
Discussion on Comparing Machine Learning Models
for Health Outcome Prediction
Janusz Wojtusiak
a
and Negin Asadzadehzanjani
Health Informatics Program, Department of Health Administration and Policy,
George Mason University, Fairfax, VA, U.S.A.
Keywords: Machine Learning, Health Data, Model Comparison.
Abstract: This position paper argues the need for more details than simple statistical accuracy measures when comparing
machine learning models constructed for patient outcome prediction. First, statistical accuracy measures are
briefly discussed, including AROC, APRC, predictive accuracy, precision, recall, and their variants. Then,
model correlation plots are introduced that compare outputs from two models. Finally, a more detailed
analysis of inputs to the models is presented. The discussions are illustrated with two classification problems
in predicting patient mortality and high utilization of medical services.
1 INTRODUCTION
There is a significant recent growth in the use of
Machine Learning (ML) methods in medical,
healthcare and health applications. These applications
span from patient risk stratification (Tseng et al.,
2020; Beaulieu-Jones et al., 2021), to differential
diagnosis (Castellazzi et al., 2020; Vaccaro et al.,
2021), to image recognition and analysis (Rahane et
al., 2018; Saha et al. 2021). The application of ML
methods achieved incredible results that often
outperform human experts. This growing interest is
followed by strong opposition to the use of ML and
criticism of the methods. Among the most often cited
criticisms are lack of reproducibility of results
(McDermott et al., 2021), biases in constructed
models, sometimes referred to as lack of fairness
(Mehrabi, 2021), and lack of transparency.
The context of the presented work is supervised
learning from medical or health data applied to
prediction of patient outcomes. Such data can consist
of medical claims, electronic medical records,
registries, surveys, and all other types of data, but can
be applied beyond healthcare. In our 2021
HEALTHINF presentation (Wojtusiak, 2021), we
argued the need for detailed reporting of results when
presenting outcomes of ML modeling of health-
related problems. That work identified ten MLI
a
https://orcid.org/0000-0003-2238-0588
criteria for reporting results: (1) experimental design,
(2) statistical model evaluation, (3) model calibration,
(4) top predictors, (5) global sensitivity analysis, (6)
decision curve analysis, (7) global model explanation,
(8) local prediction explanation, (9) programming
interface and (10) source code. Although not always
sufficient, these criteria are argued to be necessary to
describe constructed models and allow for
reproducibility of results.
Different questions arise when one needs to
compare two or more models. Is it sufficient to
compare models based on their performance
(accuracy) only? Should one use all of the above
ten criteria to compare models? Are some
additional tests needed to understand differences
between models? And most importantly: what does
it actually mean that one model is better than
another?
There is surprisingly little literature that present
frameworks for comparing ML models. When
searching for published works on comparison of ML
models, all papers that appear are comparing specific
models (or algorithms) for solving specific problems
at hand. Virtually all of them report only some
statistical measures discussed here in Section 3.
Similarly, large number of “data science” websites
discuss practical aspects of comparing models,
including examples of source code, but also limit
these comparisons to statistical accuracy measures.
Wojtusiak, J. and Asadzadehzanjani, N.
Discussion on Comparing Machine Learning Models for Health Outcome Prediction.
DOI: 10.5220/0010916600003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 711-718
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
711

There are approaches available in other fields. Lee
and Sangiovanni-Vincentelli (1998) presented a
general framework for comparing computation
methods. While their work is applicable to the
problems presented here, they do not provide
practical insights.
The concepts presented in this paper are described
in context of classification learning problems, and
most often binary classification, but can be
generalized to multiclass problems as well as
regression.
The discussed approaches are illustrated in terms
of two supervised learning problems, which are
constructed to predict patient outcomes using medical
claims data. More specifically, medical claims for
five percent control set of the linked Medicare
beneficiaries from the Surveillance, Epidemiology,
and End Results (SEER)-Medicare dataset between
1995 and 2013 were used to construct the models.
The Medicare claims collected by Medicare and
Medicaid Services (CMS) provide one of the largest
longitudinal datasets for the Medicare eligible
population (aging population) in the United States.
Two classification problems were established for
predicting one-year mortality (Problem 1), and high
utilization of medical services (Problem 2). For both
problems, the inputs were derived from data before
year 2013 and the outcomes were calculated in 2013.
Participants in the cohort were at least 70 years old
and alive on January 1
st
, 2013. Patients’ demographic
information including age and race as well as their
diagnosis codes collected over the course of 18 years
were used to create models. In this study, diagnosis
codes in the form of nineth revision of the
International Classification of Diseases (ICD-9)
codes were transformed into 282 clinically relevant
categories using Clinical Classification Software
(CCS) codes. A binary outcome was created for
Problem 1 indicating whether or not the patient died
by the end of 2013. In Problem 2, patients who had
total number of claims exceeding the 90th percentile
in 2013 were considered high utilizers in defining the
binary outcome of high utilization.
