delineation of car-
diac anatomical structures is expensive to acquire and requires expert knowledge. Recently, weakly- and
self-supervised feature learning techniques have been pro-posed to avoid or substantially reduce the effort of
manual annotation. Due to their end-to-end design, many of these techniques are hard to train. In this paper,
we propose a simple modular segmentation framework based on U-net architecture that injects class activation
maps of separately trained classification models to guide the segmentation process. In a small data setting (20-
35% of training data), our framework significantly improved the segmentation accuracy of a baseline U-net
model (5%-150%).
1 INTRODUCTION
Segmented cardiac magnetic resonance (CMR) im-
ages can computationally quantify significant mor-
phological and pathological changes, such as stroke
volume or ejection fraction. These features are essen-
tial in cardiac disease quantification and non-invasive
pre-clinical diagnosis (Peng et al., 2016). To facili-
tate the computation of such features, deep-learning-
based cardiac segmentation algorithms have been re-
cently proposed in the literature (Bernard et al., 2018;
Chen et al., 2020; Oktay et al., 2018). While
these algorithms promise the creation of (semi-) au-
tomatic segmentation tools, their successful applica-
tion is heavily conditioned on the availability of large
amounts of labeled segmented data. Unfortunately,
obtaining segmented MR images is a tedious and
time-consuming delineation task that represents a vast
challenge in the cardiac imaging domain. While re-
searchers focused on improving the performance, a
major challenge in cardiac image segmentation con-
tinues to be the scarcity of annotated data (Chen et al.,
2020).
Methods focusing on segmentation label depen-
a
https://orcid.org/0000-0003-0178-8867
b
https://orcid.org/0000-0003-0096-0143
dence reduction include data augmentation (Madani
et al., 2018), transfer learning (Tran, 2016), and
weakly or self-supervized methods (Oktay et al.,
2018; Bai et al., 2019; Ciga and Martel, 2021; Zim-
mer et al., 2020). These approaches focus on end-to-
end framework design, where easily-obtainable clas-
sification labels are encoded as an auxiliary prediction
task, which, however, can be extremely sensitive to
hyperparameter optimization. Instead, we propose a
modular design of a segmentation framework decou-
pled from a classification model in this work.
A classification model can be trained and opti-
mized separately, and its information can be injected
into a segmentation model as a separate input channel.
Our main hypothesis is that a (pre-trained) AI classi-
fication model could be used as a template for seg-
mentation labels. The segmentation framework uses
the anatomical priors extracted from a classification
model with explainable artificial intelligence (XAI)
techniques. Compared to segmentation labels, clas-
sification labels, e.g., patient’s diagnosis, are much
easier to obtain. Indeed, a cardiologist may need to
look at a few MR slices and establish the diagnosis,
whereas manual segmentation may take hours.
The proposed methodology re-purposes a pre-
trained classification model by obtaining the class ac-
Stria, A. and Agibetov, A.
Towards Reducing Segmentation Labeling Costs for CMR Imaging using Explainable AI.
DOI: 10.5220/0011531200003523
In Proceedings of the 1st Workshop on Scarce Data in Artificial Intelligence for Healthcare (SDAIH 2022), pages 11-16
ISBN: 978-989-758-629-3
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
11

Figure 1: A high-level view of our methodology. Panel A: Training of the classification model. Panel B: Explainable AI
technique extract the approximate positions (priors) of the desired anatomical region of interest in the MR image from the
classification model. Panel C: These priors are then used as additional input for the segmentation model and reduce the
overall search space of possible anatomical regions. Panel D: detailed illustration of the different used input channels for the
experiment.
tivation maps (CAMs (Selvaraju et al., 2020)) as seg-
mentation priors. CAM is an explainable AI tech-
nique that generates a localization map, which high-
lights the relevant regions of the image with respect
to the prediction of the deep learning model. In this
project, Gradient CAMs were used (Selvaraju et al.,
2020). These proxy labels are added as an additional
input channel to the segmentation model. Therefore,
increasing the model complexity and injecting spatial
information should constrain the search space. An
overview of the methodology is presented in Figure 1.
