
Simulation of the Evolution of a Virtual Patient’s Physiological Status in
the Operating Room: Application to Computer-assisted Anaesthesia
Training
Hugo Boisaubert
1
, Lucas Vincent
1
, Corinne Lejus-Bourdeau
2,3
and Christine Sinoquet
1
1
Research Laboratory of Digital Science in Nantes (LS2N) / UMR CNRS 6004, University of Nantes,
2 rue de la Houssini
`
ere, Nantes, France
2
Experimental Universitary Laboratory for Simulation in Intensive Care (LESiMU) in Nantes,
9 rue Bias Ricordeau, Nantes, France
3
Department of Anaesthesia and Intensive Care, Nantes University Hospital, 1 place Alexis Ricordeau, Nantes, France
Keywords:
Computer-assisted Medical Training, Virtual Patient, Operating Room, Anaesthesia, Reactive Scenario,
Simulation, Prediction, Case-based Reasoning, Data Mining, Pattern Recognition, E-health Record, Event
Trace, Multivariate Time Series.
Abstract:
Half a million surgeries are performed every day around the world, which places safety and quality at the
heart of global health issues. In this context, we introduce a novel approach, SVP-OR (Simulation of Virtual
Patient at the Operating Room), designed for digital training support. For this purpose, we must evolve the
physiological parameters of a virtual patient submitted to the actions of a user (trainee), and of a virtual
medical team. We formulate the problem as a case-based reasoning approach in which (i) we identify real
patients whose anaesthetic profiles show a region similar to the recent history of the virtual patient and (ii)
we predict the near future of the virtual patient (a multivariate time series) from the multivariate time series
of the most similar real patients. The first contribution in this paper consists in the design of a contextualized
multidimensional pattern recognition approach. Our second contribution is the development of a generic
framework based on the concept of contextualized multidimensional pattern, to predict the evolution of the
virtual patient. In a third contribution, we instantiate our framework, and we evaluate and compare the realism
of two predictive strategies.
1 INTRODUCTION
Over the world, the network of simulation platforms
in intensive care, hosted by university hospitals, aims
at training medical interns and nurses, as well as
more experienced physicians. In particular, the Ex-
perimental Universitary Laboratory for Simulation in
Intensive Care (LESiMU) in Nantes offers training
in seven medical specialities, including anaesthesia.
For this purpose, LESiMU uses high-fidelity patient
simulators, namely mannequins, with immersion of
the trainees in a full-scale interprofessional medical
team. Currently, the training scenarios are written in
advance, the trainer makes himself evolve the phys-
iological parameters of the mannequin ”by hand”, in
response to the actions of the medical team (including
those of the trainee immersed in this team). There is
therefore little variability in the scenarios.
Our work is motivated by a real need expressed
by LESiMU. In order to improve the safety and qual-
ity of intraoperative care, the trainers at LESiMU
wish to vary the diversity of the scenarios to be pro-
posed to anaesthesia interns and nurse anesthetists, in
initial training, as well as to more experienced prac-
titioners. We proposed to develop a digitally assisted
modality, based on the database of anaesthetic profiles
recorded by Nantes University Hospital since 2004.
In this modality, we wish the trainee to run a software
program consisting in a generator of reactive scenar-
ios. The application is parameterized with the surgery
of interest, together with the age, gender, weight and
medical history (e.g., diabetes) of the virtual patient
to be simulated. The other members of the medical
team are simulated in a very simple way (icons car-
rying out actions and transmitting information, on the
screen of trainee’s laptop). Serving this training ob-
jective also responds in the long run to the need for
228
Boisaubert, H., Vincent, L., Lejus-Bourdeau, C. and Sinoquet, C.
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted Anaesthesia Training.
DOI: 10.5220/0010981000003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 228-239
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

anticipation through prediction, to estimate a risk at
the operating room. This objective is inherent to the
emerging paradigm of personalized medicine.
To generate reactive scenarios, one has to predict
the evolution of the physiological parameters of the
virtual patient after an action has been triggered by the
medical team. More than 500,000 anaesthetic profiles
have been collected since 2004, at Nantes University
Hospital. Such anaesthetic data include a multivari-
ate time series and an action trace. The multivariate
time series describes the evolution of the physiolog-
ical parameters (e.g., blood pressure, oxygen satura-
tion, heart rate) of a real patient, during their monitor-
ing in the operating room. Hereinafter, we will also
use the term ”variables” to refer to the physiological
parameters. The action trace records the actions trig-
gered by the medical team. Given a cohort of patients
operated on for the same surgery, and in the same
”class” as the virtual patient to be simulated (age, gen-
der, weight and medical history), the time series and
action traces of the cohort contain knowledge to pre-
dict the evolution of the virtual patient after a given
action has been triggered (For instance, a class may
correspond to a 25-35 year old male weighting be-
tween 75 and 85 kg and with no medical antecedents).
We can capture this knowledge envisaging either a
modelling approach or a data mining approach.
In this paper, we present a case-based reasoning
(CBR) approach. We consider the available appropri-
ate cohort of real patients, that is patients with same
surgery, age, gender, weight and medical history as
the virtual patient to be simulated. We call reactive
simulation scenario the dynamical process in which
a user (the trainee) makes evolve a virtual patient. At
the end of the reactive scenario, we obtain a multi-
variate time series together with the trace of actions
carried out by both trainee and medical team until
surgery completion.
In a case-based reasoning framework, we have to
identify real patients whose historical data show a re-
gion similar to the recent history of the virtual patient.
The historical data of a patient embraces a multivari-
ate time series and an action trace. We have to han-
dle two pattern recognition problems, one where the
query is a sequence of time-stamped actions, the other
one where the query is a multivariate time series. Fi-
nally, for a reactive simulation scenario, we must meet
a real-time constraint.
In this paper, our first contribution consists in the
design of a contextualized multidimensional pattern
recognition approach, to identify real patients most
similar in some region of their anaesthetic profiles to
the simulated patient. Our second contribution is the
design of a generic method to predict the evolution
of the virtual patient, based on the concept of con-
textualized multidimensional pattern. In a third con-
tribution, we instantiate our generic framework, and
we describe, evaluate and compare the realism of two
predictive strategies.
The remainder of the paper is organized as fol-
lows. Section 2 briefly mentions some related work.
Section 3 describes the two main components of our
CBR approach, that is (i) a contextualized pattern
recognition method and (ii) a prediction strategy of
short-term evolution for the virtual patient. This sec-
tion ends with the description of the general CBR-
based algorithm SVP-OR (Simulation of Virtual Pa-
tient at the Operating Room). Section 4 presents an
evaluation of two instantiations of SVP-OR. Section 5
concludes and opens up future directions of research.
From now on, we denote p the number of vari-
ables described in any anaesthetic profile of the pa-
tient cohort.
2 RELATED WORK
The motivation for our work is to implement short-
term prediction on the fly of multivariate time series,
in order to handle reactive scenarios simulating a sur-
gical operation. Although other works in the literature
may be related to our proposal, no such approach has
ever been proposed.
