
Counting or Localizing? Evaluating Cell Counting and Detection in
Microscopy Images
Luca Ciampi
a
, Fabio Carrara
b
, Giuseppe Amato
c
and Claudio Gennaro
d
Institute of Information Science and Technologies, National Research Council, Pisa, Italy
Keywords:
Automatic Cell Counting, Biomedical Image Analysis, Deep Learning, Deep Learning for Visual
Understanding, Convolutional Neural Networks, Counting Objects in Images, Visual Counting.
Abstract:
Image-based automatic cell counting is an essential yet challenging task, crucial for the diagnosing of many
diseases. Current solutions rely on Convolutional Neural Networks and provide astonishing results. However,
their performance is often measured only considering counting errors, which can lead to masked mistaken
estimations; a low counting error can be obtained with a high but equal number of false positives and false
negatives. Consequently, it is hard to determine which solution truly performs best. In this work, we investigate
three general counting approaches that have been successfully adopted in the literature for counting several
different categories of objects. Through an experimental evaluation over three public collections of microscopy
images containing marked cells, we assess not only their counting performance compared to several state-of-
the-art methods but also their ability to correctly localize the counted cells. We show that commonly adopted
counting metrics do not always agree with the localization performance of the tested models, and thus we
suggest integrating the proposed evaluation protocol when developing novel cell counting solutions.
1 INTRODUCTION
Microscopy medical images analysis comprises sev-
eral challenging Computer Vision problems involving
a wide variety of tasks. Among them, cell localization
(Lugagne et al., 2019) and counting (Falk et al., 2018)
are essential steps for basic research, like disease di-
agnosis via the evaluation of cell growth kinetics, the
estimation of cytotoxicity (i.e., the quality of being
toxic to cells) (Kotoura et al., 1985), the quantification
of perineuronal nets (Fawcett et al., 2019), the discov-
ery of the role of particular genes in cell biology, mi-
crobiology, and immunology (Zhang et al., 2015), and
many more. Manual cell counting is still conducted
in many laboratories, often with the aid of a hemocy-
tometer and its variants, which has been commonly
used due to its low cost and versatility (Johnston,
2010). However, the procedure is time-consuming
and error-prone, being subject to inter-user variation
depending on the degree of expertise of the analyst
(Altman et al., 1993). Therefore, there is a need to
a
https://orcid.org/0000-0002-6985-0439
b
https://orcid.org/0000-0001-5014-5089
c
https://orcid.org/0000-0003-0171-4315
d
https://orcid.org/0000-0002-3715-149X
count cells automatically to facilitate this tedious and
challenging task.
Recently, several vision models (mostly based on
Convolutional Neural Networks) have been success-
fully adopted to count cells and other biological struc-
tures from microscopy images. However, the per-
formance of these techniques is often measured only
considering the counting errors occurring at inference
time (i.e., the difference between the predicted and
the actual cell numbers), which often leads to masked
mistaken estimations. Indeed, counting errors do not
take into account where the cells have been local-
ized in the images and, consequently, counting mod-
els might achieve low values of errors while providing
wrong predictions (e.g., a high number of false posi-
tives and false negatives). Therefore, it is hard to per-
form a fair comparison between the different state-of-
the-art cell counting approaches to determine which
performs best.
In this work, we investigate three baseline
solutions belonging to the three main counting
methodologies — a segmentation-based approach, a
localization-based approach, and a count-density es-
timation approach — that have been successfully ex-
ploited for counting several different categories of ob-
jects, such as people and vehicles, and that repre-
Ciampi, L., Carrara, F., Amato, G. and Gennaro, C.
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images.
DOI: 10.5220/0010923000003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
887-897
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
887

sent the conceptual basis also for the cell counting
techniques. We conduct experiments on three public
datasets containing different cell types and character-
ized by distinct peculiarities. In addition to compar-
ing the performance of investigated methods against
state-of-the-art cell counters using established count-
ing evaluation metrics, we also measure the ability
of the models to localize the counted cells correctly.
Specifically, we adopt two additional metrics; a) the
Grid Average Mean absolute Error (GAME) metric,
a hybrid metric that simultaneously considers errors
in the object count and in their coarse location, and
b) the mean Average Precision (mAP), that summa-
rizes the cell precise localization performance. We
show that commonly adopted counting metrics (like
mean absolute error) do not always agree with the lo-
calization performance of the tested models, and thus
we suggest measuring both whenever possible to fa-
cilitate the practitioner in picking the most suitable
solution.
We organize the paper as follows. We review re-
lated work in Section 2. In Section 3, we describe
the datasets used for our experiments. Section 4 de-
scribes the investigated methodologies, while Sec-
tion 5 outlines the performed experiments and the ob-
tained results. Finally, Section 6 concludes the pa-
per. The code and the trained models are publicly
available at https://github.com/ciampluca/counting
perineuronal nets/tree/visapp-counting-cells.
2 RELATED WORKS
This section reviews some works concerning the
counting task in its generality and specifically tailored
to estimating the number of cells in microscopy im-
ages.
Visual Counting. The goal of the visual counting
task is to estimate the number of object instances
in still images or video frames (Lempitsky and Zis-
serman, 2010). Due to its interdisciplinary and
widespread applicability to many real-world applica-
tions, like calculating the number of people present
at an event (Boominathan et al., 2016), evaluating
the number of vehicles in urban scenarios (Ciampi
et al., 2021a), or counting animals in ecological
surveys (Arteta et al., 2016b), visual counting has
recently drawn the attention of researchers. Cur-
rent solutions address this task as a supervised deep
learning-based process. They fall into two main cat-
egories: counting by detection (Amato et al., 2019;
Amato et al., 2018; Laradji et al., 2018; Ciampi et al.,
2018) that requires prior detection or segmentation of
the single instances of objects, and counting by re-
gression (O
˜
noro-Rubio and L
´
opez-Sastre, 2016; Li
et al., 2018; Ciampi et al., 2020; Ciampi et al., 2021b)
that instead tries to establish a direct mapping be-
tween the image features and the number of objects
in the scene, either directly or via the estimation of a
density map (i.e., a continuous-valued function). Re-
gression techniques have demonstrated superior per-
formance in crowded scenarios where the objects’ in-
stances are sometimes not well visible due to occlu-
sions and clumps. However, they cannot precisely lo-
calize the objects present in the scene, eventually pro-
viding only a coarse position of the area in which they
are distributed.