2 METHODS
2.1 Image Data and Preprocessing
The Automatic Cardiac Diagnosis Challenge (ACDC)
dataset from the University Hospital of Dijon, pub-
lished in 2018, was used for development and eval-
uation (Bernard et al., 2018). We took cine-MRIs
of 100 patients, uniformly distributed over five di-
agnostic groups (healthy, previous myocardial infarc-
tion, dilated cardiomyopathy, hypertrophic cardiomy-
opathy, abnormal right). Segmentation ground truth
masks for all CMRs consists of four classes: back-
ground, left ventricle, myocardium, and right ventri-
cle. To prevent information leak-age between training
and evaluation, the image data was split into training,
validation, and test set (1150, 382, 370 images, re-
spectively).
2.2 Model Specifications
Different convolutional neural network (CNN) ar-
chitectures were used for the multiclass classifica-
tion task, including the most common architectures,
VGG16 (Simonyan and Zisserman, 2015), Dense-Net
(Huang et al., 2017), and ResNet (He et al., 2016).
For segmentation models, we used the baseline U-Net
architecture (Ronneberger et al., 2015).
Classification models were trained by minimizing
categorical cross-entropy loss, where RELU was the
used activation function for the convolutional layers.
On the final fully-connected layer, the softmax activa-
tion function was used. U-net was trained to minimize
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
12

intersection over union (IoU) metric. VGG16 was
optimized with Stochastic gradient descent (SGD),
all other classification models with ADAM. For op-
timization we used learning rate scheduler. On all the
models, early stopping was triggered after ten epochs
without improvement in the validation loss. To fur-
ther reduce the risk of overfitting, improve general-
ization and performance of the classification models,
different hyperparameter optimization methods were
tested. They include the usage of augmentation on the
training set, class weights for the slightly imbalanced
classes, different image sizes for more efficient train-
ing. We performed a grid search over dropout rates of
0.1, 0.2, and 0.25, number and size of filters in the 11
layered architecture. Different base learning rates in-
cluded 0.1, 0.01, and 0.001. To reduce the number of
filters and the search space, the CMRs were zoomed
in to exclude the background.
2.3 Evaluation of Model Performances
Since our interpatient splits were slightly imbalanced,
we used the area under the receiver-operating charac-
teristic curve (ROC AUC) as our classification met-
ric. Additionally, we used accuracy and F1 scores.
CAMs from our classification network were extracted
and primarily compared to the ground truth using the
IoU score. In addition, we used Dice similarity coef-
ficient, and Specificity and Sensitivity metrics.
2.4 Sample Size Dependence
The sample size dependency was analyzed by reduc-
ing the number of patients in the training and valida-
tion set by 5% increments, starting from 100% until
15% sample sizes. The U-Net performance was eval-
uated by using the mean IoU score. There were differ-
ent experiments performed to test the impact of input
channels and the quality of injected in-formation of
our extracted priors: 3 Channel CMR (3Ch), 3 Chan-
nel CMR + 1 Channel CAM (3Ch + CAM), 3 Chan-
nel CMR + 1 Channel post-processed CAM (3Ch
+ CAM*), 4 Channel CMR (4Ch), 1 Channel CMR
(1Ch), 1 Channel CMR + 1 Channel CAM (1Ch +
CAM), 1 Channel CMR + 1 Channel post-processed
CAM (1Ch + CAM*), 2 Channel CMR (2Ch).
1Ch/2Ch/3Ch/4Ch are baseline models, each rep-
resenting a single/double/triple/quadruple grayscale
image(s). The 3Ch + CAM and the 1Ch + CAM
represent our proposed methodology. The post-
processed CAMs refer to CAMs where a mode filter
was applied to smooth the edges of the areas, which
is the case for 1Ch + CAM* and 3Ch + CAM*. The
mode pixel filter selects the most common pixel value
from a box with a specified size — in this case, 12
pixels. Pixel values that occur only once or twice are
disregarded. The original pixel value is maintained if
there are no pixel values that appear more than twice.