These recent years, digital training assistance
has entered the hospital. The range of assistance of-
fered extends from the provision of scenarios to be
replayed on computer from a selection of real scenar-
ios, to the immersion in a virtual operating room (OR)
(Nagendran et al., 2013; Qi et al., 2021).
Beside manual annotation, artificial intelligence
has much to contribute to knowledge extraction from
OR data. Machine learning techniques allow the
automatic detection and prediction of surgical ac-
tivities, for instance using Hidden Markov models
(Meißner et al., 2014) or random forests (Stauder
et al., 2014). In particular, deep learning can recog-
nize various types of surgical procedures from videos
(Khalid et al., 2020).
In the field of data mining, the literature reports
work on the exploitation of low-level surgical tasks
to predict the possible surgeons’ subsequent tasks
(Forestier et al., 2017), as well as interventional time
(Franke et al., 2013). Works in the line of (Erdogan
and Tarhan, 2018) apply process mining techniques to
logs obtained in a healthcare context, with a focus on
process discovery, conformance checking and process
enhancement.
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
229

Much work has been done on multivariate time se-
ries forecasting (see for instance the recent survey in
(Liu et al., 2021)). In our application, medical actions
may exert synergistic or antagonistic effects on some
of the patient’s physiological parameters. Under these
conditions, designing and training a prediction model
based on the joint modelling of time series and event
traces are highly challenging.
The case-based reasoning approach described in
this paper relies on the concept of local similarity be-
tween real patients and a digital patient. A number
of works reported in the literature deal with the no-
tion of similarity between patients. Importantly, the
massive and systematic use of electronic medical data
has laid the foundation for personalized medicine. In
this field, various definitions of global and local simi-
larities have therefore been proposed; they leverage a
variety of components of patient data, to apply clus-
tering or classifying on patients, such as in (Ng et al.,
2015; Brown, 2016; Parimbelli et al., 2018; Wang
et al., 2019; Fang et al., 2021). However, in such
works, prediction is focused on risk, survival time, for
instance, and not on the dynamic behaviour of physi-
ological parameters, as required by our application.
Among various artificial intelligence tracks, case-
based reasoning can be considered as a form of
similarity-based or analogical reasoning. Case-based
reasoning (CBR) has been used extensively for di-
agnosis, classification, recommendation and therapy
planning in medicine. The reader is directed to
(Choudhury and Begum, 2016), specifically pages
138 to 140, for illustrations. The reader is also re-
ferred to (Goel and Diaz-Agudo, 2017) for a recent
comprehensive overview on the developments in the
field of CBR. So far, few works have exploited knowl-
edge on patients’ temporal data in the CBR frame-
work. For instance, the work reported in (Ganzinger
et al., 2019) organizes patient temporal data in a time-
graph structure to calculate temporal similarities of
disease progress among patients. In this case, the tem-
poral data consist of medical events. In (Sha et al.,
2016), the authors adapt a temporal similarity mea-
sure to the case of irregularly measured data. The
recent work in (M
¨
ul
ˆ
ayim and Arcos, 2020) tackles
CBR in presence of millions of patients’ longitudi-
nal records. This approach implements an anytime
lazy k-nearest neighbor (kNN) algorithm by avoid-
ing unnecessary neighbor assessments. For the situ-
ations when this speed-up may not suffice, the CBR
approach can be interrupted earlier and it returns best-
so-far kNNs. The positioning of our proposal is dis-
ruptive in the field of CBR: on the one hand, our ap-
proach allows the prediction of temporal subseries in-
stead of more classical outcomes; on the other hand,
the prediction task is iterated throughout a reactive
scenario, in response to external sollicitations (med-
ical actions).
3 CASE-BASED REASONING
APPROACH
To make evolve the virtual patient during a surgery,
we simulate their multivariate time series between
two consecutive action triggers, and we iterate this
process until the end of the surgery.
We propose a Contextualized Multidimensional
Pattern Retrieval approach which relies on two tasks:
(i) identification of real patients most similar to the
virtual patient, in the cohort of patients in the same
class as the virtual patient (Section 3.2); (ii) predic-
tion of the near future of virtual patient from the pre-
vious real patients (Section 3.3). This second task is
based on three subtasks : off-line annotation of the co-
hort of patients (Subsection 3.2.1), similarity search
for action traces (Subsection 3.2.2), similarity search
for time series (Subsection 3.2.3).
3.1 Contextualized Multidimensional
Pattern
Our case-based reasoning (CBR) approach relies on
Contextualized Multidimensional Pattern retrieval, in
a set of multivariate time series each annotated with a
trace of time-stamped triggers of actions.
We consider that (i) each variable is influenced by
one action at least, (ii) an action may impact several
variables, (iii) a variable impacted by several actions
is subject to the combined effect of these actions, ini-
tiated at different time-steps or not.
The objective is to predict the evolution of the vir-
tual patient subsequent to the triggering of an action
A. The recent history of the virtual patient provides
the context of the action triggering. Such context in-
cludes the ”most recently triggered” actions (C and B
on Figure 1), together with the recent evolution of the
virtual patient’s variables, prior the initiation of action
A. On the example of Figure 1, the actions of the con-
text are C, B and A, and a contextual time window is
defined, that extends from the triggering of action C
to that of action A. A Contextualized Multidimen-
sional Pattern (CMP) can therefore be defined for
the virtual patient. It is composed of (i) the restric-
tion of the virtual patient’s multivariate time series to
the contextual time window and of (ii) the sequence
of actions in the context, annotated with the durations
between two successive triggers of actions. The lat-
HEALTHINF 2022 - 15th International Conference on Health Informatics
230
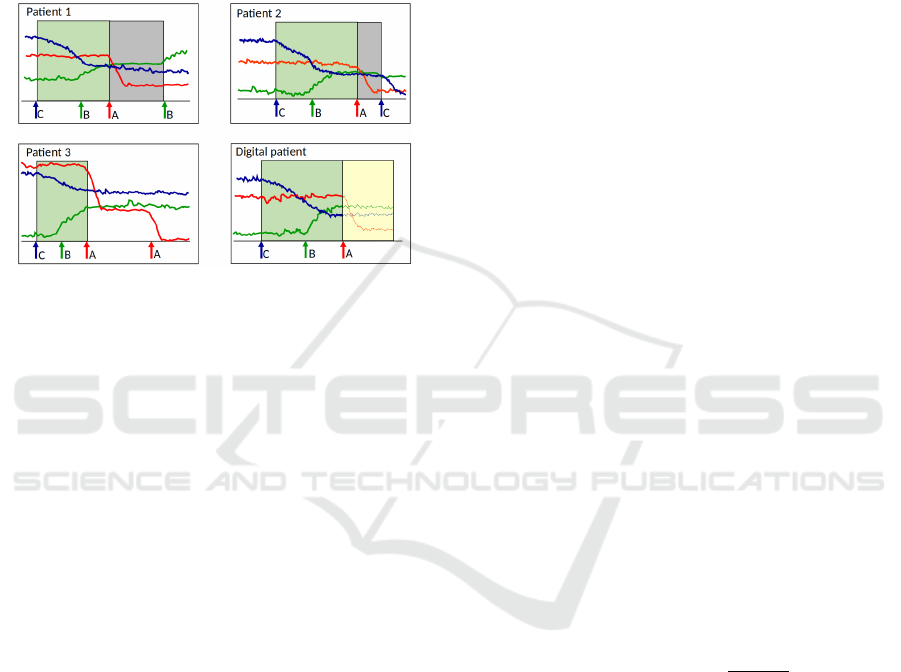
ter sequence of intertwined actions and durations is
called action-signature in the remainder of the paper.