Microscope Cell Counting. Because of its
paramount importance, several cell counting deep
learning-based methods have been proposed in the
last years. They belong to both the detection-based
and the regression-based approaches, each having
the advantages and the drawbacks already discussed
above. A relevant example belonging to the for-
mer category is (Paulauskaite-Taraseviciene et al.,
2019), where authors exploited the popular Mask
R-CNN (He et al., 2017) instance segmentation
framework to detect overlapping cells. On the other
hand, a notable regression-based work is (Aich
and Stavness, 2018), where the authors regulated
activation maps from the final convolutional layer of
the network by exploiting coarse ground-truth acti-
vation maps generated from simple dot annotations.
Authors in (Xie et al., 2016), instead, introduced a
CNN-based regression approach that maps the image
features with an associated density map, providing
also a coarse localization of the cells by finding
its peak values. Another example is represented
by (Segui et al., 2015), where the authors proposed a
regression-based technique and explored the features
that are learned to understand their underlying
representation. In (Cohen et al., 2017), another
regression-based deep neural network architecture
(named Count-ception) is presented, inspired by
the Inception family (Szegedy et al., 2015). More,
in (Guo et al., 2021), another density-based deep net-
work framework designed to solve the cell counting
task is introduced. Specifically, the authors propose
SAU-Net, extending the segmentation network U-
Net (Ronneberger et al., 2015) with a Self-Attention
module. Finally, in (He et al., 2021) the authors
exploited auxiliary CNNs to assist the training of the
intermediate layers of a density regressor. Hybrid
strategies have also been devised to deal with densely
concentrated cells but still generating individual cell
detections, such as (Falk et al., 2018; Xie et al.,
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
888

2018). These approaches first generate intermediate
maps that indicate the likelihood of each pixel being
the center of a cell in the image. Then, they convert
these maps into detections by applying some form of
Non-Maximum Suppression (NMS).
Most of these works measure the counting perfor-
mance by computing the error between the predicted
and the actual cell number, hiding potentially mis-
taken localization. In this work, we consider three
general counting approaches on which cell-specific
techniques rely, and we also evaluate the quality of
the produced detections.
3 DATASETS
In this section, we describe the datasets employed in
this work, summarized in Table 1; in particular, we
consider three publicly available collections of mi-
croscopy images widely used in the context of the cell
counting task, presenting different peculiarities and
challenges.
3.1 VGG Cells Dataset
The VGG Cells dataset, introduced in (Lempitsky
and Zisserman, 2010), comprises 200 RGB highly-
realistic synthetic emulations of fluorescence mi-
croscopy images of bacterial cells. Images have a
fixed size of 256 × 256 × 3 pixels, and the cells are
clustered in specific regions and occluded with each
other. It is worth noting that the annotation procedure
is performed automatically and so labels are free of
errors. We show a sample of this dataset in Figure 1.
3.2 MBM Cells Dataset
The Modified Bone Marrow (MBM) Cells has been
initially collected by the authors of (Kainz et al.,
2015) from 11 RGB microscopy images (having a
fixed size of 1200 × 1200 × 3 pixels) of the human
bone marrow tissues pertaining to 8 different pa-
tients. The marked cells belonging to this dataset
have a significant shape variance; furthermore, non-
homogeneous tissue background makes their local-
ization more difficult. In a subsequent work (Cohen
et al., 2017), the authors divided each image into four
patches of 600 × 600 × 3 pixels, for a total of 44 im-
ages. A sample of this dataset is reported in Figure 1.
3.3 Nuclei Cells Dataset
This dataset has been presented in (Sirinukunwattana
et al., 2016) and comprises 100 RGB microscopy
H&E stained histology images of colorectal adenocar-
cinomas having a common size of 500×500× 3. The
images refer to 9 different patients. They have been
cropped from non-overlapping areas representing a
variety of tissue appearances from normal and ma-
lignant regions. Still, they also comprise areas with
artifacts, over-staining, and failed autofocussing to
simulate realistic outliers. Another peculiarity of this
dataset is that the nuclei of the cells belong to four dif-
ferent categories, presenting different visual charac-
teristics; some experts have manually annotated them
by putting a dot over the centroids of each biologi-
cal structure for a total of 29,756 nuclei marked. In
the following, we refer to this dataset as Nuclei Cells
dataset. We report a sample of this dataset in Figure 1.
4 METHOD
We assume to have a labeled collection of N mi-
croscopy images X = {(I
1
,
ˆ
L
1
), . . . , (I
N
,
ˆ
L
N
)}, where
ˆ
L
i
is the set of 2D-point annotations associated to the
i-th image I
i
. Each image has been manually anno-
tated by a human expert, and the annotations are in
the form of dots, i.e., coordinates localizing the cen-
troids of the cells present in the region of interest, as
is usually the case in the counting task.
We define a localization model f
θ
as a Deep
Learning-based algorithm that takes as input an im-
age I and produces as output an associated set of co-
ordinates L = {p
1
, . . . , p
C
| p
j
∈ R
2
} localizing the
centroids of the cells to be counted. This model is
trained using location data X and can be implemented
following several different strategies; here, we test
three successful approaches from the literature, that
are segmentation, detection, and density estimation,
described below.
4.1 Foreground/Background
Segmentation
Proposed by (Falk et al., 2018), in this approach we
locate cells on the basis of a binary segmentation map
S ∈ {0, 1}
H×W
where ones represent pixels of objects
of interest, while zeros are considered background.
