
Par-VSOM: Parallel and Stochastic Self-organizing Map Training
Algorithm
Omar X. Rivera-Morales and Lutz Hamel
Department of Computer Science, University of Rhode Island, College Road, South Kingstown, Rhode Island, U.S.A.
Keywords:
SOM, VSOM, GPU, Parallel Computing, Self-organizing Map, Stochastic Training, Vector Optimization.
Abstract:
This work proposes Par-VSOM, a novel parallel version of VSOM, a very efficient implementation of stochas-
tic training for self-organizing maps inspired by ideas from tensor algebra. The new algorithm is implemented
using parallel kernels on GPU accelerators. It provides performance increases over the original VSOM algo-
rithm, PyTorch Quicksom parallel version, Tensorflow Xpysom parallel variant, as well as Kohonen’s classic
iterative implementation. Here we develop the algorithm in some detail and then demonstrate its performance
on several real-world datasets. We also demonstrate that our new algorithm does not sacrifice map quality for
speed using the convergence index quality assessment.
1 INTRODUCTION
The self-organizing map (SOM) is a neural network
designed for unsupervised machine learning (Koho-
nen, 2001). The generated maps are powerful data
analysis tools applied to diverse areas such as atmo-
spheric science, nuclear physics, pattern recognition,
medical diagnosis, computer vision and other data do-
mains (Barney, 2018; Li et al., 2018a; Ramos et al.,
2017). See reference (Kohonen, 2001) for a more
comprehensive literature survey. Here we introduce
the Parallel VSOM (Par-VSOM), a parallel imple-
mentation of the efficient VSOM algorithm (Hamel,
2019). The novel approach presented here, replaces
all iterative constructs of the SOM algorithm with
kernels running in a hardware accelerator to perform
vector and matrix operations in parallel. The al-
gorithm kernels provide substantial performance in-
creases over Kohonen’s SOM iterative algorithm, the
XpySom(Mancini et al., 2020), and Quicksom (Mallet
et al., 2021b; Mallet et al., 2021a) parallel BatchSOM
implementations.
The training of the SOM is computationally de-
manding, but a great advantage of SOMs is that the
computations can be parallelize with algorithm modi-
fications like in the BatchSOM or using hardware vec-
torization. Currently, various types of hardware ac-
celerators are easily available, allowing us to process
Big-Data (Mor
´
an et al., 2020) datasets using high-
performance computers (HPC), Graphical Processing
Units (GPU), and Field Programmable Gate Arrays
(FPGA)(Richardson and Winer, 2015; Abadi et al.,
2016).This research provides an alternative efficient
SOM algorithm to accelerate the training of highly
complex rectangular maps.
Our experiments demonstrate that our parallel al-
gorithm is better suited for highly computational de-
manding maps, such as the maps generated with large
SOMs. Using a large number of neurons provides
a higher resolution clustering of the data and facil-
itates the pattern recognition during the analysis, as
shown in Figure 1. Furthermore, the maps produced
by the Par-VSOM are equivalent in quality to the
maps produced by the original SOM iterative algo-
rithm. The current Par-VSOM model is parallel and
multi-threaded, and therefore well suited as a replace-
ment for other parallel algorithms to train the self-
organizing maps.
The paper is organized as follows: In Section
2, we start our discussion with an overview of the
SOM and a brief introduction to the VSOM (Hamel,
2019) vectorized rules, which can be viewed as an im-
plementation of a competitive learning scheme com-
prised of a competitive step and an update step with
vector and matrix training. The relevant details about
related research work are included in Section 3. As
part of Section 4, we develop the Par-VSOM vector-
based parallel training and examine the data level par-
allelisms achievable using vectorized single instruc-
tion with multiple data (SIMD) registers and discuss
the limitations. Under Section 5, we included the
study of the performance of our parallel vectorized
Rivera-Morales, O. and Hamel, L.
Par-VSOM: Parallel and Stochastic Self-organizing Map Training Algorithm.
DOI: 10.5220/0011377700003332
In Proceedings of the 14th International Joint Conference on Computational Intelligence (IJCCI 2022), pages 339-348
ISBN: 978-989-758-611-8; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
339

training implementation by comparing it to various
CPU and GPU SOMs variants. Finally, in Section 6,
we conclude our discussion with a summary of the ob-
servations and some future research ideas under con-
sideration.
2 THE SOM AND VSOM
ALGORITHMS
The origins of the self-organizing maps model can be
traced back to the Vector Quantization (VQ) method
(Kohonen, 2001). The VQ is a signal-approximation
algorithm that approximates a finite “codebook” of
vectors m
i
∈ R
n
, i = 1, 2, ..., k to the distribution of the
input data vector x ∈ R
n
. In the SOM context, the
approximated codebook allows us to categorize the
nodes and forms an “elastic network,” which becomes
a meaningful, coordinated map or grid system.
From a computational perspective, the SOM can
be described as a mapping of high dimensional in-
put data onto a low dimensional neural network pro-
jected as a 2D or three-dimensional (3D) constrained
topological map (Hastie et al., 2001). The map-
ping is accomplished by assuming that the input data
set is a real vector such as x
k
= [ξ
1
, ξ
2
, ..., ξ
n
]
T
∈
R
n
. The SOM neuronal map can be defined as a
model containing the parametric real vector m
i
=
[u
i1
, u
i2
, ..., u
in
]
T
∈ R
n
associated with the neurons’
weights. If we consider the distance between the in-
put vector x
k
and the neuron vector m
i
then we can es-
tablish an initial minimum distance relation between
the input and the neurons by calculating the Euclidean
distances. Then, these distances are used to identify
the best matching unit (BMU) index with equation
(1).
c = argmin
i
(||m
i
− x
k
||
2
) (1)
To define the SOM in terms of matrix and vector
operations it is assumed that the map’s neurons are
stored in a n×d matrix M where each row i represents
the neuron m
i
with d components,
M[i, ] = m
i
= (m
1
, . . . , m
d
)
i
, (2)
with i = 1, . . . , n. The training data x consists of a
set D= {x
1
, . . . , x
l
}. The set can be defined as a l × d
matrix where each row k represents the training vector
x
k
with d components,
D[k, ] = x
k
= (x
1
, . . . , x
d
)
k
, (3)
with k = 1, . . . , l.
Essential details to consider include (1) the dimen-
sionality d for the input, and (2) the neuron vectors
Figure 1: IRIS 15x10 small SOM and IRIS 100x100 large
SOM, neuronal heatmaps patterns with different resolu-
tions.
are required to be the same size for well-defined ma-
trix operations.