The number of actions in the action-signature, further
referred to as nba, is specified through the expertise
of anaesthesiologists. For instance, if nba equals 4,
”D <10> C <35> B <72> A” represents an action-
signature in which the time intervals between two suc-
cessive triggers are respectively 10, 35 and 72 number
of time-steps.
Figure 1: Concept of contextualized multidimensional pat-
tern and use to simulate the evolution of the virtual patient.
The mutivariate time series is composed of the three uni-
variate time series respectively associated to the variables
represented in red, green and blue. In this example, actions
A, B and C respectively exert an impact on the variables
represented in red, green and blue. These impacts are re-
spectively a sharp decline, a slow growth, and a slow de-
cline (until stabilization in all three cases). Patients 1, 2 and
3 are real patients. The multivariate time series of the four
patients represented have all been left-truncated, as previ-
ous actions are assumed to have no effect on the variables
beyond a certain duration. In this toy example, real patients
1, 2 and 3 have each their action-signature similar to that of
the virtual patient. The mutivariate time series of patients 1
and 2 are supposed to be sufficiently similar to that of the
virtual patient. We use the multivariate time series of pa-
tients 1 and 2 (gray rectangles truncated on the right when
the action following A is triggered) to predict short-term
evolution of the virtual patient (yellow rectangle).
3.2 Identifying Real Patients Most
Similar to the Virtual Patient
To note, we only have to examine the real patients
on targeted regions of their event traces and on mul-
tivariate time subseries. However, real-time identifi-
cation of patients most similar to the virtual patient
is challenging: (i) at the hospital, the available cohort
of patients sharing the same characteristics as the vir-
tual patient (surgery, age, gender, weight, pathologi-
cal history) may be large, (ii) we deal with multivari-
ate time series.
The current CMP of the virtual patient will be fur-
ther referred to as the query. The number of actions in
a CMP is a constant set by the training managers (i.e.,
anaesthesia experts). This number is denoted nba, and
includes the last action (act) triggered by the user (i.e.,
the trainee).
3.2.1 Off-line Annotation
The first subtask is off-line and is dedicated to the an-
notation of the cohort. Based on this annotation, at
each novel query, we wish to make available an index
whose entries are actions. Such index will allow to ef-
ficiently retrieve real action-signatures (ASs) similar
to the AS of the virtual patient.
3.2.2 Similarity for Action Traces
As already mentioned, the second (on-line) subtask in
CMP retrieval involves index-based AS recognition.
We add flexibility in two ways. Thanks to experts,
we are able to categorize actions: for instance, it may
be possible to replace an anaesthetic by another. Real
patients found similar to the virtual patient (VP) are
expected to have been anaesthetized with the same
product as the VP, but product replacement is allowed.
For small cohorts, this relaxation is crucial to raise the
probability to identify patients similar to the VP. On
the other hand, we recall that ASs are time-stamped.
AS recognition must therefore take into account du-
rations between any two consecutive actions. Again,
relaxation is necessary. This time, for more realism,
it is wise to gradually penalize time deviations from
the VP’s action-signature: we care that pairs of con-
secutive actions matched between VP and real patient
are not too different for the most recent pairs; in con-
strast, we accept a lower similarity in time intervals if
the actions are older. For a given relaxation percent-
age ∆, the deviation allowed gradually decreases as
follows:
Dev
time
(i) = ∆
1 −
i − 1
nba − 1
, (1)
where i stands for the rank of the pair of consecu-
tive actions under examination in the action-signature.
For instance, in action-signature ”D <10> C <35>
B <72> A”, the rank of (C,B) is 2. When applied
to an action-signature of 4 actions and ∆ = 5%, Eq.
(1) yields the three following relaxation percentages:
5%, 3.3%, 1.6%. In practice, specifying parameter ∆
will automatically discard real patients showing devi-
ation from the VP, at rank i, above the corresponding
threshold Dev
time
(i).
3.2.3 Similarity for Time Series
Downstream AS recognition, the third substask con-
sists in the retrieval of patients with multivariate time
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
231

series similar to those of the virtual patient, and is far
more challenging. Computing a multivariate dissim-
ilarity measure from the outset, for instance a mul-
tivariate Dynamic Time Warping (DTW) distance is
expensive, even for a number p of variables equal to
3. As an alternative, we propose to rank the real pa-
tients relying on p univariate dissimilarities: for each
of the p variables in our problem, we compute the dis-
similarity between the univariate time series of virtual
patient and real patient. We recall that we only have to
compute these dissimilarities for restrictions of time
series to the contextual window defined by the current
CMP.
We denote D a dissimilarity measure dedicated
to univariate time series comparison. Subsection 4.2
will briefly indicate how we chose D in the context of
our application. We average the p univariate D mea-
sures computed for the p variables:
D =
1
p
p
∑
i=1
D
i
, (2)
with D
i
the dissimilarity obtained using D for i
th
vari-
able. Thereinafter, D will be referred to as the multi-
variate dissimilarity.
On this basis, the similarity score used to rank the
real patients (already selected based on ASs) com-
bines three similarity scores:
Sim
AS+T S
= κ
1
Sim
AS
match
+ κ
2
Sim
AS
time
+ κ
3
Sim
T S
,
(3)
in which Sim
AS
match
rewards pairwise matching of ac-
tions in query AS and real patient’s AS, Sim
AS
time
re-
wards pairwise matching of time intervals in the pre-
vious ASs, and Sim
T S
is a normalized similarity score
for time series. These three scores are defined as fol-
lows:
Sim
AS
match
=
1
nba
nba
∑
i=1
sim(ASa
(V P)
i
,ASa
(RP)
i
), (4)
where ASa
(V P)
i
and ASa
(RP)
i
respectively stand for
i
th
action in the action-signature of virtual patient
(VP) and i
th
action in the action-signature of real pa-
tient (RP), and sim(s,s
0
) = 1 if s = s
0
and 0 otherwise.
Sim
AS
time
=
ε
max
− ε
ε
max
, (5)
ε =
nba−1
∑
i=1
|ASt
(V P)
i
− ASt
(RP)
i
|, (6)
where ASt
(X)
i
denotes the time interval between i
th
and i + 1
th
actions in the action-signature of patient
X, X being either the VP or a real patient, and ε
max
is the largest value of ε over all AS-selected patients
examined.
Sim
T S
=
D
max
− D
D
max
, (7)
where Sim
T S
is equal to 1 for the query, as the dissim-
ilarity of the query against itself is null, and Sim
T S
equals 0 for the real patient with largest multivariate
dissimilarity D
max
, amongst all AS-selected patients
examined.