Each connected component in the segmentation map
represents a single object; the positions of the ob-
jects L are set to the coordinate of the centroids of
the connected components. As the implementation
of the model f
θ
, we adopt the original U-Net archi-
tecture (Ronneberger et al., 2015) commonly used in
segmentation tasks. The model is trained to produce a
real-valued segmentation map
ˆ
S = f
θ
(I) ∈ [0, 1]
H×W
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images
889
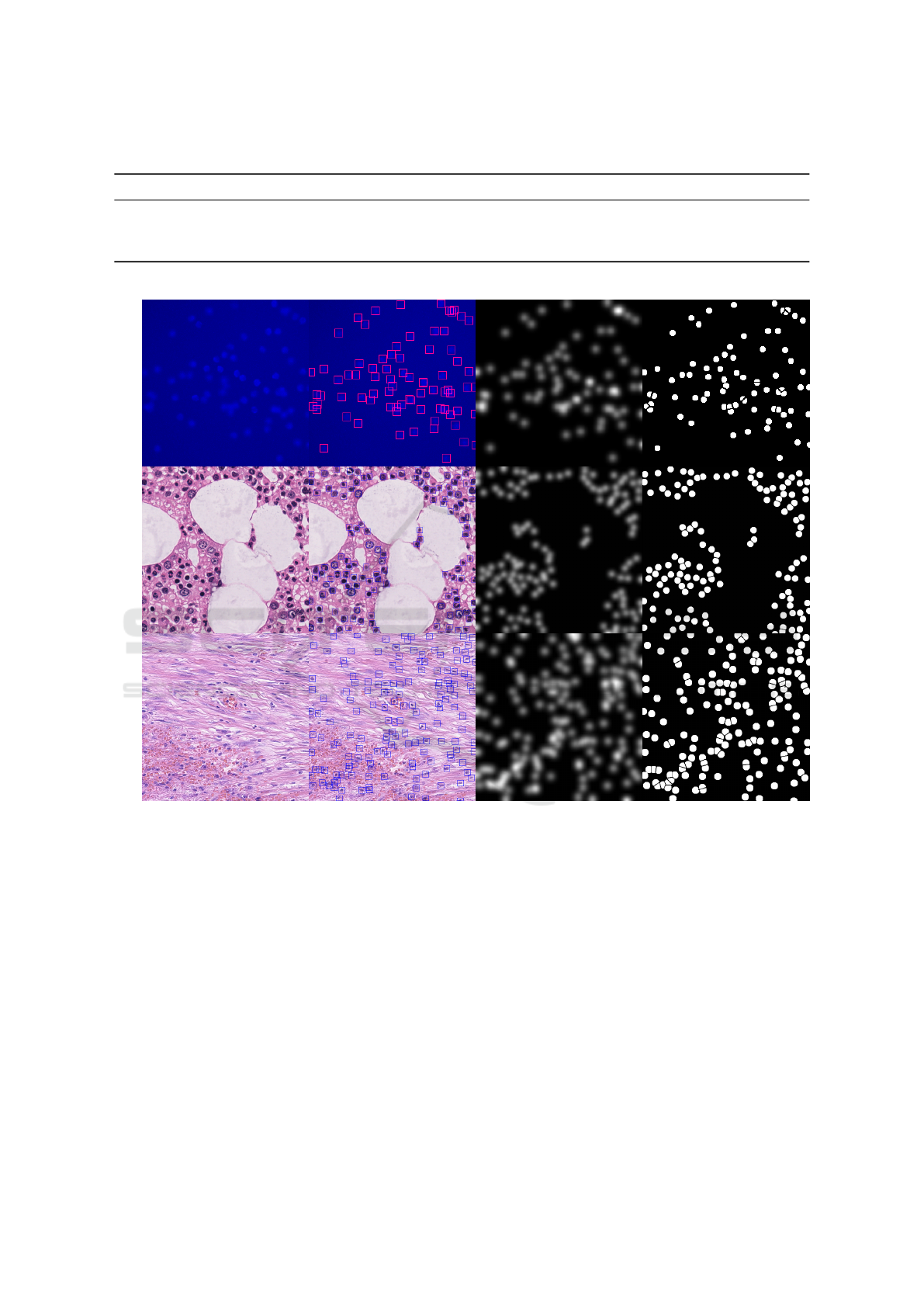
Table 1: Summary of datasets. We show the different peculiarities that characterize the three datasets exploited in this work.
Dataset N.Img Size N.Objs Objs/Img
VGG (Lempitsky and Zisserman, 2010) 200 256×256 35,192 176 ± 61
MBM (Kainz et al., 2015; Cohen et al., 2017) 44 600×600 5,553 126 ± 33
Nuclei (Sirinukunwattana et al., 2016) 100 500×500 29,756 297 ± 218
Sample Detection Target Density Target Segmentation Target
VGGMBM
Nuclei
Figure 1: Samples and Targets. We show a dataset sample (1
st
column) and the corresponding targets used when train-
ing i) the detection-based method FRCNN (2
nd
column), ii) the density-based method D-CSRNet (3
rd
column), and iii) the
segmentation-based method S-UNet (4
th
column).
that is then thresholded to obtain S. The target seg-
mentation maps are generated drawing discs at the
annotated positions and carefully separating overlap-
ping discs with a background ridge (see the fourth
column of Figure 1 for examples of targets). We min-
imize the weighted binary cross-entropy between pix-
els of the output and target maps as specified in (Falk
et al., 2018); more important pixels (near ridges and
foreground objects) are given an increased weight in
the total loss computation. We will refer to this ap-
proach as S-UNet.
4.2 Bounding Box Regression
For this approach, we employ the standard Faster-
RCNN detector (Ren et al., 2017). This deep neural
network takes images as input and produces a list of
bounding boxes localizing the objects as output. The
detection pipeline follows the two-stage paradigm. In
the first stage, the network generates a bunch of re-
gion proposals likely to contain objects, exploiting a
set of anchors (i.e., pre-defined boxes) that are sliced
over the image; in the second stage, these priors are
refined and, for each of them, a score is assigned ex-
pressing the likelihood to really containing the object.
We consider the centers of the final boxes as the lo-
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
890

calization of the entities we want to consider. We
produce the targets by generating squared bounding
boxes centered in the dot-annotated data and having
fixed sides, again, depending on the typical object size
in the dataset. A sample of a target is shown in the
second column of Figure 1. We implement f
θ
as a
Faster-RCNN network with a Feature Pyramid Net-
work module and a ResNet-50 backbone. From now
on, we will refer to this method as FRCNN.
4.3 Density Estimation
We also account for density-estimation approaches
that have shown superior counting performances in
very “crowded” scenarios. In this case, the goal is
to learn a regression between the features of an in-
put image having height H and width W to a density
map D = f
θ
(I) ∈ R
H×W
. The notion of density map
is close to the physical/mathematical notion of den-
sity; specifically, each pixel of D corresponds to the
quantity of the objects present at that precise location.