2.1 The SOM and VSOM Competitive
Step
In the competitive step, we find the BMU for a partic-
ular training instance x
k
. In the classic SOM we use
an iterative process to find the BMU using 1. Here
the i = 1, 2, ..., n represents the index of the neurons
in the map and m
i
represents the neuron in index i.
The argmin is a function that returns the minimum
value and c contains the index of the BMU.
In the VSOM context this step requires us to cal-
culate the Euclidean distance as a set of vector and
matrix operations. These operations find the c in-
dex associated with the neuron with the minimum dis-
tance to the training instance. The BMU c index cor-
responds to the neuron in the map with the highest
resemblance to the particular x
k
selected for training
during the epoch.
The first step to calculate the BMU requires us
to compute a matrix X to hold a randomly selected
training vector. The matrix X in equation (4) is de-
fined with a component sizes of n × d, where each
row is holding the current epoch training vector x
k
=
(x
1
, x
2
, . . . , x
d
)
k
, which is randomly selected from ma-
trix D,
X = 1
n
⊗ x
k
. (4)
Here, the symbol ⊗ represents the outer product and
1
n
is a column vector defined as,
1
n
= (1, 1, . . . , 1
| {z }
n
)
T
. (5)
Since 1
n
is a column vector and x
k
is a row vector the
operation in (4) is well defined. After populating the
instance matrix X with the duplicated x
k
values, equa-
tions (6), (7) and (8) are used to compute the square of
the Euclidean distances between all the map neurons
and the selected input vector,
∆ = M − X (6)
Π = ∆ ◦ ∆ (7)
s = Π × 1
d
(8)
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
340

In equation (6) we calculate the difference between
the matrices with an element-by-element matrix sub-
traction. In equation (7) we use the Hadamard product
to allow us to calculate the Π matrix, in this context
◦ represents the element-by-element matrix product
and X, M, ∆ and Π are all n × d matrices.
Lastly, in equation (8) we use a ‘row sum’ matrix
reduction to compute the vector s of size n. Here, 1
d
is
a column vector similar to (5) with the dimensionality
defined by the value of d.
2.2 The SOM and VSOM Update Step
In the classic stochastic SOM, after completing
the BMU calculations, the updates to the neuronal
weights are accomplished using the training instance
x
k
to influence the best matching neuron and its sur-
rounding neighborhood.
m
i
← m
i
− η(m
i
− x
k
)h(c, i) (9)
The weights update step in equation (9), affects
every neuron inside the neighborhood radius of influ-
ence. Here, the learning rate η serves as a scaling
factor between 0 and 1. The h(c, i) acts as the loss
function , where i = 0, 1, ... , n and it can be defined
as,
h(c, i) =
1 if i ∈ Γ(c),
0 otherwise,
(10)
where Γ(c) is the neighborhood of the best matching
neuron m
c
with c ∈ Γ(c). In the classic SOM, the
learning factor and the loss function both decreased
monotonically over time (Kohonen, 2001).
In the VSOM, the update step for all the neurons
in the map is accomplished with matrix operations
and is defined as,
M ← M − η∆ ◦ Γ
c
. (11)
Here, η is the learning rate, ∆ contains the calcula-
tions of the difference between the neurons and the
selected training instance as computed in (6), and
the symbol ◦ represents the Hadamard product. The
Hadamard product represent by ◦ is the element-by-
element matrix product. Similarly to the SOM, in
the VSOM, the learning rate η is linearly reduced as
epochs increase.
However, our experimental results demonstrate
that a constant learning rate η generates higher quality
convergence indexes in large map instances. Initially,
the update rule for each best matching neuron has
a very large radius of influence and includes all the
neurons on the map. After multiple training epochs,
the neighborhood radius around the BMU gradually
Figure 2: SOM preserving the neighborhood topology in
3D space (Hastie et al., 2001).
shrinks to the point that the field of influence only in-
cludes the best matching neuron m
c
as shown in (12).
Γ(c)|
t0
= {c}. (12)
The competitive and the update steps are com-
puted during each epoch using the randomly selected
training instances until some convergence criterion is
fulfilled. After reaching a maximum convergence, ev-
ery data point will be assigned to an specific data neu-
ron forming clusters in the grid and preserving the
neighborhood topology as shown in Figure 2.
Algorithm 1 and 2 summarizes the matrix and
vector operations required for the parallel Par-VSOM
training. For a more detailed explanation of the SOM
and VSOM algorithms, see reference (Hamel, 2019).
3 RELATED WORK
In this section, we look at prior work related to par-
allel SOM algorithms and its applications. Recent
parallel self-organizing maps research has demon-
strated promising improvements using various paral-
lel methods. Some of the methodology mentioned in
current scientific publications on this topic include:
combining data and network partitioning techniques
(Richardson and Winer, 2015; Silva and Marques,
2007), exploiting cache effects (Rauber and Merkl,
2000), using map-reduce programming paradigm
(Sarazin et al., 2014; Sul and Tovchigrechko, 2011;
Schabauer and Weishaupl, 2005), replacing the SOM
iterative construct with vector and matrix operations
(Hamel, 2019), and using various types of acceler-
ated architectures for parallelism (Davidson, 2015;
Moraes et al., 2012; Abadi et al., 2016; Mancini et al.,
2020; Wittek et al., 2013). In addition, recent publi-
cations demonstrate how to utilize SOM as a pattern
recognition tool (Kim et al., 2020; Li et al., 2018b;
Lokesh et al., 2019). In general, recent research pub-
lications share similars goals such as: finding new
applications, improving optimal performance and in-
creasing speed-up using different SOM approaches.
Par-VSOM: Parallel and Stochastic Self-organizing Map Training Algorithm
341

3.1 SOM Parallel Hybrid Methods
The combination of data and network partitioned par-
allel methods develop by Richarson et al. (Richard-
son and Winer, 2015) splits up the map to compute
the best matching calculation and nodes update on
separate threads. This hybrid methodology also di-
vides the data amongst individual threads for data par-
tition parallelism. As part of their research findings,
they concluded that parallelizing the classic SOM al-
gorithm using such techniques in a GPU can save
computation time and increase the speed-up by nearly
15X in maps with 10,000 points and 5 dimensions. A
similar method was proposed by (Silva and Marques,
2007), achieving a performance increases of 1.27X
training large maps on a small HPC cluster.