3.3 Prediction Task
This prediction task is not straightforward, since (i)
the time windows of the similar patients are not equal
(gray rectangles in Figure 1), (ii) we do not know to
which extent we must predict the evolution of the VP,
(iii) the shapes of the univariate time series in real pa-
tients might be similar to that of the VP but with dis-
cordances in variable values at the triggering of the
action of interest (action A in Figure 1). In the fol-
lowing, the prediction window of a similar patient is
defined as the time window left- and right-bounded by
the action just triggered and the next action’s trigger.
In Figure 1, such prediction windows are represented
in gray rectangles. For simplicity, the action just trig-
gered will be referred to ”action A” in this section.
To simulate the multivariate time series over the
largest possible time interval, we propose a strat-
egy that iterates through the ordered right bounds
r
1
,r
2
,·· ·r
n
s
of the prediction windows of the n
s
simi-
lar patients, considering these patients in parallel. At
iteration i, for each variable v, we generate a (univari-
ate) time series fragment by averaging over the uni-
variate time subseries of the similar patients whose
prediction windows extend after right bound r
i
. Thus,
at first iteration, we average over all subseries left-
bounded by action A’s trigger and right-bounded by
r
1
; at second iteration, we average over all remain-
ing subseries that can be left-bounded by r
1
and right-
bounded by r
2
, and so on. The principle is shown in
Figure 2 (a). The n
s
fragments thus obtained are con-
catenated to produce U
+
v
. Afterwards, for each vari-
able v, the novel predicted univariate time series U
+
v
is connected to the corresponding time series U
v
gen-
erated so far for the virtual patient. The connection
generally requires shifting along the y-axis, to start
U
+
v
from the value where U
v
ended in. The princi-
ple is shown in Figure 2 (b). When repeated over the
p variables, this process makes evolve the p physio-
logical parameters of the virtual patient for some time
after an action is triggered.
Importantly, in the scenario that is being played
out, the next action after action A might be triggered
by the trainee beyond the farthest time-step we were
able to predict for. In this case, in the first version
of our reactive scenario generator, we generate stable
univariate time series to wait until the next action is
triggered by the trainee or a timeout is reached.
HEALTHINF 2022 - 15th International Conference on Health Informatics
232
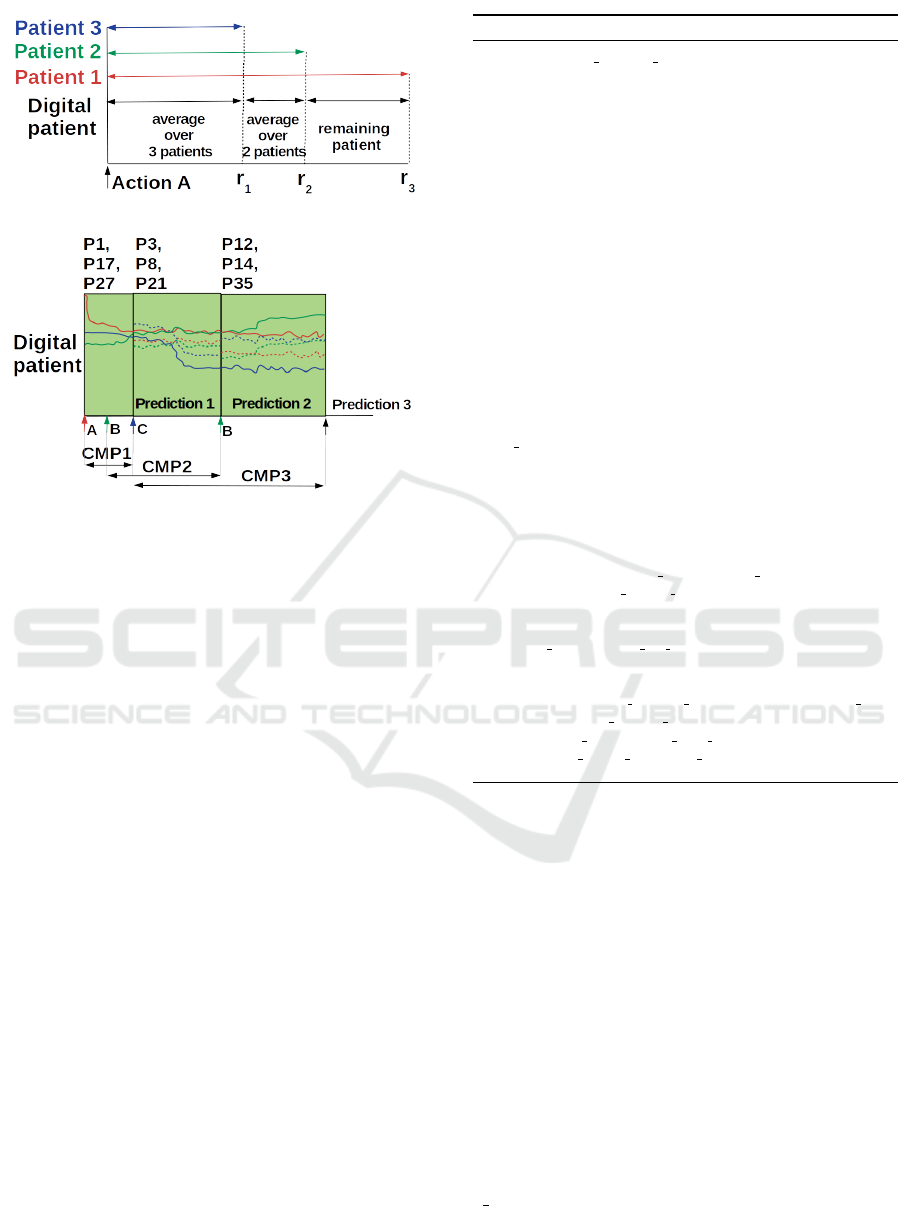
(a)
(b)
Figure 2: Incremental generation of the virtual patient’s
multivariate time series, from the real patients’ multivari-
ate time series, at action A triggering. In this example,
n
s
= 3 (number of real patients most similar to virtual pa-
tient), p = 3 (number of variables) and nba = 3 (number
of actions in the Contextualized Multidimensional Pattern).
(a) Averaging over less and less real patients for one vari-
able produces a set of n
s
= 3 univariate fragments. These
fragments are concatenated to yield a synthetic time series
running from action A to the farthest right bound (r
3
). (b)
The Contextualized Multidimensional Pattern CMP
i
drives
Prediction
i
. For example, in the prediction frame Predic-
tion2, P12, P14 and P35 are the real patients showing high-
est similarity with the CMP2 of the virtual patient. The pro-
cess explained in (a) provides p = 3 univariate fragments
(dotted lines, one line per variable). A shift along the y-axis
allows to concatenate each of these fragments with the cor-
responding univariate time series, in the multivariate time
series simulated so far (solid lines).
3.4 Sketch of Algorithm
Algorithm 1 presents the main lines of our approach.