The number of the objects n present in an image sub-
region P ⊆ I is estimated by summing up pixel val-
ues in the region of interest, i.e., n =
∑
p∈P
D
p
. Al-
though these approaches are not suited for precisely
localize objects, a coarse localization can be obtained
by analyzing the estimated density map, in particular
by finding the top-n maximum local peaks of it, as al-
ready done in (Xie et al., 2016). We train the model
by minimizing the mean squared error loss between
target and predicted density maps. Following previ-
ous works, we generate the target density maps by su-
perimposing Gaussian kernels G
σ
centered in the dot-
annotated locations; the spread parameter σ is fixed,
and it has been estimated depending on the typical
object size in the considered dataset. We show an
example of a target density map in the third column
of Figure 1. We implement f
θ
exploiting the Con-
gested Scene Recognition Network (CSRNet), pro-
posed in (Li et al., 2018), a CNN for accurate den-
sity estimation of congested scenes, comprising two
major components. Specifically, it uses a modified
version of the popular VGG-16 network (Simonyan
and Zisserman, 2015) to extract the image features;
stacked upon this, the authors built a back-end com-
posed of dilated convolutional (Yu and Koltun, 2016)
layers to extract deeper information of saliency and,
at the same time, maintain the output resolution. We
will refer to this method as D-CSRNet.
5 EXPERIMENTS AND RESULTS
In this section, we describe the experiments per-
formed to validate our approach and discuss the ob-
tained results. First, we evaluate the three adopted
general counting solutions, i.e., the segmentation-
based S-UNet, the detection-based FRCNN, and the
density-based D-CSRNet approaches, over the three
standard cell counting benchmarks described above
to verify that the obtained counting errors are compa-
rable with the ones provided by state-of-the-art cell-
specific counting methods. Then, we perform addi-
tional experiments evaluating the quality of the local-
ization of the cells, an aspect that is not taken into
account by counting metrics.
5.1 Comparison with the
State-of-the-Art
We evaluate the three adopted counting methodolo-
gies over the VGG Cells, the MBM Cells, and the
Nuclei Cells counting benchmarks described in Sec-
tion 3, and we compare their performances with other
state-of-the-art approaches. For the VGG Cells and
the MBM Cells datasets, we follow the evaluation
protocol introduced by (Lempitsky and Zisserman,
2010) and adopted by most subsequent works. Specif-
ically, we consider a testing subset fixed for all the
experiments (100 and 10 images for VGG Cells and
MBM Cells, respectively) and training and validation
subsets of varying size (N images for each subset) to
simulate lower or higher numbers of labeled exam-
ples. This evaluation protocol simulates the real sce-
nario in which scientists often have a significant vari-
ance regarding the number of available microscopy
images. Following previous work, we set N to 16, 32,
and 50 for VGG Cells and to 5, 10, 15 for MBM Cells.
Concerning the Nuclei Cells dataset, we instead use
two-fold cross-validation, with 50 images for testing,
according to (Sirinukunwattana et al., 2016) and sub-
sequent works. Following standard counting bench-
marks, we use the Mean Absolute Error (MAE) to
measure the counting performance. Specifically, it is
defined as:
MAE =
1
N
N
∑
n=1
c
n
gt
− c
n
pred
, (1)
where N is the number of test images, c
n
gt
is the ac-
tual count (i.e., the ground truth), and c
n
pred
is the pre-
dicted count of the n-th image. For the VGG Cells
and the MBM Cells, we repeat the experiment 10
times, randomly sampling ten different splits for each
configuration, and we report the mean and standard
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images
891

deviation of the MAE computed between the differ-
ent runs. On the other hand, concerning the Nuclei
dataset, we report the mean and the standard devia-
tion of the MAE calculated between the 100 images
comprising the two test splits.
Table 2 reports the obtained results. The density-
based solution performs best among the VGG Cells
dataset and, more strongly, with the Nuclei Cells
dataset, comparably to the state of the art. The other
two adopted methods, i.e., the segmentation-based S-
UNet and the detection-based FRCNN, show larger
errors, according to their intrinsic limitations when
employed in highly “crowded” scenarios with oc-
cluded objects like the VGG Cells dataset and, espe-
cially, the Nuclei Cells dataset. On the other hand,
considering the MBM Cells dataset, characterized by
challenges more related to the object shape variations,
all the approaches show competitive results, in some
cases also outperforming state-of-the-art solutions.
5.2 Localization Analysis
Although the MAE is a fair metric for establishing a
comparative in terms of counting, it can often lead to
masking erroneous estimations. The reason is that the
MAE does not take into account where the estima-
tions have been done in the images. In other words,
the MAE does not capture localization errors; mod-
els might achieve low values of MAE while provid-
ing wrong predictions (e.g., a high number of false
positives and false negatives in detection-based tech-
niques, or a bad allocation of density values in pre-
dicted maps of density-based methods). Hence, pick-
ing up the best counting model basing the decision
only on the MAE metric can lead to blunders.
In this section, we conduct experiments to assess
the ability of the three adopted solutions to local-
ize the counted cells correctly. Specifically, we con-
sider two additional metrics described in the follow-
ing paragraphs.
Grid Average Mean absolute Error (GAME).
(Guerrero-G
´
omez-Olmedo et al., 2015) is a hybrid
metric that simultaneously considers the object count
and the estimated locations of the cells. Specifi-
cally, it is computed by sub-dividing the image in 4
L
non-overlapping regions and summing the MAE com-
puted in each of these sub-regions. Formally:
GAME(L) =
1
N
N
∑
n=1
(
4
L
∑
l=1
|c
l
gt
− c
l
pred
|), (2)
where N is the total number of test images, c
l
pred
is the
estimated count in a region l of the n-th image, and
c
l
gt
is the ground truth for the same region in the same
image. The higher L, the more restrictive the GAME
metric will be. Note that the MAE can be obtained as
a particular case of the GAME when L = 0.