3.2 SOM Vectorization
The VSOM by (Hamel, 2019) replaced all the iter-
ative constructs of the standard stochastic SOM al-
gorithm with vector and matrices operations. The
VSOM implementation resulted in a performance in-
crease of up to 60X faster after running 10000 itera-
tions in a 25 X 20 map. Since the VSOM seems to be
offering the highest speed-up increase of all the cur-
rent SOM research publications, our research is focus
on the parallelization of the VSOM algorithm and its
implementation in hardware accelerators.
3.3 SOM in Multiple Parallel
Architectures
Among the SOM parallel approaches previously dis-
cussed, not many offer an available open source
repository to validate the research findings or con-
tinue with further investigations. In this paper, we
decided to compare our proposed parallel implemen-
tation with some of the widely available parallel SOM
projects packages. As part of the GPU comparisons
we utilize, Quicksom (Mallet et al., 2021a) which
offers a parallel GPU Batch-SOM algorithm imple-
mented using the Python PyTorch framework and
speed-ups results showed at least a 20 speed-up over
the CPU version using bioinformatics datasets (Mal-
let et al., 2021b). In addition, we also included a com-
parison with XpySom (Mancini et al., 2020) a paral-
lel Batch-SOM variant implemented using the Google
Tensorflow 2.0 framework and Python Numpy li-
brary. The XpySom package is based on the Mini-
som(Vettigli, 2021), a non-parallel, minimalistic and
Numpy based widely know implementation of the
SOM. The XpySom research paper (Mancini et al.,
2020) indicates their parallel variants outperforms the
popular SOM GPU package Somoclu by two and
three orders of magnitude.
4 Par-VSOM: PARALLEL
VECTORIZED SOM
4.1 Hardware for Parallel Vectorization
Our novel parallel implementation is based on the
VSOM algorithm proposed by (Hamel, 2019). On the
VSOM, the stochastic SOM training is redefined to
execute as a set of vector and matrix operations. Since
all the matrix data elements are independent of each
other, they can be executed as coarse-grained “embar-
rasingly parallel” tasks to exploit multiple hardware
threads (or cores) available in the devices (Jaaske-
lainen, 2019). In the VSOM context, the vectoriza-
tion of the calculations can be implemented as vector
instructions, which are also known as SIMD instruc-
tions and are a form of Data-Level Parallelism. These
vector instructions apply the same operation over
multiple data elements (like integers and floating-
point values) concurrently, given that these items are
stored contiguously in vector/SIMD registers (Pilla,
2018). In modern Intel and AMD CPU architectures,
these vector instructions are known as Advance Vec-
tor Extensions (AVX), AVX2, AVX-512 instructions
set and Streaming SIMD Extensions (SSE4).
In contrast, the GPUs with their substantial
amount of nodes allows for the creation of thousands
of threads to perform vector calculations simultane-
ously. Furthermore, the current NVIDIA GPUs can
access there memory much faster when accessing ad-
jacent data concurrently. This is optimized when
groups of 32 GPU threads or warps do the request
simultaneously, causing “memory coalescing” (Dick-
son et al., 2011).
4.2 Par-VSOM Algorithm
In the classic SOM with iterative operations, the op-
erations per column are solved in a loop structure
sequentially. This serial approach results in high
overhead and additional latency during every training
epoch. Conversely, the VSOM vector and matrix op-
erations are vectorized by the compiler, and they are
executed in the CPU as vector operations. To illus-
trate, in a data set with 32 instances, the VSOM using
vectorized operations will need to execute a total of
four “minus” operations to compute a ∆ matrix en-
tirely. Using the VSOM vectorization, the ∆ matrix
“minus” operation can be completed with a speed-up
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
342

Figure 3: The time comparison of ∆ calculation during
the competitive step for SOM, VSOM, and PAR-VSOM
demonstrate modern architecture advances in vectorization
capability increases the primitive operations’ overall speed-
up performance.
increase of 4X compared to the SOM, as illustrated in
Figure 3.
In the Par-VSOM, the vector and matrix opera-
tions of the original VSOM are replaced with parallel
computational kernels executing in a hardware accel-
erator architecture. The parallel kernels manipulate
the matrices columns in a unified vector V
u
as shown
in equation (13). In the kernel, the matrices are ex-
pressed as tuples of column vectors and encapsulated
into one unifying vector. Based on the data of our ex-
ample in Figure 3, the unifying vector technique will
result in executing the 32 elements operation in one
single vectorized operation, providing a performance
increase of 128x.
V
u
[i ∗ n] = (t
1
, . . . ,t
n
)
1
∪ (t
1
, . . . ,t
n
)
2
. . . ∪ (t
1
, . . . ,t
n
)
i
(13)
In the unifying vector equation (13), we have
shown how the matrices can be express in terms of
union of tuples. Here, ∪ represents the union oper-
ator. In the Par-SOM algorithm (1) and (2), we are
assuming all the matrices of the VSOM are imple-
mented as a data structure consisting of multiple tu-
ples (t
1
,t
2
, ...,t
n
) where each column is represented
by the tuple (t
n
)
i
with i representing the dimensional-
ity of the matrix and n the number of instances. This
technique allows the data-level parallelism to occur
by executing all the matrix operations as optimized
vector operations inside the Φ kernels as presented in
algorithm 1 and 2.
In our GPU implementation, we decided to use
CUDA Thrust. Considering that the Par-VSOM is a
parallel and vectorized implementation of the VSOM
algorithm, the Thrust template is an ideal candidate
Algorithm 1: The Par-VSOM training algorithm.
1: Given:
2: D ← {training instances, a l x d matrix}
3: M ← {neurons, a n x d vector of tuples}
4: η ← {learning rate 0 < η < 1}
5: Γ(c) ← {neighborhood function for some neuron c}
6: minIndex(s) ← {returns location of min. val in s}
7: Φ ← {Vectorized kernel operation, with all matrices
8: columns unified as tuples in a single column vector.}
9: Repeat
10: /***Select a matrix training instance as vector
11: for some k = 1, ..., l and f = 1, ..., d : ***/
12:
13: x
k
← D[k][1] ∪ D[k][2]... ∪ D[k][ f ]
14:
15: /***Find the winning neuron using kernels ***/
16: X ← Φ
x
(1
n
⊗ x
k
)
17: ∆ ← Φ
∆
(M − X)
18: Π ← Φ
Π
(∆ ◦ ∆)
19: /***Sum of vector subsections (rowsum) ***/
20: s ← Φ
s
(Π
1...(n∗1)
+ Π
(n∗1)...(n∗2)
+
21: ... Π
(n∗(d−1)...(n∗d)
)
22: c = minIndex(s)
23:
24: /***Update neighborhood with vector operations ***/
25: Γ
c
← Φ
Γ
(Γ(c))
26: M
new
← Φ
M
new
(M
current
− η∆ ◦ Γ
c
)
27: Until done
28: return M
new
Algorithm 2: The Par-VSOM Neighborhood Function Γ(c)
as mentioned in equation (10) and (11).