In line 1, a cohort of real patients is built: it is
composed of the patients who shared the same charac-
teristics Char (age, gender, weight, medical history)
as the virtual patient when they underwent surgery
Surg; besides, the cohort is indexed by sequences
of nba actions, for fastest similarity search of action-
signatures in the patients’ action traces. To start the
simulation corresponding to the first nba − 1 actions,
we proceed as follows: at the trigger of the first ac-
tion of the scenario, for instance action A’s trigger, we
select n
s
patients in Cohort who underwent action A,
Algorithm 1: Simulation of virtual patient.
FUNCTION play reactive scenario(Su
u
ur
r
rg
g
g, Ch
h
ha
a
ar
r
r, n
n
nb
b
ba
a
a, ∆
∆
∆
`
, ∆
∆
∆
m
,
∆
∆
∆
h
, n
s
)
INPUT:
• Surg, surgery of interest
• Char, characteristics of virtual patient (VP)
• nba, number of actions in contextualized multidimensional
pattern (CMP)
• ∆
`
, ∆
m
, ∆
h
, respectively low, medium and relaxation
percentages to guide three CMP retrievals in parallel
• n
s
, number of real patients most similar to virtual patient
OUTPUT:
• T, event trace of VP
• M, multivariate time series of VP
VARIABLES:
• Cohort, cohort of real patients sharing same surgery Surg and
characteristics Char as virtual patient
• Index, indexation of Cohort by sequences of nba actions
• CMP = (T
∗
,M
∗
), contextualized multidimensional pattern of
VP, with
- e time, time ellapsed between the penultimate action and the
current action act triggered by user
- T
∗
, action-signature of VP, of size nba, including last action
act
- M
∗
, restriction of M to the current contextual time window
defined by T
∗
1: (Cohort, Index) ← select preannotated cohort (Surg,Char)
2: (T, M) ← initialize digital patient(nba − 1)
3: while (scenario is not over)
4: (act,e time) ← wait for action()
5: if no action is triggered then break end if
6: (T
∗
,M
∗
) ← build current cmp(T, M,nba − 1, act, e time)
7: Pat ← identify similar patients(T
∗
,M
∗
,∆
`
,∆
m
,∆
h
,n
s
)
8: M ← grow multivariate time series(M, Pat)
9: T ← grow action trace(T, e time,act)
10: end while
and we predict virtual patient’s evolution from them.
At the occurrence of the second action of the sce-
nario, say action B, the action-signature (AS) is ”A
<duration> B”, and the prediction is made using this
AS. Up to the occurrence of nba − 1
th
action, we can
only operate following this initialization mode (line
2). At the end of the initialization, an action trace
T containing nba − 1 actions has been grown and the
corresponding multivariate time series M has been
simulated for the digital patient.
From nba
th
action occurrence onwards (line 3 to
10), we successively use ASs of full length (i.e., with
nba actions). In line 6, once a novel action act has
been triggered, the AS T
∗
is constructed by (i) con-
sidering the nba − 1 latest actions in T and the cor-
responding time intervals, (ii) adding the time inter-
val between penultimate action and current action act,
e
time, (iii) and adding the current action act. The
multivariate time subseries M
∗
corresponding to the
temporal scope of T
∗
is then obtained. It is a suf-
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
233

fix of the multivariate time series T simulated so far.
Together with T
∗
, M
∗
forms the current CMP.
In line 7, we identify the n
s
real patients most sim-
ilar to the virtual patient. We recall that all patients
with ASs similar to the digital patient’s AS T
∗
, given
a specified threshold ∆, are first identified (Subsec-
tion 3.2.2). To note, this identification is accelerated
thanks to the off-line annotation mentioned in Subsec-
tion 3.2.1. Then, the patients thus obtained are ranked
considering the score in Eq. (3), which takes into ac-
count the similarity with T
∗
and the similarity with
subseries M
∗
(Subsection 3.2.3). If threshold ∆ (see
Subsection 3.2.2) is two low, we run the risk of not de-
tecting any similar patient, and of having to restart a
search with a higher ∆ value. Therefore, we designed
a procedure where three searches are run in parallel,
with different ∆ values. This way, we expect to obtain
n
s
similar patients.
In line 8, these n
s
patients are used to predict the
evolution of the digital patient as from current action
act. The multivariate time series M is grown accord-
ingly. The action trace T of the virtual patient is up-
dated in line 9.
This iterative simulation stops when no more ac-
tion is triggered.
4 APPLICATION
The SVP-OR framework has been implemented in
Python 3.10.0. Subsection 4.1 first describes the
data used to evaluate the SVP-OR approach. Then,
Subsection 4.2 provides some information about the
comprehensive comparative analysis of 8 dissimilar-
ity measures which has been conducted separately to
identify the most relevant measure for our application.
Next, Subsection 4.3 describes two instantiations of
SVP-OR. In Subsection 4.4, we finally evaluate these
two instantiations.
4.1 Data
In the current part of our research project, the SVP-
OR approach has been evaluated through realistic data
for reasons of health data protection. These realis-
tic data are produced by the DBLBS data generator
that we have implemented, based on the expertise of
anaesthesiologists. In a nutshell, this generator runs
two steps.
The first step is driven by a grammar that pro-
vides a hierarchical description of a surgery, for a tar-
geted patient (age, gender, weight, medical history).
When fed with a user-specified number of patients n
p
,
this first step produces as many action traces. Expert
knowledge is required to construct such grammar. In
our work, we focused on inguinal hernia operation un-
der laparoscopy, for a 30 year old male patient, with
weight 80 and with no medical history. Figure 3 pro-
vides a simplified view of this surgery.
Based on additional expert knowledge supple-
mented by consultation of the literature, we were able
to draw rules about the impacts of the actions on
the physiological parameters (growth, decay, no im-
pact), the delay, duration and intensity of these im-
pacts. When fed with these rules, a number n
p
of ac-
tion traces produced by first step, and initial values set
for the parameters of each simulated patient, the sec-
ond step produces n
p
multivariate time series. In this
first version of the DBLBS generator, the number p
of variables considered equals 4 (heart rate; diastolic,
systolic and average blood pressures). Thus, we gen-
erated 1000 simulated patients, that is 1000 realistic
multivariate time series of length in the order of 200
time-steps, with measure points spaced every 30 s.
An incremental process based on expert feedback
allowed a first validation of the DBLBS generator.
Furthermore, we conducted a statistical analysis to
compare the time series of real patients against the
time series of patients simulated through DBLBS. Be-
tween 2002 and 2019, 418 patients underwent laparo-
scopic surgery with prosthetics setting, for an inguinal
hernia, at Nantes University Hospital. Amongst these,
170 offered a complete anaesthetic profile. The corre-
sponding patients are referred to as the reference co-
hort hereafter.
In a nutshell, we evidenced that for each variable,
once the effect of the initial values has been erased
through differencing, the empirical distribution func-
tions of reference and simulated cohorts are close.
Differencing is the operation that produces the times
series {X
t
−X
t−1
}
t=2,...,n
from the original time series
{X
t
}
t=1,...,n
of length n. The empirical distribution
function is an estimate of the cumulative distribution
function that generated the values in a given sample.