The final dataset included 83,590 patients in
which 10% of the population were high healthcare
utilizers and about 7% of the cohort died in 2013. The
majority of patients in the cohort were white, and the
average age was 79. All patients were male.
In the presented work, the requirement (1) for
model comparison is the ability to apply models to the
same cases in order to compare results. In practice,
this means that the test sets used to evaluate models
are derived from the same database, or different
databases linked by a common identifier. For
example, one model can be built from EHR data and
another from claims data. The models can still be
compared if the EHR and claims data are linked to the
same cases using common identifier. Further, this
means that (2) the models need to be based on the
same unit of analysis, i.e., each row in the test datasets
corresponding to the same objects. Finally, (3)
outputs from the models need to be the same. For
example, when one model predicts the probability of
high utilization of medical services, the other model
must predict the same output (and not for example
time to the next hospitalization). While one can argue
about the possibility of relaxing the requirement (3)
to a certain degree for simplicity this paper assumes
that all three conditions are held.
The content of Sections 2 and 3 of this paper is
mainly informative and reflect comparisons
presented in most published works. The methods
described in Section 4 has been used by the authors
for several years but are not present in mainstream
literature. Finally, the comparisons described in
Section 5 have not been previously discussed.
2 MODEL CONSTRUCTION
The presented work focuses on models constructed by
supervised machine learning. Construction of such
models follows two main steps: data preprocessing
and application of ML algorithms as shown in Figure
1. Even small changes at any step of model
construction can have a drastic impact on the results.
The process of finding the best set of settings in model
construction is called model tuning or
hyperparameter tuning. Interestingly, most
researchers only consider hyperparameter tuning as
optimization of learning algorithm, while keeping
fixed preprocessing steps (or optimizing them
separately/manually). Changes to the representation
space resulting from data preprocessing result in very
different types of changes to the models than those
resulting from tuning hyperparameters.
Figure 1: From raw data to preprocessed data to model.
Let’s consider models M
1
, M
2
, .. M
k
that classify
or predict the same targets (i.e., the models are
intended to solve the same problem). For simplicity,
let’s assume that these models are intended for binary
HEALTHINF 2022 - 15th International Conference on Health Informatics
712
classification.
𝑀
(𝑋
)→
{
0,1
}
For each representation space 𝑋
, there needs to
be a transformation Φ
(
𝐷𝐵
)
→𝑋
from the original
data to that specific representation. That
transformation involves all the steps required to
convert raw data into one that can be directly used by
ML algorithms and models.
There are several cases to consider. When 𝑋
=
𝑋
, the same preprocessing can be applied to both
representations, and the differences in models 𝑀
and
𝑀
is in the learning algorithms or their
hyperparameters. Similarly, when 𝑋
𝑋
, different
preprocessing steps were applied to obtain the two
representations. For example, raw claims data can be
transformed into binary representation in which
presence/absence of diagnoses is represented as {0,1}
or into Temporal Min-Max representation in which
time from the first known and the most recent
occurrence of a diagnosis are represented (Wojtusiak
et al., 2021a,b).
3 STATISTICAL MEASURES
The standard in model comparison is based on
statistical accuracy measures of the models. For
classification problems, Area Under Receiver-
Operator Curve, denoted as AROC or sometimes
simply AUC (Hanley & McNeil, 1982; Fawcett,
2006) is the most frequently used measure in
biomedical and health informatics literature, followed
by predictive accuracy, recall (sensitivity) and
precision (Powers, 2020). In biomedical literature
sensitivity and specificity are typically used instead
of recall and precision. Many authors combine
precision and recall into a single F1-score (Goutte &
Gaussier, 2005). In the published literature, these
measures are used to report results of modeling
efforts, but also when performing hyperparameter
tuning to achieve the highest accuracy, AUC or F1-
score. Other metrics are also used such as Area Under
Precision-Recall Curve (AUCPR) (Boyd et al., 2013),
Kappa Statistic (McHugh, 2012), relative entropy,
mutual information and others (Baldi et al., 2000).
Most notably these measures are not typically
used by the learning algorithms whose “internal”
measure of fit is defined by a loss function. Loss
functions combine some metrics of accuracy with
additional terms used for regularization. Large
number of loss functions have been investigated in
ML
and
recently
most
often
in
the
context
of
neural
networks (Wang et al., 2020).