1: Given:
2: c ← {index of winning neuron}
3: n ← {the number of neurons on the map}
4: nsize ← {neighborhood radius}
5: P ← {an n × 2 vector with p
i
= P[i, ] = (x
i
, y
i
)}
6: 1
n
← {constant column vector with value 1}
7: 0
n
← {constant column vector with value 0}
8: Φ ← Vectorized kernel operation, with all matrices
9: columns unified as tuples in a single column vector.
10: x ← { x values in first section: 1,...,(
n
2
− 1)}
11: y ← { y values in second section:
n
2
, ..., (n × 2)}
12:
13: P
c
← Φ
pc
(P[c, ])
14: C ← Φ
C
(1
n
⊗ p
c
)
15: ∆ ← Φ
∆
(P − C)
16: Π ← Φ
Π
(∆ ◦ ∆)
17: /***Perform rowsum with vector subsections
18: d ← Φ
d
(Π
x
+Π
y
)
19: hood ← Φ
hood
(ifelse(d < (nsize × 1.5)
2
, 1
n
, 0
n
))
20: return hood
due to the vast number of vector functions available.
In addition, Thrust manages all the CUDA kernel
initialization, memory transfers and allocation in the
background, and provides highly optimized libraries
for vector operations (Nvidia.com, 2020).
Par-VSOM: Parallel and Stochastic Self-organizing Map Training Algorithm
343

Since most of the VSOM algorithm consists of
matrix operations, we utilized Thrust specialized
transformation and reduction functions to process the
matrices as vectors. In the case of a matrix with three
columns, storing 3d points as an array of float3 in
CUDA is generally a bad idea, since array accesses
are not properly coalesced (Nvidia.com, 2020). To
address this memory access issue, the number of rows
n was used as a delimiter to identify the beginning and
the end of each column in the unifying vector Vu. The
column-wise encapsulation of the matrix transforms
the three-dimensional columns in to one Vu vector.
This allows coalesced memory access and faster op-
eration execution.
One of the important differences between the orig-
inal VSOM algorithm and the Par-VSOM algorithm ,
is the data structure manipulation during the selection
of the D matrix random training instance algorithm 1
(line 13). Here, the training computation transforms
the selection into an X
k
vector that includes all the
matrix D columns and allows us to find the BMU us-
ing vector operations. To be able to use the optimized
“minIndex(s)” function in line 22, we reduced the Π
vector with length n∗d into a vector of length n, using
operations equivalent to a rowsum across d dimen-
sions in line 20.
Similarly, the Par-VSOM neighborhood Function
Γ in algorithm 2, emulates the rowsum operations of
algorithm 1 in line 18 by utilizing the vector elements
representing the x and y columns accordingly and re-
turns one vector that includes the distances of the neu-
rons in the grid. In lines 19 to 20 using the computed
distances, the vector neighborhood determination is
performed and return a hood vector that activates the
neurons considered to be part of the neighborhood by
flipping to “1” their corresponding neurons index.
4.3 Limitations
4.3.1 Large Computational Workloads
The Par-VSOM is recommended for clustering prob-
lems requiring high computational workloads. To ob-
tain our experimental results, we tested with multiple
datasets and various map sizes. The results demon-
strated the Par-VSOM is not suitable for small maps,
low-dimensional datasets, or minimal computational
workloads. Here, we assume the users will have a
GPU hardware accelerator available as part of their
setup.
In general, Big data and other extensive datasets
analysis requires generating large neuronal maps as
part of the pattern analysis and clusters visualizations.
The GPUs have become one of the default tools to
process high complexity problems and are easily ac-
cessible in cloud environments, but we are aware that
not everyone may have access to one.
5 EXPERIMENTS
5.1 Hardware Setup
All the Par-VSOM, Xpysom and Quicksom parallel
experiments were performed using the Amazon AWS
cloud service instances with Linux and Deep Learn-
ing Amazon Machine Images (AMI). The sequential
CPU experimental setting included an Intel I7-7700K
running at 4.20 GHz/ 4.50GHz turbo with four cores
and capable of executing eight threads. The GPU tests
were performed in an AWS P3.2xlarge with 18 virtual
Intel Xeon E5 2686 CPU operating at 2.7 GHz/ 3.0
GHz turbo and an NVIDIA Tesla V100. The Tesla
V100 contains 5120 NVIDIA Cuda cores with 16 Gb
of HBM2 memory. The Tesla V100 memory clock
setting was 877 Mhz with memory graphics clocked
at 1530 Mhz.
5.2 Par-VSOM Setup and
Hyper-parameters
The experimental setup utilized the default values
of the SOM and VSOM Popsom (Hamel et al.,
2016). For the Quicksom(Mallet et al., 2021b)
and Xpysom(Mancini et al., 2020) BatchSOM pack-
ages, we maintained the learning rate constant to ob-
tain higher convergence indexes and tune the hyper-
parameters as defined in Table 1.
Table 1: Par-VSOM Hyper-Parameters.
**Hyper-Parameters** **Values**
Training Iterations 1 × 10
0
... 1 × 10
5
Learning Rate η 0.7
Neighborhood Radius Bubble, Gaussian(for Quicksom)
Map sizes 15x10, 150x100, 200x150
Datasets Iris, Epil, WDBC
As part of our tests, we compared the performance
and the quality of the maps generated by our parallel
Par-VSOM with two CPU SOM and two GPU SOM
variants. The quality of the maps is based on the con-
vergence index as define in (Tatoian, 2018). The CPU
single-node tests used the SOM and the VSOM al-
gorithms included as part of the R language Popsom
package with C bindings applications. In contrast, the
parallel comparisons were done using the two GPU-
based SOM packages; Quicksom with Python 3, Py-
torch 1.4 and Xpysom using Tensorflow 2.0 in their
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
344

Table 2: Times and Speed-up gains of the Par-VSOM for different training algorithms using a 200 × 150 map.