Moreover, we observed that the autocorrelation func-
tions of the two cohorts are close. Autocorrelation is
the correlation between a time series variable and its
lagged version in time. Due to space limitations, these
results are not detailed here.
4.2 Choice of Dissimilarity Measure
To select a dissimilarity measure D (see Subsection
3.2.3) appropriate to our application, we conducted
the extensive comparative study of 8 pre-selected uni-
variate dissimilarities. The details of this study will
be published elsewhere. In the rest of this section,
we just mention some information about the context
HEALTHINF 2022 - 15th International Conference on Health Informatics
234
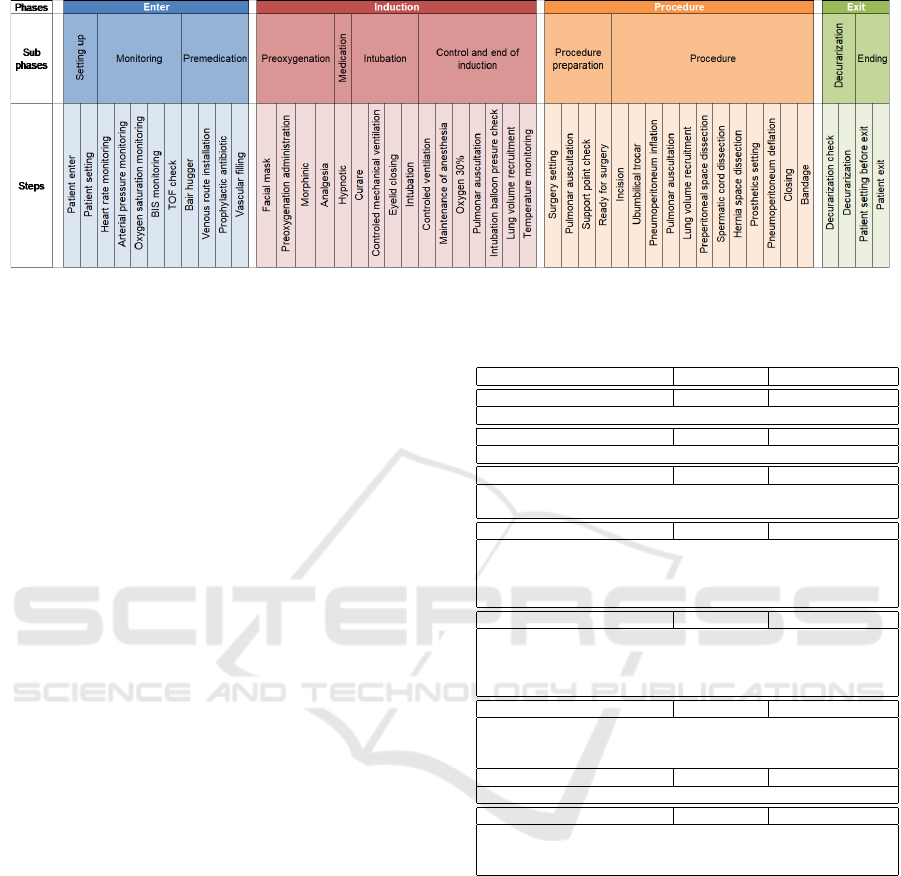
Figure 3: Surgery for the laparoscopic inguinal hernia operation with prosthetics setting.
of this study. Finally, we indicate which dissimilarity
measure was chosen for our application.
Several studies are reported in the literature, that
compare performances or properties of various simi-
larity measures applied to time series. However, none
of these studies deals with short time series, as re-
quired by our application. In a nutshell, we used 28
data sets in our experiments. Twenty-six data sets
were from the UCR repository (Dau et al., 2018).
The 27
th
data set was simulated using the PHMC-
LAR (Partially Hidden Markov Chain Linear Au-
toRegressive) model described in (Dama and Sino-
quet, 2021): the 4-state PHMC-LAR model with au-
toregressive order 2 used is described in the Section
7.1.1 of this latter document. The 28
th
data set was
generated through the DBLBS generator we have de-
signed, based on expert specification (see Subsection
4.1).
In each time series of these 28 data sets, subseries
of length 10, 20 and 30 time-steps were drawn at ran-
dom. The aim was to check whether our conclusions
held for the three lengths. We perturbed each such
subseries using 6 types of perturbations (for instance,
by increasing amplitude at each time-step, depending
on initial amplitude). We computed the dissimilari-
ties between originals and perturbed versions for the
8 dissimilarity measures.
Table 1 briefly describes the 8 pre-selected uni-
variate dissimilarities.
Our experiments allowed us to discard MPDist
and the tsfresh-based measure. Moreover, we checked
that DTW variants are relevant in presence of very
short to short time series, as is the case in the SVP-
OR framework. This result is not trivial. Amongst
these variants, DTWdtai was chosen for its greatest
speed.
We were compelled to run a comparative analysis
focused on short time series, to fill a gap in the litera-
ture. Interestingly, other researchers and practitioners
can rely on our results to apply some framework sim-
Table 1: The 8 dissimilarity measures compared. DTW:
Dynamic Time Warping.
measure abbreviation library
classical DTW DTWc pyts
global alignement of two time series (Sakoe and Chiba, 1971)
fast exact DTW DTWdtai dtaidistance
exact calculus of DTW implemented in Cython language
fast DTW DTWf pyts
whole alignment of the two time series by repeatedly aligning
subseries of the former (Salvador and Chan, 2007)
Sakoe-Chiba DTW DTWsc pyts
similarity assessment between the two time series limited within
a warping path region, the Sakoe-Chiba band, defined as a region
of same width along the diagonal of the alignment matrix
(Sakoe and Chiba, 1978)
Itakura DTW DTWi pyts
warping path region in the shape of a parallelogram, defined
through two parameters controlling the maximum warping width
and the warping extent increase from beginning of warping path
to maximum warping width (Itakura, 1975)
multiscale DTW DTWm pyts
downsampling of the two time series, optimal path thus obtained
projected on the original scale, to be used as the warping path region
(M
¨
uller et al., 2006)
MPDist MPDist MatrixProfile
feature-based dissimilarity measure (Gharghabi et al., 2018)
transformed-based measure tsfresh
Euclidian distance applied to the numerical representations of the
two time series, obtained through the tsfresh feature extraction
package (Christ et al., 2018)
ilar to SVP-OR: the DTW measure is appropriate.
4.3 The Two Instantiations of the
SVP-OR Algorithm
On expert advice, we set nba at 4 (see Subsection 3.1).
We ran three similarity searches in parallel: we chose
∆
`
= 1%, ∆
m
= 5% and ∆
h
= 10% (see Subsection
3.4).
To rank the real patients, we recall that score
Sim
AS+T S
(Eq. 3) is a weighted combination of simi-
larity scores on event traces and time series. We fixed
the weights as follows: κ
1
= κ
2
= 1 (event traces),
κ
3
= 2 (time series).