2.5 Experimental Setting
This approach (including data preprocessing, model
training, and model evaluation) is developed in
Python (version 3.8.12). For deep learning, Ten-
sorFlow (version 2.5) was used. All models were
trained on the Vienna Scientific Cluster (VSC) with a
NVIDIA GTX1080 GPU (8 GB GDDR5) and locally
with a NVIDIA GTX 1660 TI (6 GB GDDR5). The
medical image segmentation library MISeval (version
1.2) was used for generating CAM tf-keras-vis (ver-
sion 0.8.1).
3 RESULTS
3.1 The focus of CAM on Important
Regions
We trained classification models that look at the
whole CMR image as input and predict one of the
multiple diagnostic groups. The best performing
classification model chosen for the generation of the
CAM was a VGG16 architecture that achieved a
weighted F1 score of 0.23 and a mean ROC AUC of
0.57. The average IoU score for the CAM of the test
set was 0.18, i.e., 18% overlap on all cine MR image
slices. When using the post-processed CAM, the IoU
was 23%. While the classification performance was
low, the IoU in some slices was up to 80%. By visu-
ally examining the CAMs of this model, we noticed
that it was attending closer to the heart region. The
produced segmentation maps from the model were of
satisfactory quality, focusing on essential structures
(Figure 3). Using different techniques described in
the methods section, as well as pre-trained classifica-
tion models (on ImageNet) yielded no performance
gain.
3.2 Extracted Priors from Classification
Model Influence Segmentation
Performance
Various scenarios of input channels were tested to an-
alyze the influence of injected information on the seg-
mentation performance and what type of content is
injected. The best segmentation performance over all
different channel combinations Table 1 was obtained
Towards Reducing Segmentation Labeling Costs for CMR Imaging using Explainable AI
13
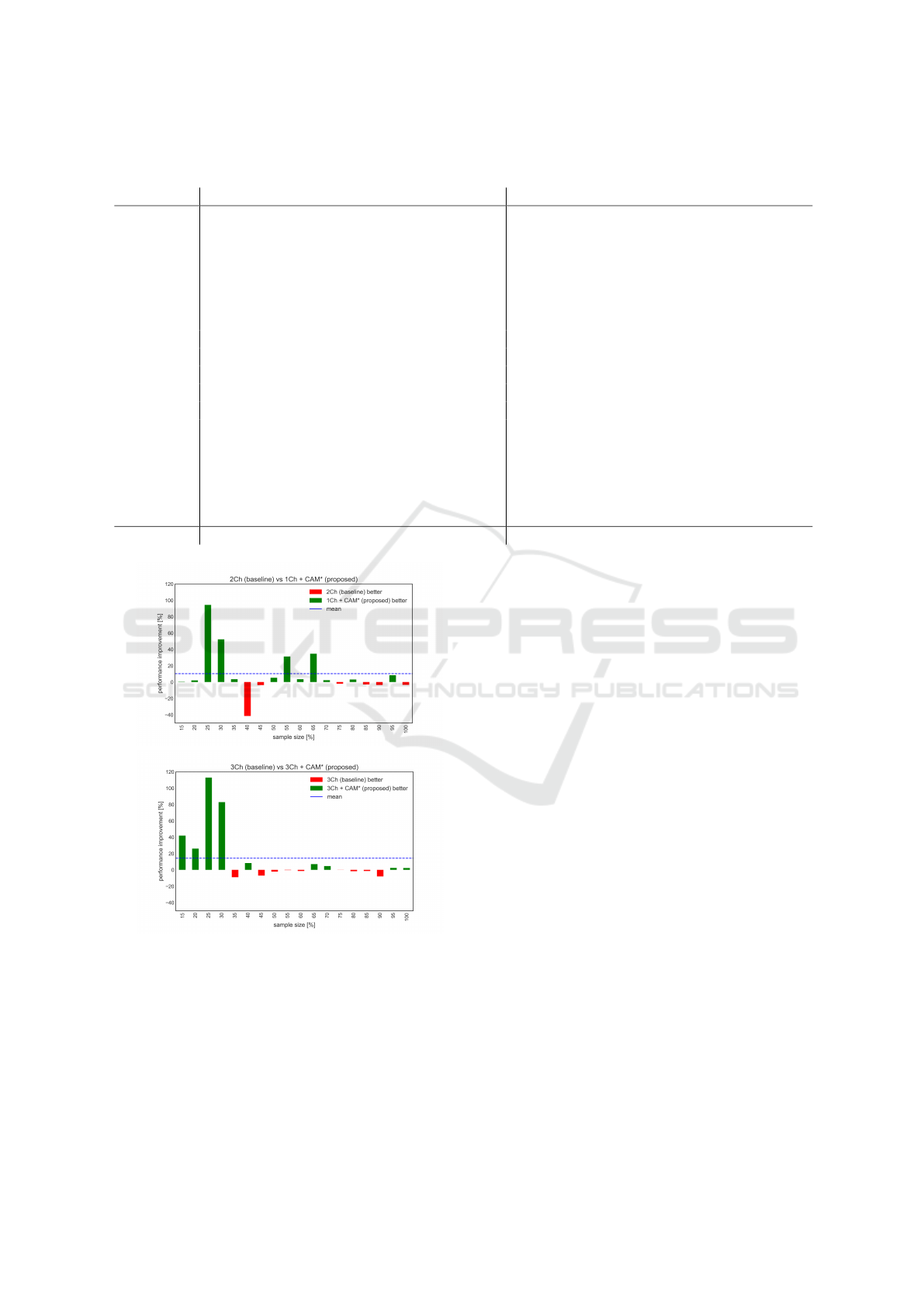