iter Time Time Time Time Time Speed-up Speed-up Speed-up Speed-up
SOM(s) VSOM(s) P-VSOM(s) Xpysom(s) Quicksom(s) Par-VSOM/ Par-VSOM/ Par-VSOM/ Par-VSOM/
CPU CPU GPU CPU-GPU CPU-GPU SOM VSOM Xpysom Quicksom
R\C R\Fortran Cuda Thrust TensorFlow PyTorch
*** Iris D=4***
1 1.148 0.035 0.027 0.301 0.257 42.5 1.3 11.1 9.5
10 1.350 0.046 0.029 0.319 0.257 46.6 1.6 11.0 8.9
100 2.362 0.067 0.049 0.414 0.434 48.2 1.4 8.4 8.9
1000 13.447 0.324 0.235 1.408 2.32 57.2 1.4 6.0 9.9
10000 124.011 2.756 1.925 10.742 21.456 64.4 1.4 5.6 11.1
100000 1228.811 26.210 18.275 110.900 212.791 67.2 1.4 6.1 11.6
*** Epil D=8***
1 1.831 0.053 0.046 0.300 0.262 39.8 1.2 6.5 5.7
10 1.949 0.058 0.049 0.313 0.259 39.8 1.2 6.9 5.3
100 3.125 0.108 0.072 0.412 0.643 43.4 1.5 5.7 8.9
1000 14.854 0.554 0.294 1.411 4.667 50.5 1.9 4.8 15.8
10000 132.193 4.928 2.577 10.660 46.755 51.3 1.9 4.1 18.1
100000 1306.793 47.560 22.535 115.372 462.908 58.0 2.1 5.1 20.5
*** WDBC D=30***
1 0.966 0.152 0.125 0.303 0.262 7.7 1.2 2.4 2.0
10 1.167 0.165 0.130 0.319 0.256 9.0 1.3 2.5 2.0
100 3.161 0.342 0.174 0.416 0.762 18.2 2.0 2.5 4.4
1000 23.236 2.076 0.601 1.387 6.386 38.7 3.5 2.3 10.6
10000 222.034 19.105 4.712 11.389 63.871 47.1 4.1 2.4 13.6
100000 2224.134 188.080 46.114 111.223 634.687 48.2 4.1 2.4 13.8
Table 3: Quality of maps produced by the different training algorithms (SOM=Classic SOM, VSM=VSOM, P-V=Par-VSOM,
X-P=Xpysom, Q-S=Quicksom and D=Dimensions).
iter 15x10 150x100 200x150
10
x
SOM VSM P-V X-P Q-S SOM VSM P-V X-P Q-S SOM VSM P-V X-P Q-S
*** Iris, D=4***
1 |0.50 0.15 0.09 0.50 0.45| |0.41 0.00 0.00 0.50 0.12| |0.40 0.00 0.00 0.50 0.08|
2 |0.43 0.53 0.48 0.37 0.49| |0.02 0.45 0.49 0.50 0.50| |0.34 0.45 0.49 0.47 0.50|
3 |0.92 0.95 0.93 0.88 0.48| |0.42 0.79 0.49 0.40 0.50| |0.12 0.85 0.77 0.32 0.50|
4 |0.93 0.91 0.91 0.92 0.37| |0.92 0.91 0.96 0.28 0.48| |0.92 0.93 0.91 0.29 0.48|
5 |0.95 0.94 0.94 0.87 0.27| |0.96 0.99 0.95 0.26 0.41| |0.90 0.99 0.97 0.32 0.37|
*** Epil, D=8***
1 |0.03 0.14 0.15 0.72 0.40| |0.12 0.00 0.00 0.46 0.06| |0.12 0.00 0.0 0.46 0.13|
2 |0.70 0.56 0.40 0.60 0.48| |0.03 0.45 0.45 0.49 0.50| |0.07 0.38 0.40 0.50 0.50|
3 |0.92 0.92 0.94 0.81 0.80| |0.31 0.68 0.53 0.36 0.50| |0.27 0.40 0.64 0.41 0.50|
4 |0.94 0.92 0.93 0.65 0.79| |0.45 0.48 0.68 0.29 0.86| |0.85 0.60 0.56 0.40 0.56|
5 |0.96 0.91 0.93 0.95 0.78| |0.85 0.97 0.96 0.40 0.84| |0.91 0.98 0.93 0.38 0.54|
*** WDBC, D=30***
1 |0.31 0.14 0.11 0.68 0.37| |0.00 0.00 0.00 0.62 0.13| |0.07 0.00 0.00 0.50 0.00|
2 |0.50 0.53 0.50 0.67 0.66| |0.08 0.51 0.45 0.53 0.55| |0.27 0.55 0.44 0.50 0.50|
3 |0.90 0.92 0.88 0.50 0.80| |0.30 0.48 0.64 0.40 0.66| |0.40 0.60 0.63 0.40 0.50|
4 |0.92 0.90 0.90 0.69 0.67| |0.47 0.81 0.80 0.43 0.89| |0.52 0.85 0.85 0.44 0.50|
5 |0.89 0.92 0.93 0.68 0.68| |0.88 0.90 0.91 0.37 0.76| |0.81 0.97 0.98 0.37 0.50|
implementation.
For our experiments we used three real-world
datasets to train our algorithms:
1. Iris (Fisher, 1936) - a dataset with 150 instances
and 4 attributes that describes three different
species of Iris.
2. Epil (Thall and Vail, 1990) - a dataset on two-
week seizure counts for 59 epileptics. The data
consists of 236 observations with 8 attributes. The
dataset has two classes - placebo and progabide, a
drug for epilepsy treatment.
3. Wisconsin Breast Cancer Dataset (wdbc) (Street
et al., 1993) - a dataset with 30 features and 569
instances related to breast cancer in Wisconsin,
for our experiment we generated a random nor-
malized sample of 100 instances. The dataset has
two classes: malignant and benign.
These datasets are purposely selected to test the
algorithm performance by increasing the dimension-
ality complexity of the input data. To measure the Par-
VSOM performance, we ran each timing test three
times and took the average time over these runs. The
times reported are the time required for the CPU to
perform the calculations and it is given in CPU sec-
onds. Similarly, the quality tests were done by aver-
aging three quality measurements using the conver-
gence index (CI) explain in detail in (Tatoian, 2018)
and included as part of the R Popsom Package (Hamel
et al., 2016). The CI provides a 0 to 1 numbering scale
to measure the maps’ quality, with 0 represents the
Par-VSOM: Parallel and Stochastic Self-organizing Map Training Algorithm
345
lowest quality and 1 the highest quality. Furthermore,
three map sizes were considered for these experi-
ments, 15×10 (small), 150×100 (medium), 200×150
(large), to see how the different implementations per-
form on different map sizes. In addition, we trained
with various number of training iterations (in pow-
ers of 10) to discover what type of effect a change of
training duration had on the implementations.