Mean Average Precision (mAP). is an established
metric for the localization performances of object de-
tectors. We compute the average precision for an im-
age as follows. i) We assign a score to each detected
cell in the image. Detection scores are obtained dif-
ferently for the three tested methods. For the S-UNet
model, the detection score of an object is set to the
value of the predicted segmentation map
ˆ
S at the lo-
cation of the centroid of the corresponding connected
component. For D-CSRNet, the location of an object
is set to a local peak in the predicted density map, and
its score is set to the value at its location. For FR-
CNN, the detection score is already part of the output
of the Faster-RCNN model. ii) We filter out weak
detections using a threshold on the detection scores.
iii) We match the filtered detections with the ground-
truth object positions using the Hungarian algorithm
with a constraint on the maximum accepted displace-
ment in pixels between predicted and real locations;
once matches are found, we obtain the number of true
positives (matched detection and ground-truth pairs),
false positives (unmatched detections), and false neg-
atives (unmatched ground-truth positions) locations.
iv) We repeat these steps for several threshold values
to obtain the precision-recall curve and the average
precision (i.e., the area under the curve).
In Table 3, we report the MAE, the GAME, and
the mAP metrics for all the tested solutions and the
adopted datasets. Here, we consider only the splits
having N to 50 and 15 for the VGG and the MBM
datasets, respectively, and the same two-fold cross-
validation with 50 images for testing concerning the
Nuclei dataset. Note that the density-based solution
D-CSRNet shines in the Nuclei benchmark where
very dense regions of overlapped cells are common
and strain non-density solutions, obtaining the best
counting metrics (MAE, GAME) among the tested
models. However, the denser the cells in the bench-
mark, the less the density-based solution can recover
the exact locations of the counted cells, thus achieving
lower mAP values. On the other hand, the detection-
based solution FRCNN performs sufficiently well
only when counting cells in the less crowded MBM
and VGG benchmarks. Still, it is able to recover
the exact position of more counted cells, as can be
seen from the higher mAP values obtained. Last, the
segmentation-based model sits in the middle of these
two extremes, providing intermediate counting and
localization performance.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
892

Table 2: Comparison on Standard Benchmarks. For VGG and MBM datasets, we vary the training and validation subsets
(N images for each subset), repeating the experiments 10 times. For Nuclei, we perform 2-fold cross-validation (N = 50
images per fold). Mean±st.dev. of MAE is reported.
VGG Cells (Lempitsky and Zisserman, 2010). (200 images in total - 100 test images)
Method N = 16 N = 32 N = 50
(Arteta et al., 2016a) N/A 5.06 ± 0.2 N/A
GMN (Lu et al., 2019) N/A 3.6 ± 0.3 N/A
(Lempitsky and Zisserman, 2010) 3.8 ± 0.2 3.5 ± 0.2 N/A
VGG-GAP-HR (Aich and Stavness, 2018)
∗
N/A 2.95
∗∗
2.67
SAU-Net (Guo et al., 2021) N/A N/A 2.6 ± 0.4
†
FCRN-A (Xie et al., 2016) 3.4 ± 0.2 2.9 ± 0.2 2.9 ± 0.2
‡
Count-Ception (Cohen et al., 2017) 2.9 ± 0.5 2.4 ± 0.4 2.3 ± 0.4
CCF (Jiang and Yu, 2020) 2.8 ± 0.1 2.6 ± 0.1 2.6 ± 0.1
C-FCRN+Aux (He et al., 2021) 2.3 ± 2.2
$
S-UNet (Falk et al., 2018) 8.3 ± 2.3 5.6 ± 1.1 4.5 ± 0.5
D-CSRNet (Li et al., 2018) 4.0 ± 0.2 3.2 ± 0.2 3.0 ± 0.1
FRCNN (Ren et al., 2017) 9.3 ± 0.7 8.2 ± 0.6 7.4 ± 1.0
* They did not report standard deviation. ** They used a validation subet of 100− N images. † They did not use a test
subset, but only a 100 − N images validation subset. ‡ Reported in their work as N = 64. $ They used a 5-fold cross
validation-based evaluation protocol considering the whole dataset.
MBM Cells (Kainz et al., 2015; Cohen et al., 2017). (44 images in total - 10 test images)
Method N = 5 N = 10 N = 15
(Xie et al., 2018) 36.3 ± 19.4
$
FCRN-A (Xie et al., 2016) 28.9 ± 22.6 22.2 ± 11.6 21.3 ± 9.4
(Marsden et al., 2018)
∗
23.6 ± 4.6 21.5 ± 4.2 20.5 ± 3.5
Count-Ception (Cohen et al., 2017) 12.6 ± 3.0 10.7 ± 2.5 8.8 ± 2.3
CCF (Jiang and Yu, 2020)
∗
9.3 ± 1.4 8.9 ± 0.9 8.6 ± 0.3
C-FCRN+Aux (He et al., 2021) 6.5 ± 5.2
∗∗
SAU-Net (Guo et al., 2021) N/A N/A 5.7 ± 1.2
†
S-UNet (Falk et al., 2018) 9.0 ± 1.9 7.0 ± 1.6 6.7 ± 2.5
D-CSRNet (Li et al., 2018) 10.8 ± 2.5 8.0 ± 1.3 7.0 ± 1.3
FRCNN (Ren et al., 2017) 8.8 ± 1.4 9.9 ± 1.5 8.3 ± 1.9
* They used 14 test images. ** They used a 5-fold cross validation-based evaluation protocol considering the whole dataset.
† They did not use a test subset, but only a 44 − N images validation subset.
$
They used a train/test split of 8/3 using
full-size images.
Nuclei Cells (Sirinukunwattana et al., 2016). (100 images in total - 50 test images)
Method N = 50
DeepFeat (Segui et al., 2015) 71.8 ± 51.4
(Lempitsky and Zisserman, 2010) 51.4 ± 39.8
StructRegNet (Xie et al., 2018) 45.9 ± 47.9
FCRN-A (Xie et al., 2016) 42.5 ± 33.5
Count-Ception (Cohen et al., 2017) 34.1 ± 29.0
C-FCRN+Aux (He et al., 2021) 29.3 ± 25.4
S-UNet (Falk et al., 2018) 62.4 ± 55.4
D-CSRNet (Li et al., 2018) 37.3 ± 41.0
FRCNN (Ren et al., 2017) 96.5 ± 128.0
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images
893

Table 3: Counting and Localization Performance. The MAE measures global counting performance independently of lo-
calization. The mAP summarizes localization performances in terms of precision and recall of localized cells. The GAME(L)
measure counting performance while being aware of the location of cells; the higher L, the more localization errors are penal-
ized. Regarding the VGG and the MBM datasets, we consider the splits having N to 50 and 15, respectively.