We instantiated the framework presented in Sec-
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
235

Figure 4: Comparison of real and simulated time series obtained through strategy #1 (most similar patient). The time series
describe heart rate. Simulation has been performed as from the action consisting in the installation of a Bair Hugger (heating
blanket) (black vertical line). Medical actions relevant to anaesthesia are indicated in capital letters. In black: real time series.
In blue: simulated time series.
tion 3. The two variants considered, #1 and #2, differ
by the value of n
s
. We recall that n
s
denotes the num-
ber of real patients most similar to the virtual patient,
used to predict short-term evolution of the latter (see
Algorithm 1, line 8). #1 relies on the most similar
patient, whereas #2 uses the 10 most similar patients.
4.4 Assessing the Realism of Simulated
Trajectories
In this work, our aim is to evaluate how simulated tra-
jectories (i.e., simulated multivariate time series) de-
part from real trajectories. In this evaluation, each
simulated trajectory is paced by the sequence of med-
ical actions (i.e., scenario) of a real trajectory against
which it is compared. In this Subsection, we first out-
line the evaluation protocol used. Then we present
and discuss the results.
4.4.1 Evaluation Protocol
To assess the realism of the trajectories provided by
each of variants #1 and #2, we implemented the fol-
lowing protocol. For i
th
patient in data set DBLBS
(1 ≤ i ≤ 1000), we selected at random a time-step τ
i
where an action was performed. We then simulated
the rest of the surgery from time-step τ
i
, which means
that we enforced the user to trigger the same acts of
anaesthesia as in the real scenario, and at the same
time-steps.
To note, under the normal conditions of use of our
generator, the user is a human being. However, for
the assessment purpose, we modified the interface of
our generator: we enforced the generator to play the
role of the user (trainee), following the real scenario
of i
th
patient as from time-step τ
i
, that is the genera-
tor triggers the same anaesthesia actions as in the real
scenario. Importantly, this particular context of use of
the generator eliminates the problem of having to trig-
ger the actions that are not perpetrated by the trainee.
In our context, the real scenario is simply fed to the
modified interface of our generator, as from time-step
τ
i
.
Figure 4 shows a real time series and the predicted
subseries.
We introduce the following notations and defini-
tions:
Notation 1. For each patient i, we call M
(R)
τ
i
the real
multivariate time series between time-step τ
i
and the
end of the real surgery.
Notation 2. For each patient i, M
(P)
τ
i,# j
denotes the pre-
dicted multivariate time series as from τ
i
to the end of
the surgery, when variant # j is used for prediction.
Definition 1. D
i,# j
is defined as the dissimilarity D
(see Eq. 2) computed between the multivariate time
series M
(R)
τ
i
and M
(P)
τ
i,# j
. In other words, D
i,# j
mea-
sures the dissimilarity between the observed physio-
logical evolution (real trajectory) and predicted phys-
iological behavior (simulated trajectory) of i
th
real
patient, when variant #j is applied. A similarity
score is obtained through normalization: Sim-g
i,# j
=
D
max,# j
−D
i,# j
D
max,# j
where D
max,# j
is the largest value ob-
tained across the 1000 patients.
Notation 3. The distribution of normalized similari-
ties Sim-g
i,# j
computed for the 1000 patients in data
set DBLBS is denoted D
Sim−g
# j
.
HEALTHINF 2022 - 15th International Conference on Health Informatics
236

For each variant # j ∈ {#1, #2}, we computed the
distribution of normalized similarities D
Sim−g
# j
over
the 1000 patients of data set DBLBS.
4.4.2 Results and Discussion
Figure 5 shows the boxplots for the distributions
D
Sim−g
#1
and D
Sim−g
#2
. We conclude that for the
surgery of interest, the time series generated by strat-
egy #2 are less similar to the real time series (average
around 0.78) than those generated through strategy #1
(average around 0.82). Besides, the simulation from
10 real patients (#2) creates more variability than the
simulation from a single real patient (#1). This result
is not trivial as variabilily could arise from the contin-
ual change of the single patient in strategy #1.
Figure 5: Prediction abilities of strategies #1 (most simi-
lar patient) and #2 (10 most similar patients). Each boxplot
represents the distribution of similarities D
Sim−g
i,# j
between
real and predicted multivariate time series of patient i, for
the 1000 realistic patients of data set DBLBS, and for strat-
egy j. For each patient, the prediction was made under the
same left-truncated scenario (same sequence of actions) as
in the real patient. The scenario was left-truncated at ran-
dom over the 1000 patients, to assess the prediction ability
along the whole surgery. The 1000 random truncations were
the same for the two strategies.
We used a Wilcoxon / Mann-Whitney (WMW)
test together with a Kolmogorov-Smirnov (KS) test to
compare the distributions D
Sim−g
#1
and D
Sim−g
#2
. We
obtained a p-value equal to 4.69 × 10
−130
(WMW)
and a p-value equal to 1.11 × 10
−251
(KS). We con-
clude that the distributions of time series simulated
through #1 and #2 significantly differ.
The main conclusion to draw is that the two vari-
ants assessed produce trajectories sufficiently similar
to real trajectories, when constrained by real scenar-
ios. In the context of the application targeted, our
simulation software will have to trigger the actions
that are not perpetrated by the trainee. A synthetic
representation of the steps, substeps, and possibly al-
ternative steps will be constructed off-line for a given
surgery. This representation will be leveraged on the
fly each time an action by the medical team is plau-
sible. In this case, assessing whether the simulated
trajectories are still realistic seems far more challeng-
ing than in the present situation: it seems a priori that
the only way to assess the realism of the simulation
will require analysis by human experts.
5 CONCLUSION AND
PERSPECTIVES
In this work, we have introduced a generalist frame-
work, SVP-OR (Simulation of Virtual Patient at the
Operating Room), which adopts a case-based ap-
proach to make evolve a virtual patient in response
to the actions of a user and of a virtual medical team.
The aim is to provide a digital training support for
the user (trainee). SVP-OR lies on two corner stones,
contextualized multidimensional pattern retrieval and
short-term prediction from real patients’ histories.
The ability to handle scenarios responsive to ex-
ternal sollicitations offers possibilities that go beyond
the training objective considered here. First, our
CBR-based SVP-OR framework allows to simulate
the evolution of a patient’s physiological status in a
simple but efficient way. Therefore, without the need
to learn a complex model in machine learning, we are
able to develop a digital twin for a patient undergoing
a surgery. In this way, for instance, we can anticipate
the risks in the operating room for patients with med-
ical antecedents. A second notable potentiality is of
primary importance for developing research collabo-
rations between University Hospitals and the outside
world. Protected data cannot be easily shared with the
outside world; a variant can be instantiated from our
SVP-OR framework, to implement a simple but effec-
tive anonymization procedure capable of preserving
the dependencies between the dynamics of variables,
as well as between the variables and a trace of actions.
One of our next works will be to design a model
to subsume all traces of actions ever observed for a
given surgery. This synthetic representation will al-
low SVP-OR to automatically generate the actions of
the medical team. Moreover, it will offer the way to
identify inappropriate actions by the trainee.
A step further, we aim at integrating in the SVP-
OR framework factors that may mislead the trainee.