5.3 Results
In the large map environment results included in Ta-
ble 2, we see the recurrent speed-up gains of the al-
gorithm with larger maps. The large size of data
buffers require for the calculations, the CPU cache
memory size limitations and DDR4 lower clock rate
does present an performance impact for the SOM
and VSOM CPU variants. The large workload and
substantial computational resources available in the
GPU, allows the Par-VSOM performance scale fur-
ther. Here, the Par-VSOM achieves a speed-up of 67
in comparison to the SOM. The table results demon-
strates, the Par-VSOM achieves superior speed-up
in all the three datasets comparisons, surpassing the
speed rates of all the other algorithm implementa-
tions. In this large map environment, the Par-VSOM
surpassed the SOM with a 67, the VSOM with a 4.1,
Xpysom with 6.1 and the Quicksom by 20 speed-up
increase.
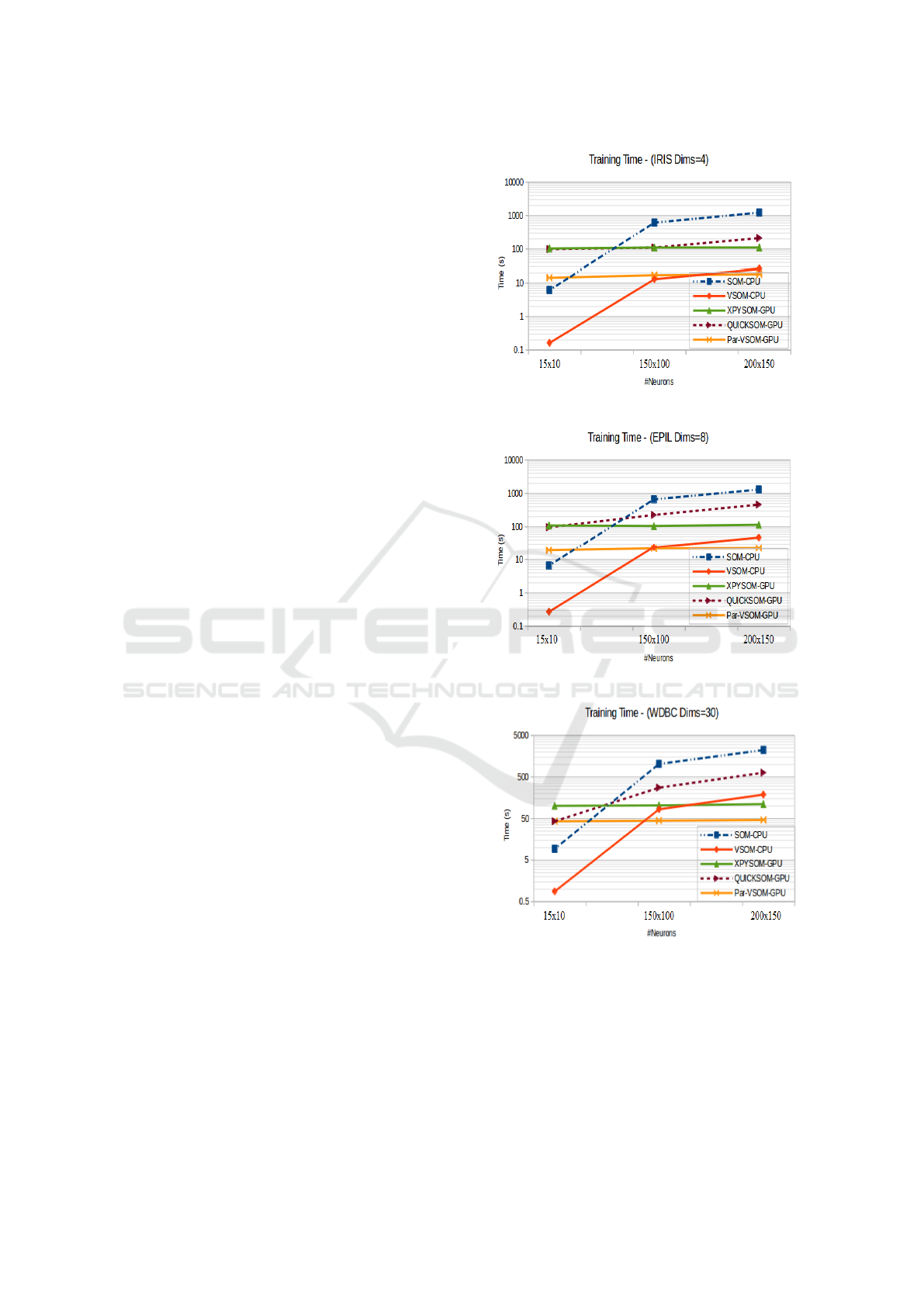
The training time charts included in Figure 4, cap-
ture a generalize representation of the overall results.
The Par-VSOM offers speedup performance increases
for the three datasets in medium and larger size maps
instances. The obtained results allows us to estab-
lish a direct relation between large neuronal maps and
better achievable times using the Par-VSOM. That is,
with a higher number of neurons an scalable speed up
can be achieved.
The Table 3 illustrates the baseline quality of orig-
inal algorithms using our three datasets. The results
present us with a recurring behaviour in most of the
maps, their is a pattern to decrease the convergence
quality when the datasets dimensionality increases.
However, we also identified as the size of the maps in-
creases, there is tendency for the vectorized variants
(VSOM and Par-VSOM) to generate higher quality
maps. Furthermore, our testing demonstrates Xpysom
and Quicksom SOM parallel versions can not reach
a high convergence index when larger map sizes are
used.