VGG Cells (Lempitsky and Zisserman, 2010). (200 images in total - 100 test images)
GAME(L) ↓
Method MAE ↓ L = 1 L = 2 L = 3 L = 4 mAP (%) ↑
S-UNet 4.5 ± 0.5 7.7 ± 1.3 12.8 ± 1.5 21.6 ± 2.4 38.0 ± 4.1 75.3 ± 15.8
D-CSRNet 3.0 ± 0.1 6.5 ± 0.2 11.3 ± 0.4 18.9 ± 0.6 28.7 ± 1.0 43.2 ± 1.6
FRCNN 7.4 ± 1.0 11.1 ± 0.9 18.3 ± 1.3 29.7 ± 2.0 43.3 ± 3.2 93.3 ± 0.6
MBM Cells (Kainz et al., 2015; Cohen et al., 2017). (44 images in total - 10 test images)
GAME(L) ↓
Method MAE ↓ L = 1 L = 2 L = 3 L = 4 mAP (%) ↑
S-UNet 6.7 ± 2.5 10.4 ± 2.5 17.3 ± 1.9 27.6 ± 2.0 40.9 ± 3.1 53.5 ± 5.3
D-CSRNet 7.0 ± 1.3 10.8 ± 1.2 16.7 ± 1.3 27.0 ± 1.6 41.5 ± 2.2 67.9 ± 1.2
FRCNN 8.3 ± 1.9 12.7 ± 2.4 20.4 ± 3.9 32.5 ± 4.7 47.2 ± 8.7 87.4 ± 1.9
Nuclei Cells (Sirinukunwattana et al., 2016). (100 images in total - 50 test images)
GAME(L) ↓
Method MAE ↓ L = 1 L = 2 L = 3 L = 4 mAP (%) ↑
S-UNet 62.4 ± 55.4 66.9 ± 51.7 75.1 ± 50.6 95.3 ± 54.1 138.4 ± 75.2 66.8 ± 11.7
D-CSRNet 37.3 ± 41.0 45.7 ± 38.8 58.2 ± 38.5 77.6 ± 39.8 100.5 ± 45.0 27.7 ± 8.5
FRCNN 96.5 ± 128.0 103.8 ± 125.3 112.6 ± 121.9 133.9 ± 118.7 168.2 ± 123.2 57.9 ± 10.8
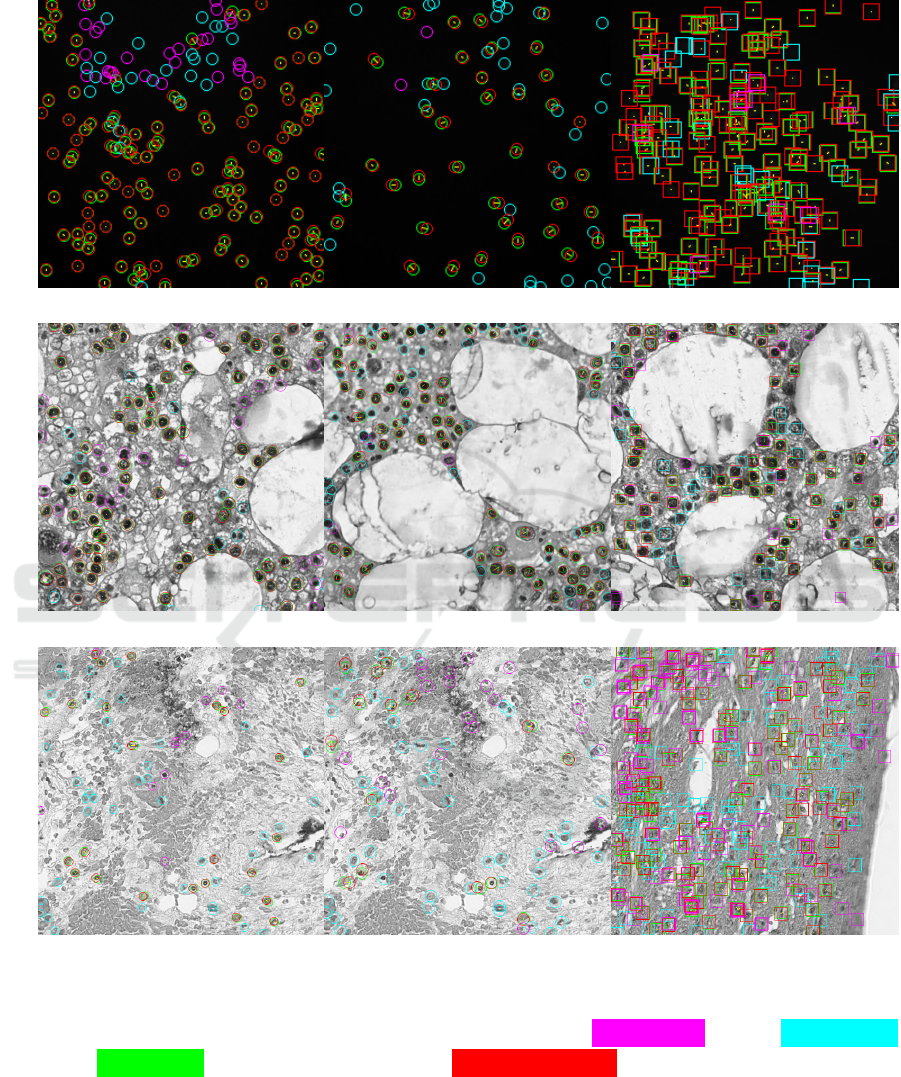
Finally, in Figure 2, we show some examples of
predictions with very low absolute counting errors
(suggesting good performance) but in which the aver-
age precision metric indicates erroneous predictions
instead. Note that in the detection-based solution
FRCNN, the disagreement between the two metrics
is less pronounced, as this methodology is usually
adopted to optimize AP. Thus, we suggest integrat-
ing the mean average precision, or at least a GAME-
L metric with a high-enough L, when optimizing and
evaluating novel cell counting solutions. We deem the
additional evaluation protocol would help practition-
ers to better characterize the performance of devel-
oped solutions.