Such factors are for instance an abnormal situation,
stress, interactions within the medical team that may
be disruptive to the trainee.
ACKNOWLEDGEMENTS
H. Boisaubert is supported by a PhD scholarship
granted by the Pays de la Loire regional research
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
237

project RFI OIC EXAN. The internship work of
L. Vincent was supported by the FAME research
cluster (Human Factors for Medical Technologies,
NExT/ANR-16-IDEX-0007). All authors would like
to thank F. Dama, PhD student, whose contribution in
the development of the DBLBS generator is invalu-
able. The software development and execution of ex-
periments were carried out at the CCIPL (Centre de
Calcul Intensif des Pays de la Loire, Nantes, France).
REFERENCES
Brown, S.-A. (2016). Patient similarity: emerging concepts
in systems and precision medicine. Frontiers in Phys-
iology, 7:561.
Choudhury, N. and Begum, S. (2016). A survey on case-
based reasoning in medicine. International Jour-
nal of Advanced Computer Science and Applications,
7(8):132–136.
Christ, M., Braun, N., Neuffer, J., and Kempa-Liehr, A.
(2018). Time Series FeatuRe Extraction on basis of
Scalable Hypothesis tests (tsfresh – A Python pack-
age). Neurocomputing, 307:72–77.
Dama, F. and Sinoquet, C. (2021). Partially Hidden Markov
Chain Linear Autoregressive model: inference and
forecasting. arXiv:2102.12584.
Dau, H., Keogh, E., Kamgar, K., Yeh, C.-C., Zhu, Y.,
Gharghabi, S., Ratanamahatana, C., Yanping, Hu,
B., Begum, N., Bagnall, A., Mueen, A., Batista,
G., and Hexagon-ML (2018). The UCR time se-
ries classification archive. https://www.cs.ucr.edu/ ea-
monn/time
series data 2018.
Erdogan, T. and Tarhan, A. (2018). A goal-driven evalu-
ation method based on process mining for healthcare
processes. Applied Sciences, 8(6):894.
Fang, H., Tan, N., Tan, W., Oei, R., Lee, M., and Hsu,
W. (2021). Patient similarity analytics for explainable
clinical risk prediction. BMC Medical Informatics and
Decision Making, 21(1):207.
Forestier, G., Petitjean, F., Riffaud, L., and Jannin, P.
(2017). Automatic matching of surgeries to pre-
dict surgeons’ next actions. Artificial Intelligence in
Medicine, 81:3–11.
Franke, S., Meixensberger, J., and Neumuth, T. (2013).
Intervention time prediction from surgical low-level
tasks. Journal of Biomedical Informatics, 46(1):152–
09.
Ganzinger, M., Schrodt, J., and Knaup-Gregori, P. (2019).
A concept for graph-based temporal similarity of pa-
tient data. Studies in Health Technology and Informat-
ics, 264:138–142.
Gharghabi, S., Imani, S., Bagnall, A., Darvishzadeh, A.,
and Keogh, E. (2018). Matrix Profile XII: MPdist: a
novel time series distance measure to allow data min-
ing in more challenging scenarios. In IEEE Inter-
national Conference on Data Mining (ICDM), pages
965–970.
Goel, A. and Diaz-Agudo, B. (2017). What’s hot in case-
based reasoning. In Thirty-first AAAI Conference on
Artificial Intelligence (AAAI), pages 5067–5069.
Itakura, F. (1975). Minimum prediction residual principle
applied to speech recognition. IEEE Transactions on
Acoustics, Speech, and Signal Processing, 23(1):67–
72.
Khalid, S., Goldenberg, M., Grantcharov, T., Taati, B., and
Rudzicz, F. (2020). Evaluation of Deep Learning
models for identifying surgical actions and measuring
performance. JAMA Network Open 3(3):e201664.
Liu, Z., Zhu, Z., Gao, J., and Xu, C. (2021). Forecast meth-
ods for time series data: a survey. IEEE Access, 606–
617:3091162.
Meißner, C., Meixensberger, J., Pretschner, A., and Neu-
muth, T. (2014). Sensor-based surgical activity recog-
nition in unconstrained environments. Minimally In-
vasive Therapy & Allied Technologies, 23:198–205.
M
¨
ul
ˆ
ayim, M. and Arcos, J. (2020). Fast anytime retrieval
with confidence in large-scale temporal case bases.
Knowledge-Based Systems, 206:106374.
M
¨
uller, M., Mattes, H., and Kurth, F. (2006). An effi-
cient multiscale approach to audio synchronization.
In International Conference on Music Information Re-
trieval (ISMIR), pages 192–197.
Nagendran, M., Gurusamy, K., Aggarwal, R., Loizidou, M.,
and Davidson, B. (2013). Virtual reality training for
surgical trainees in laparoscopic surgery. Cochrane
Database of Systematic Reviews, 8:CD00657.
Ng, K., Sun, J., Hu, J., and Wang, F. (2015). Personal-
ized predictive modeling and risk factor identification
using patient similarity. MIA Joint Summits on Trans-
lational Science proceedings, pages 132–136.
Parimbelli, E., Marini, S., Sacchi, L., and Bellazzi, R.
(2018). Patient similarity for precision medicine: a
systematic review. Journal of Biomedical Informat-
ics, 83:87–96.
Qi, D., Ryason, A., Milef, N., Alfred, S., Abu-Nuwar, M.,
Kappus, M., De, S., and Jones, D. (2021). Virtual
reality operating room with AI guidance: design and
validation of a fire scenario. Surgical Endoscopy,
35(2):779–786.
Sakoe, H. and Chiba, S. (1971). A dynamic programming
approach to continuous speech recognition. In ICA,
Paper 20 CI3.
Sakoe, H. and Chiba, S. (1978). Dynamic programming
algorithm optimization for spoken word recognition.
IEEE Transactions on Acoustics, Speech and Signal
Processing, 26(1):43–49.
Salvador, S. and Chan, P. (2007). Toward accurate dynamic
time warping in linear time and space. Intelligent Data
Analysis, 11(5):561–580.
Sha, Y., Venugopalan, J., and Wang, M. (2016). A novel
temporal similarity measure for patients based on ir-
regularly measured data in electronic health records.
In Seventh ACM International Conference on Bioin-
formatics, Computational Biology, and Health Infor-
matics (BCB), pages 337–344.
Stauder, R., Okur, A., Peter, L., Schneider, A., Kranzfelder,
M., Feussner, H., and Navab, N. (2014). Random
HEALTHINF 2022 - 15th International Conference on Health Informatics
238

forests for phase detection in surgical workflow anal-
ysis. In International Conference on Information
Processing in Computer-Assisted Interventions, pages
148–157.
Wang, N., Huang, Y., Liu, H., Fei, X., Wei, L., Zhao, X.,
and Chen, H. (2019). Measurement and application
of patient similarity in personalized predictive model-
ing based on electronic medical records. BioMedical
Engineering OnLine, 18:98.
Simulation of the Evolution of a Virtual Patient’s Physiological Status in the Operating Room: Application to Computer-assisted
Anaesthesia Training
239