Table 1: Mean IoU segmentation results of all different experiments with varying sample sizes. The values in green represent
the best performance for each sample size increment.
percentage 1Ch 1Ch + CAM 1Ch + CAM* 2Ch 3Ch 3Ch + CAM 3Ch + CAM* 4Ch
15 0.250 0.244 0.242 0.241 0.252 0.241 0.342 0.245
20 0.242 0.241 0.246 0.241 0.242 0.246 0.305 0.320
25 0.241 0.560 0.601 0.309 0.299 0.385 0.637 0.316
30 0.343 0.575 0.387 0.254 0.330 0.649 0.604 0.298
35 0.616 0.442 0.635 0.613 0.648 0.695 0.590 0.661
40 0.483 0.553 0.398 0.679 0.604 0.626 0.655 0.665
45 0.434 0.660 0.624 0.648 0.689 0.418 0.641 0.369
50 0.475 0.666 0.689 0.654 0.626 0.697 0.613 0.397
55 0.675 0.681 0.663 0.505 0.678 0.678 0.676 0.468
60 0.617 0.712 0.662 0.639 0.694 0.682 0.685 0.713
65 0.364 0.676 0.326 0.242 0.639 0.663 0.684 0.687
70 0.692 0.700 0.741 0.724 0.703 0.727 0.736 0.661
75 0.663 0.655 0.701 0.715 0.720 0.704 0.719 0.715
80 0.684 0.693 0.708 0.686 0.709 0.701 0.698 0.707
85 0.572 0.718 0.708 0.730 0.723 0.735 0.713 0.679
90 0.684 0.711 0.727 0.754 0.737 0.730 0.678 0.730
95 0.730 0.719 0.747 0.689 0.730 0.676 0.748 0.727
100 0.721 0.723 0.707 0.732 0.724 0.707 0.741 0.681
Average 0.527 0.607 0.584 0.559 0.597 0.609 0.637 0.558
(a)
(b)
Figure 2: Relative performance difference of proposed
against the baseline. The x-axes are the percentages, and
the y-axis is the relative difference between the models. (a)
In the first panel, the 2Ch is the baseline, and the 1Ch +
CAM* is the proposed model. (b) The baseline model 3Ch
is compared against the proposed model 3Ch + CAM*.
with the model using 3Ch + CAM*, with a mean IoU
score of 0.637. All Models using the CAM as an addi-
tional input channel performs better than the baseline.
The highest difference of 0.49 was between the 3Ch +
CAM* and the 4Ch. The lowest performance increase
was with 3Ch + CAM over the 3Ch with an increase
of 0.012 IoU score. Overall, the models with fewer
channels (one or two) show a decreased performance
compared to those using three or four channels.