In terms of the quality of the maps, Figure 5 cap-
tures all the algorithm convergence indexes for the
three datasets. As illustrated, the Par-VSOM main-
tains relatively the same quality as the original SOM
(a) Iris Total Training Time at Convergence
(b) Epil Total Training Time at Convergence
(c) WDBC Total Training Time at Convergence
Figure 4: Total training time for all datasets with multiple
map sizes at the convergence iteration 100000.
and the VSOM variants in all the maps. In contrast,
the parallel GPU BatchSOM variants (Xpysom and
Quicksom) only obtained good quality indexes with
smaller maps (15 x 10). In both of these parallel pack-
ages, the convergence index quality starts decreas-
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
346
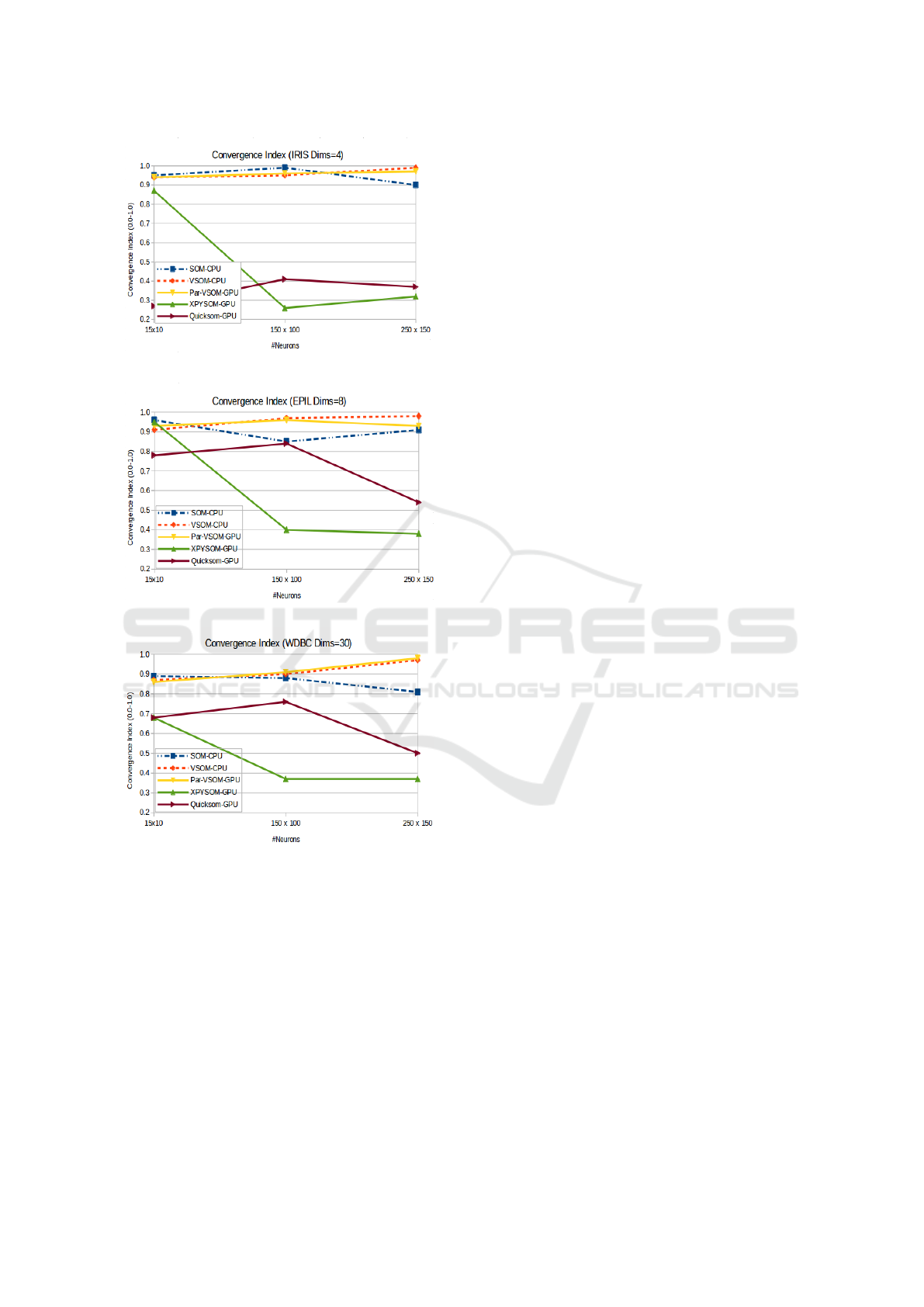
(a) Iris Convergence Index (Map Quality)
(b) Epil Convergence Index (Map Quality)
(c) WDBC Convergence Index (Map Quality)
Figure 5: Convergence Index for the three datasets with var-
ious map sizes.
ing drastically after trying to organized medium and
larger SOM maps.
6 CONCLUSIONS
This work introduced the Par-VSOM, a highly par-
allel, vectorized and matrix-based implementation of
stochastic training for self-organizing maps. The
novel implementation presented here provides sub-
stantial performance increases over Kohonen’s itera-
tive SOM algorithm (up to 67 times faster), the CPU
based vectorized VSOM (up to 4 times faster), the
GPU Xpysom (up to 6.1 times) and Quicksom’s GPU
(up to 20 times) in large maps environments. The re-
sults clearly indicate the parallel BatchSOM approach
provided by Xpysom and Quicksom’s are no longer the
most optimal parallel option in newer architectures
due to the overhead and latency added by search for
winner in the batch algorithm. The performance gains
follow a direct relation with the increment of the map
sizes, as shown in Figure 4. Furthermore, the results
obtained by increasing the dimensionality and maps
sizes demonstrated the Par-VSOM provides a scal-
able speed-up performance when the neuronal map
size increases. In terms of the quality of the maps,
the maps produced by Par-VSOM approximates the
high quality values generated by the VSOM iterative
algorithms and original Kohonen’s SOM algorithm.
In the proposed design, the Par-VSOM is a multi-
threaded algorithm running in a GPU and therefore is
an adequate replacement for iterative stochastic train-
ing of SOM and parallel SOM variants. We are cur-
rently investigating how the Par-VSOM can be im-
plemented in an FPGA and what kind of performance
increase we can expect from this type of hardware ar-
chitecture. Based on our results, the Par-VSOM can
be viewed as an alternative to parallel SOM and a new
alternative for other parallel algorithms for clustering
and pattern recognition. In summary, since the train-
ing algorithms results demonstrate the produce maps
are roughly the same quality, the Par-VSOM provides
a parallel and high-performance alternative to SOM
algorithms. The Par-VSOM source code is available
at (Rivera-Morales, 2022).
REFERENCES
Abadi, M., Jovanovic, S., Ben Khalifa, K., Weber, S., and
Bedoui, M. (2016). A scalable flexible som noc-based
hardware architecture. Advances in Self-Organizing
Maps and Learning Vector Quantization, pages 164–
175.
Barney, B. (2018). Introduction to Parallel Computing.
Lawrence Livermore National Laboratory.
Davidson, G. (2015). A parallel implementation of the self
organising map using opencl. University of Glasgow.
Dickson, N. G., Karimi, K., and Hamze, F. (2011). Impor-
tance of explicit vectorization for cpu and gpu soft-
ware performance. Journal of Computational Physics,
230(13):5383–5398.