6 CONCLUSIONS
In this work, we consider the cell counting task in
microscopy images, investigating the ability of three
general counting methodologies not only in estimat-
ing the number of the biological structures but also
in localizing them. Indeed, most state-of-the-art so-
lutions tailored to cell counting are evaluated merely
considering the difference between the predicted and
the actual number of the cells, skipping a further anal-
ysis focused on the quality of the provided estima-
tions. We show that relying only on the counting met-
rics can lead to models producing incorrect cell lo-
calization. We performed experiments on three cell
counting benchmarks, and we assessed that counting
errors do not always agree with the localization per-
formance. Thus, we suggest measuring and reporting
also the mean average precision (or at least a grid av-
erage mean absolute error) whenever possible to help
practitioners developing better models and to guide
users to choose the model most tailored to their sce-
nario.
ACKNOWLEDGEMENTS
This work was partially funded by: AI4Media -
A European Excellence Centre for Media, Society
and Democracy (EC, H2020 n. 951911), Exten-
sion (ESA, n. 4000132621/20/NL/AF), and AI-MAP
(CNR4C program) - Tuscany POR FSE 2014-2020
(CUP B15J19001040004).
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
894
S-UNet D-CSRNet FRCNN
VGG
AE = 0 AP = 36.9% AE = 2 AP = 50.8% AE = 0 AP = 88.3%
MBM
AE = 1 AP = 34.3% AE = 3 AP = 61.9% AE = 0 AP = 79.1%
Nuclei
AE = 4 AP = 36.7% AE = 1 AP = 17.4% AE = 2 AP = 43.3%
Figure 2: Absolute Error (AE) can be misleading. For each considered model (one per column), we show predictions
obtaining a low AE, but also a low Average Precision (AP) due to high numbers of false positive and false negatives. The AP
can discern cases where the MAE fails to capture poor model outputs. We indicate false positives in purple, false negatives
in cyan, and true positives in green, with the corresponding ground-truth position drawn in red and connected via a thin
yellow line. (Best viewed in electronic format.)
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images
895

REFERENCES
Aich, S. and Stavness, I. (2018). Improving object counting
with heatmap regulation. CoRR, abs/1803.05494.
Altman, S. A., Randers, L., and Rao, G. (1993). Com-
parison of trypan blue dye exclusion and fluorometric
assays for mammalian cell viability determinations.
Biotechnology Progress, 9(6):671–674.
Amato, G., Bolettieri, P., Moroni, D., Carrara, F., Ciampi,
L., Pieri, G., Gennaro, C., Leone, G. R., and Vairo, C.
(2018). A wireless smart camera network for parking
monitoring. In 2018 IEEE Globecom Workshops (GC
Wkshps), pages 1–6. IEEE.
Amato, G., Ciampi, L., Falchi, F., and Gennaro, C. (2019).
Counting vehicles with deep learning in onboard UAV
imagery. In 2019 IEEE Symposium on Computers and
Communications (ISCC), pages 1–6. IEEE.
Arteta, C., Lempitsky, V., Noble, J. A., and Zisserman,
A. (2016a). Detecting overlapping instances in mi-
croscopy images using extremal region trees. Medical
Image Analysis, 27:3–16.
Arteta, C., Lempitsky, V. S., and Zisserman, A. (2016b).
Counting in the wild. In Computer Vision - ECCV
2016, volume 9911, pages 483–498. Springer.
Boominathan, L., Kruthiventi, S. S. S., and Babu, R. V.
(2016). Crowdnet: A deep convolutional network for
dense crowd counting. In Proceedings of the 24th
ACM international conference on Multimedia, pages
640–644. ACM.
Ciampi, L., Amato, G., Falchi, F., Gennaro, C., and Ra-
bitti, F. (2018). Counting vehicles with cameras.
In Bergamaschi, S., Noia, T. D., and Maurino, A.,
editors, Proceedings of the 26th Italian Symposium
on Advanced Database Systems, Castellaneta Marina
(Taranto), Italy, June 24-27, 2018, volume 2161 of
CEUR Workshop Proceedings. CEUR-WS.org.
Ciampi, L., Gennaro, C., Carrara, F., Falchi, F., Vairo, C.,
and Amato, G. (2021a). Multi-camera vehicle count-
ing using edge-ai. CoRR, abs/2106.02842.
Ciampi, L., Santiago, C., Costeira, J., Gennaro, C., and Am-
ato, G. (2021b). Domain adaptation for traffic den-
sity estimation. In Proceedings of the 16th Interna-
tional Joint Conference on Computer Vision, Imag-
ing and Computer Graphics Theory and Applications,
pages 185–195. SCITEPRESS - Science and Technol-
ogy Publications.
Ciampi, L., Santiago, C., Costeira, J. P., Gennaro, C., and
Amato, G. (2020). Unsupervised vehicle counting
via multiple camera domain adaptation. In Saffiotti,
A., Serafini, L., and Lukowicz, P., editors, Proceed-
ings of the First International Workshop on New Foun-
dations for Human-Centered AI (NeHuAI) co-located
with 24th European Conference on Artificial Intelli-
gence (ECAI 2020), Santiago de Compostella, Spain,
September 4, 2020, volume 2659 of CEUR Workshop
Proceedings, pages 82–85. CEUR-WS.org.
Cohen, J. P., Boucher, G., Glastonbury, C. A., Lo, H. Z.,
and Bengio, Y. (2017). Count-ception: Counting by
fully convolutional redundant counting. In 2017 IEEE
International Conference on Computer Vision Work-
shops (ICCVW), pages 18–26. IEEE.
Falk, T., Mai, D., Bensch, R.,
¨
Ozg
¨
un C¸ ic¸ek, Abdulkadir,
A., Marrakchi, Y., B
¨
ohm, A., Deubner, J., J
¨
ackel, Z.,
Seiwald, K., Dovzhenko, A., Tietz, O., Bosco, C. D.,
Walsh, S., Saltukoglu, D., Tay, T. L., Prinz, M., Palme,
K., Simons, M., Diester, I., Brox, T., and Ronneberger,
O. (2018). U-net: deep learning for cell counting, de-
tection, and morphometry. Nat. Methods, 16(1):67–
70.