Analyzing the relative performance differences
(Figure 2), it is apparent that the proposed models
significantly outperform the baseline models in the
small sample sizes (15-35%). The performance gain
is up to 150% improvement in the 1Ch + CAM* case
compared to 1Ch at 25% sample size. In the case of
the 3Ch + CAM* against the 3Ch the improvement at
25% sample size is over 110%.
4 DISCUSSION
In a small data setting (20-35% of training data),
our framework significantly improved the segmenta-
tion accuracy of a baseline U-net model (5%-150%).
These results open a promising research direction that
shows that even a far from perfect pre-trained classi-
fication model could be used to produce sensible seg-
mentation masks. Our generic methodology might
well support the creation of automatic segmentation
tools in cardiac MRI that drastically reduce the de-
pendence and thereby the cost of time-consuming de-
lineation labels. The advantage of our approach is
its simplicity and robustness to hyperparameter opti-
mization. However, such a decoupled design may pre-
vent a segmentation model of learning features from
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
14
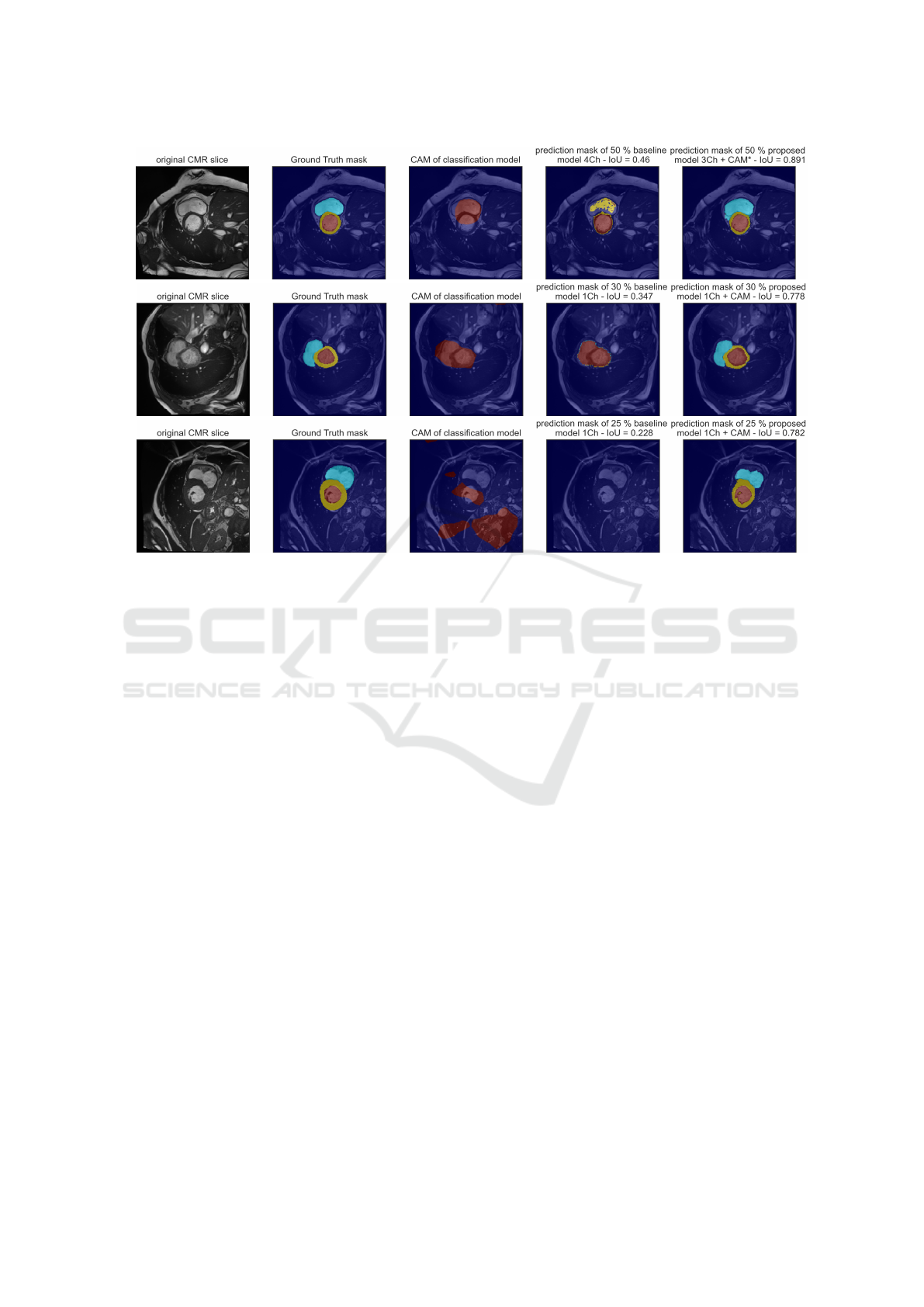
(a)
(b)
(c)
Figure 3: Example outputs of predictions masks from a baseline model and a proposed model with the CMR slice in the
background for different scenarios. Panel (a), prediction from models with 50% sample size, baseline model is 4ch, prediction
model is 3Ch + CAM*. Panel (b), prediction from models with 30% sample size, baseline model is 1ch, prediction model is
1Ch + CAM. The CAM represents the heart region and is comparable to the baseline prediction. The baseline predicts only
two classes, the IoU score is low, the prediction model outperforms the baseline model. Panel (c), prediction from models
with 25% sample size, baseline model is 1ch, prediction model is 1Ch + CAM. The CAM is dispersed without focusing on
the heart region. The proposed model outperforms the baseline model, it only predicts the background class and thereby only
achieves an IoU of 0.228. However, with the additional injected information of the CAM, the proposed model achieves an
IoU of 0.782.
auxiliary classification tasks. A thorough comparison
of our approach to end-to-end multi-task methods is