Fisher, R. A. (1936). The use of multiple measurements in
taxonomic problems. Annals of eugenics, 7(2):179–
188.
Par-VSOM: Parallel and Stochastic Self-organizing Map Training Algorithm
347

Hamel, L. (2019). Vsom efficient, stochastic self-
organizing map training. In Proceedings of the 2018
Intelligent Systems Conference (IntelliSys) Volume 2,
pages 805–821.
Hamel, L., Ott, B., and Breard, G. (2016). pop-
som: Functions for Constructing and Evaluating Self-
Organizing Maps. R package version 4.1.0.
Hastie, T., Tibshirani, R., and Friedman, J. (2001). The
Elements of Statistical Learning. Springer Series in
Statistics. Springer New York Inc., New York, NY,
USA.
Jaaskelainen, P. (2019). Task parallelism with opencl: A
case study. Journal of Signal Processing Systems,
pages 33–46.
Kim, K.-H., Yun, S.-T., Yu, S., Choi, B.-Y., Kim, M.-J.,
and Lee, K.-J. (2020). Geochemical pattern recogni-
tions of deep thermal groundwater in south korea us-
ing self-organizing map: Identified pathways of geo-
chemical reaction and mixing. Journal of Hydrology,
589:125202.
Kohonen, T. (2001). Self-organizing maps. Springer Berlin.
Li, J., Chen, B. M., and Lee, G. H. (2018a). So-net: Self-
organizing network for point cloud analysis. In Pro-
ceedings of the IEEE conference on computer vision
and pattern recognition, pages 9397–9406.
Li, T., Sun, G., Yang, C., Liang, K., Ma, S., and Huang, L.
(2018b). Using self-organizing map for coastal water
quality classification: Towards a better understanding
of patterns and processes. Science of The Total Envi-
ronment, 628-629:1446–1459.
Lokesh, S., Kumar, P. M., Devi, M. R., Parthasarathy, P.,
and Gokulnath, C. (2019). An automatic tamil speech
recognition system by using bidirectional recurrent
neural network with self-organizing map. Neural
Computing and Applications, 31(5):1521–1531.
Mallet, V., Nilges, M., and Bouvier, G. (2021a). Quicksom.
https://github.com/bougui505/quicksom.
Mallet, V., Nilges, M., and Bouvier, G. (2021b). quick-
som: Self-organizing maps on gpus for clustering
of molecular dynamics trajectories. Bioinformatics,
37(14):2064–2065.
Mancini, R., Ritacco, A., Lanciano, G., and Cucinotta, T.
(2020). Xpysom: high-performance self-organizing
maps. In 2020 IEEE 32nd International Symposium
on Computer Architecture and High Performance
Computing (SBAC-PAD), pages 209–216. IEEE.
Moraes, F. C., Botelho, S. C., Duarte Filho, N., and Gaya,
J. F. O. (2012). Parallel high dimensional self orga-
nizing maps using cuda. In 2012 Brazilian Robotics
Symposium and Latin American Robotics Symposium,
pages 302–306. IEEE.
Mor
´
an, A., Rossell
´
o, J. L., Roca, M., and Canals, V. (2020).
Soc kohonen maps based on stochastic computing. In
2020 International Joint Conference on Neural Net-
works (IJCNN), pages 1–7.
Nvidia.com (2020). Thrust quick start guide. https://
docs.nvidia.com/cuda/thrust/index.html#abstract. Ac-
cessed: 2020-04-30.
Pilla, L. L. (2018). Basics of vectorization for fortran appli-
cations. Research Report, RR-9147:1–9.
Ramos, M. A. C., Leme, B. C. C., de Almeida, L. F.,
Bizarria, F. C. P., and Bizarria, J. W. P. (2017). Clus-
tering wear particle using computer vision and self-
organizing maps. In 2017 17th International Confer-
ence on Control, Automation and Systems (ICCAS),
pages 4–8.
Rauber, Andreas, P. T. and Merkl, D. (2000). parsom: a
parallel implementation of the self-organizing map ex-
ploiting cache efects: making the som fit for interac-
tive high-performance data analysis. In Proceedings
of the IEEE-INNS-ENNS International Joint Confer-
ence on Neural Networks. IJCNN 2000, volume 6.
Richardson, T. and Winer, E. (2015). Extending paralleliza-
tion of the self-organizing map by combining data and
network partitioned methods. Advances in Engineer-
ing Software, 88:1–7.
Rivera-Morales, O. (2022). Par-vsom. https://github.com/
oxrm/Par-vsom.
Sarazin, T., Azzag, H., and Lebbah, M. (2014). Som clus-
tering using spark-mapreduce. In 2014 IEEE Interna-
tional Parallel & Distributed Processing Symposium
Workshops, pages 1727–1734. IEEE.
Schabauer, Hannes, E. S. and Weishaupl, T. (2005). Solv-
ing very large traveling salesman problems by som
parallelization on cluster architectures. In Sixth In-
ternatioanl Conference on Parallel and Distributed
Computer Applications and Technologies PDCAT’ 05,
pages 954–958. IEEE.
Silva, B. and Marques, N. (2007). A hybrid parallel som
algorithm for large maps in data-mining. New Trends
in Artificial Intelligence.
Street, W. N., Wolberg, W. H., and Mangasarian, O. L.
(1993). Nuclear feature extraction for breast tumor
diagnosis. In IS&T/SPIE’s Symposium on Electronic
Imaging: Science and Technology, pages 861–870. In-
ternational Society for Optics and Photonics.
Sul, S.-J. and Tovchigrechko, A. (2011). Parallelizing blast
and som algorithms with mapreduce-mpi library. In
2011 IEEE International Symposium on Parallel and
Distributed Processing Workshops and Phd Forum,
pages 481–489. IEEE.
Tatoian, R.and Hamel, L. (2018). Self-organizing map con-
vergence. International Journal of Service Science,
Management, Engineering, and Technology (IJSS-
MET), 9(2):61–84.
Thall, P. F. and Vail, S. C. (1990). Some covariance models
for longitudinal count data with overdispersion. Bio-
metrics, pages 657–671.
Vettigli, G. (2021). Minisom. https://github.com/
JustGlowing/minisom.
Wittek, P., Gao, S. C., Lim, I. S., and Zhao, L. (2013). So-
moclu: An efficient parallel library for self-organizing
maps. arXiv preprint arXiv:1305.1422.
NCTA 2022 - 14th International Conference on Neural Computation Theory and Applications
348