Fawcett, J. W., Oohashi, T., and Pizzorusso, T. (2019). The
roles of perineuronal nets and the perinodal extracel-
lular matrix in neuronal function. Nat. Rev. Neurosci.,
20(8):451–465.
Guerrero-G
´
omez-Olmedo, R., Torre-Jim
´
enez, B., L
´
opez-
Sastre, R., Maldonado-Basc
´
on, S., and O
˜
noro-Rubio,
D. (2015). Extremely overlapping vehicle counting. In
Pattern Recognition and Image Analysis, pages 423–
431. Springer International Publishing.
Guo, Y., Krupa, O., Stein, J., Wu, G., and Krishnamurthy,
A. (2021). SAU-net: A unified network for cell count-
ing in 2d and 3d microscopy images. IEEE/ACM
Trans. Comput. Biol. Bioinform., pages 1–1.
He, K., Gkioxari, G., Doll
´
ar, P., and Girshick, R. (2017).
Mask r-cnn. In Proceedings of the IEEE international
conference on computer vision, pages 2961–2969.
He, S., Minn, K. T., Solnica-Krezel, L., Anastasio, M. A.,
and Li, H. (2021). Deeply-supervised density regres-
sion for automatic cell counting in microscopy im-
ages. Medical Image Analysis, 68:101892.
Jiang, N. and Yu, F. (2020). A cell counting framework
based on random forest and density map. Appl. Sci.,
10(23):8346.
Johnston, G. (2010). Automated handheld instrument im-
proves counting precision across multiple cell lines.
BioTechniques, 48(4):325–327.
Kainz, P., Urschler, M., Schulter, S., Wohlhart, P., and Lep-
etit, V. (2015). You should use regression to detect
cells. In Lecture Notes in Computer Science, pages
276–283. Springer International Publishing.
Kotoura, Y., Yamamuro, T., Shikata, J., Kakutani, Y., Kit-
sugi, T., and Tanaka, H. (1985). A method for toxi-
cological evaluation of biomaterials based on colony
formation of v79 cells. Archives of Orthopaedic and
Traumatic Surgery, 104(1):15–19.
Laradji, I. H., Rostamzadeh, N., Pinheiro, P. O., Vazquez,
D., and Schmidt, M. (2018). Where are the blobs:
Counting by localization with point supervision. In
Computer Vision – ECCV 2018, volume 11206, pages
560–576. Springer International Publishing.
Lempitsky, V. S. and Zisserman, A. (2010). Learning to
count objects in images. In Advances in Neural Infor-
mation Processing Systems 23: 24th Annual Confer-
ence on Neural Information Processing Systems 2010,
pages 1324–1332. Curran Associates, Inc.
Li, Y., Zhang, X., and Chen, D. (2018). CSRNet: Dilated
convolutional neural networks for understanding the
highly congested scenes. In 2018 IEEE/CVF Con-
ference on Computer Vision and Pattern Recognition,
pages 1091–1100. IEEE.
Lu, E., Xie, W., and Zisserman, A. (2019). Class-agnostic
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
896

counting. In Computer Vision – ACCV 2018, pages
669–684. Springer International Publishing.
Lugagne, J.-B., Lin, H., and Dunlop, M. J. (2019). DeLTA:
Automated cell segmentation, tracking, and lineage
reconstruction using deep learning.
Marsden, M., McGuinness, K., Little, S., Keogh, C. E., and
O'Connor, N. E. (2018). People, penguins and petri
dishes: Adapting object counting models to new vi-
sual domains and object types without forgetting. In
2018 IEEE/CVF Conference on Computer Vision and
Pattern Recognition. IEEE.
O
˜
noro-Rubio, D. and L
´
opez-Sastre, R. J. (2016). Towards
perspective-free object counting with deep learning.
In Computer Vision – ECCV 2016, volume 9911,
pages 615–629. Springer International Publishing.
Paulauskaite-Taraseviciene, A., Sutiene, K., Valotka, J.,
Raudonis, V., and Iesmantas, T. (2019). Deep
learning-based detection of overlapping cells. In Pro-
ceedings of the 2019 3rd International Conference on
Advances in Artificial Intelligence, pages 217–220.
ACM.
Ren, S., He, K., Girshick, R., and Sun, J. (2017). Faster r-
CNN: Towards real-time object detection with region
proposal networks. IEEE Trans. Pattern Anal. Mach.
Intell., 39(6):1137–1149.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention - MICCAI 2015, vol-
ume 9351 of Lecture Notes in Computer Science,
pages 234–241. Springer.
Segui, S., Pujol, O., and Vitria, J. (2015). Learning to count
with deep object features. In 2015 IEEE Conference
on Computer Vision and Pattern Recognition Work-
shops (CVPRW). IEEE.
Simonyan, K. and Zisserman, A. (2015). Very deep convo-
lutional networks for large-scale image recognition. In
3rd International Conference on Learning Represen-
tations, ICLR 2015.
Sirinukunwattana, K., Raza, S. E. A., Tsang, Y.-W., Snead,
D. R. J., Cree, I. A., and Rajpoot, N. M. (2016). Lo-
cality sensitive deep learning for detection and clas-
sification of nuclei in routine colon cancer histol-
ogy images. IEEE Transactions on Medical Imaging,
35(5):1196–1206.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions. In
2015 IEEE Conference on Computer Vision and Pat-
tern Recognition (CVPR), pages 1–9. IEEE.
Xie, W., Noble, J. A., and Zisserman, A. (2016). Mi-
croscopy cell counting and detection with fully con-
volutional regression networks. Comput. methods
Biomech. Biomed. Eng. Imaging Vis., 6(3):283–292.
Xie, Y., Xing, F., Shi, X., Kong, X., Su, H., and Yang, L.
(2018). Efficient and robust cell detection: A struc-
tured regression approach. Medical Image Analysis,
44:245–254.
Yu, F. and Koltun, V. (2016). Multi-scale context aggre-
gation by dilated convolutions. In 4th International
Conference on Learning Representations, ICLR 2016.
Zhang, F., Qian, X., Si, H., Xu, G., Han, R., and Ni, Y.
(2015). Significantly improved solvent tolerance of
escherichia coli by global transcription machinery en-
gineering. Microbial Cell Factories, 14(1).
Counting or Localizing? Evaluating Cell Counting and Detection in Microscopy Images
897