part of our future work.
The segmentation performance increases in most
cases if the model complexity is increased. Therefore,
using additional channels and increasing the number
of variables in the model improves the performance.
However, it matters which performance is used in the
additional channels. The baseline models - with the
same amount of channels as the models where CAMs
are used as an additional input are 2Ch and 4Ch – have
a decreased performance as our proposed models. In-
jecting CAMs improve the segmentation performance
since they constrain the search space with the spacial
information given from the priors.
As this is a work in progress we are fully aware
of the current limitations of our approach. Using
the proposed methodology on a dataset with higher
classification performance could improve segmenta-
tion performance. Due to the computational complex-
ity, we have not properly quantified the uncertainty
of all models, and we only performed a shallow hy-
perparameter optimization. Finally, the CMRs were
used as single 2D images, which do not represent the
3D structure of the heart, and spatial information is
lost. Additionally, the heart regions in the end and be-
ginning slices of each cardiac phase are almost non-
existing, and thereby a segmentation model trained
only with the 2D input will perform poorly at these
slices. Future work should address these limitations
and analyze the per-class performance gain.
FUNDING
This research was funded by
¨
OKG
Forschungsstipendium ”Reducing costs of seg-
mentation labeling in cardiac MRI using explainable
AI”.
Towards Reducing Segmentation Labeling Costs for CMR Imaging using Explainable AI
15

ACKNOWLEDGEMENTS
The computational results were partly achieved by us-
ing the Vienna Scientific Cluster (VSC)
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
Bai, W., Chen, C., Tarroni, G., Duan, J., Guitton, F., Pe-
tersen, S. E., Guo, Y., Matthews, P. M., and Rueckert,
D. (2019). Self-Supervised Learning for Cardiac MR
Image Segmentation by Anatomical Position Predic-
tion. In Shen, D., Liu, T., Peters, T. M., Staib, L. H.,
Essert, C., Zhou, S., Yap, P.-T., and Khan, A., editors,
Medical Image Computing and Computer Assisted
Intervention – MICCAI 2019, volume 11765, pages
541–549. Springer International Publishing, Cham.
Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang,
X., Heng, P.-A., Cetin, I., Lekadir, K., Camara, O.,
Gonzalez Ballester, M. A., Sanroma, G., Napel, S.,
Petersen, S., Tziritas, G., Grinias, E., Khened, M.,
Kollerathu, V. A., Krishnamurthi, G., Rohe, M.-M.,
Pennec, X., Sermesant, M., Isensee, F., Jager, P.,
Maier-Hein, K. H., Full, P. M., Wolf, I., Engelhardt,
S., Baumgartner, C. F., Koch, L. M., Wolterink, J. M.,
Isgum, I., Jang, Y., Hong, Y., Patravali, J., Jain, S.,
Humbert, O., and Jodoin, P.-M. (2018). Deep Learn-
ing Techniques for Automatic MRI Cardiac Multi-
Structures Segmentation and Diagnosis: Is the Prob-
lem Solved? IEEE Transactions on Medical Imaging,
37(11):2514–2525.
Chen, C., Qin, C., Qiu, H., Tarroni, G., Duan, J., Bai, W.,
and Rueckert, D. (2020). Deep Learning for Cardiac
Image Segmentation: A Review. Frontiers in Cardio-
vascular Medicine, 7:25.
Ciga, O. and Martel, A. L. (2021). Learning to segment im-
ages with classification labels. Medical Image Analy-
sis, 68:101912.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Identity
mappings in deep residual networks. In Leibe, B.,
Matas, J., Sebe, N., and Welling, M., editors, Com-
puter Vision – ECCV 2016, pages 630–645, Cham.
Springer International Publishing.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In 2017 IEEE Conference on Computer Vision
and Pattern Recognition (CVPR), pages 2261–2269.
Madani, A., Ong, J. R., Tibrewal, A., and Mofrad, M. R. K.
(2018). Deep echocardiography: Data-efficient su-
pervised and semi-supervised deep learning towards
automated diagnosis of cardiac disease. npj Digital
Medicine, 1(1):1–11.
Oktay, O., Ferrante, E., Kamnitsas, K., Heinrich, M., Bai,
W., Caballero, J., Cook, S. A., de Marvao, A., Dawes,
T., O’Regan, D. P., Kainz, B., Glocker, B., and Rueck-
ert, D. (2018). Anatomically Constrained Neural Net-
works (ACNNs): Application to Cardiac Image En-
hancement and Segmentation. IEEE Transactions on
Medical Imaging, 37(2):384–395.
Peng, P., Lekadir, K., Gooya, A., Shao, L., Petersen, S. E.,
and Frangi, A. F. (2016). A review of heart cham-
ber segmentation for structural and functional analy-
sis using cardiac magnetic resonance imaging. Mag-
netic Resonance Materials in Physics, Biology and
Medicine, 29(2):155–195.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Navab, N., Hornegger, J., Wells,
W. M., and Frangi, A. F., editors, Medical Image Com-
puting and Computer-Assisted Intervention – MICCAI
2015, pages 234–241, Cham. Springer International
Publishing.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2020). Grad-CAM: Vi-
sual Explanations from Deep Networks via Gradient-
Based Localization. International Journal of Com-
puter Vision, 128(2):336–359.
Simonyan, K. and Zisserman, A. (2015). Very deep con-
volutional networks for large-scale image recognition.
In International Conference on Learning Representa-
tions.
Tran, P. V. (2016). A fully convolutional neural network
for cardiac segmentation in short-axis mri. ArXiv,
abs/1604.00494.
Zimmer, V. A., Gomez, A., Skelton, E., Ghavami, N.,
Wright, R., Li, L., Matthew, J., Hajnal, J. V., and
Schnabel, J. A. (2020). A Multi-task Approach Using
Positional Information for Ultrasound Placenta Seg-
mentation. In Hu, Y., Licandro, R., Noble, J. A., Hut-
ter, J., Aylward, S., Melbourne, A., Abaci Turk, E.,
and Torrents Barrena, J., editors, Medical Ultrasound,
and Preterm, Perinatal and Paediatric Image Analy-
sis, volume 12437, pages 264–273. Springer Interna-
tional Publishing, Cham.
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
16