To exemplify these most popular measures, let’s
consider four algorithms applied to prediction of 1-
year mortality (Problem 1): Random Forest
(Breiman, 2001), Gradient Boost (Friedman, 2001;
Friedman, 2002), and Logistic Regression (Hosmer et
al., 2013), and Decision Tree (CART algorithm)
(Breiman et al., 2017; Batra & Agrawal, 2018). ROC
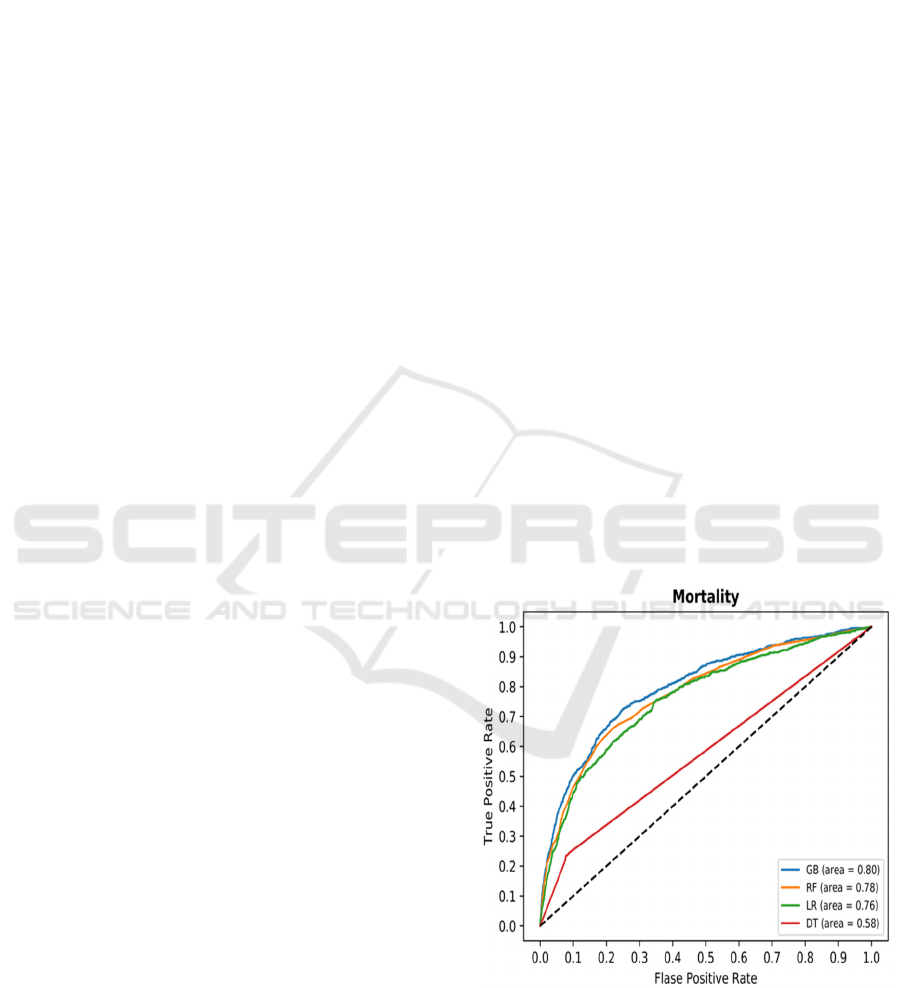
Curves for the four methods are shown in Fig. 2.
Several things are clear from the figure. The curves
for GB, RF and LR models are visually close to each
other with GB slightly dominating the other two. This
is also evident when calculating AROC for the three
models AROC(GB)=0.8, AROC(RF)=0.78, and
AROC(LR)=0.76. The differences are statistically
significant with p<0.05 for t-test over 10-fold cross-
validation. It is also clear that at certain levels of
threshold, GB and RF algorithms perform identically
when the lines overlap.
It is also clear that ROC is ill-defined and AROC
should not be calculated for decision tree-based
models that do not provide probabilistic output but
only the final decision. Literature often presents ROC
curves for decision trees or decision rules with
collected lines as in Fig 2, but this is also misleading,
as only one point exists on the curve. One can argue
that pruned DTs return scores in [0,1] range, but it is
our belief that AROC should not be used for them
either.
Figure 2: Receiver Operator Curves for four algorithms in
predicting 1-year patient mortality.
Similarly, one can compare different models
constructed with different representation spaces as
illustrated in Fig. 3. It shows ROCs for GB-based
models with Binary and Temporal Min-Max
Discussion on Comparing Machine Learning Models for Health Outcome Prediction
713
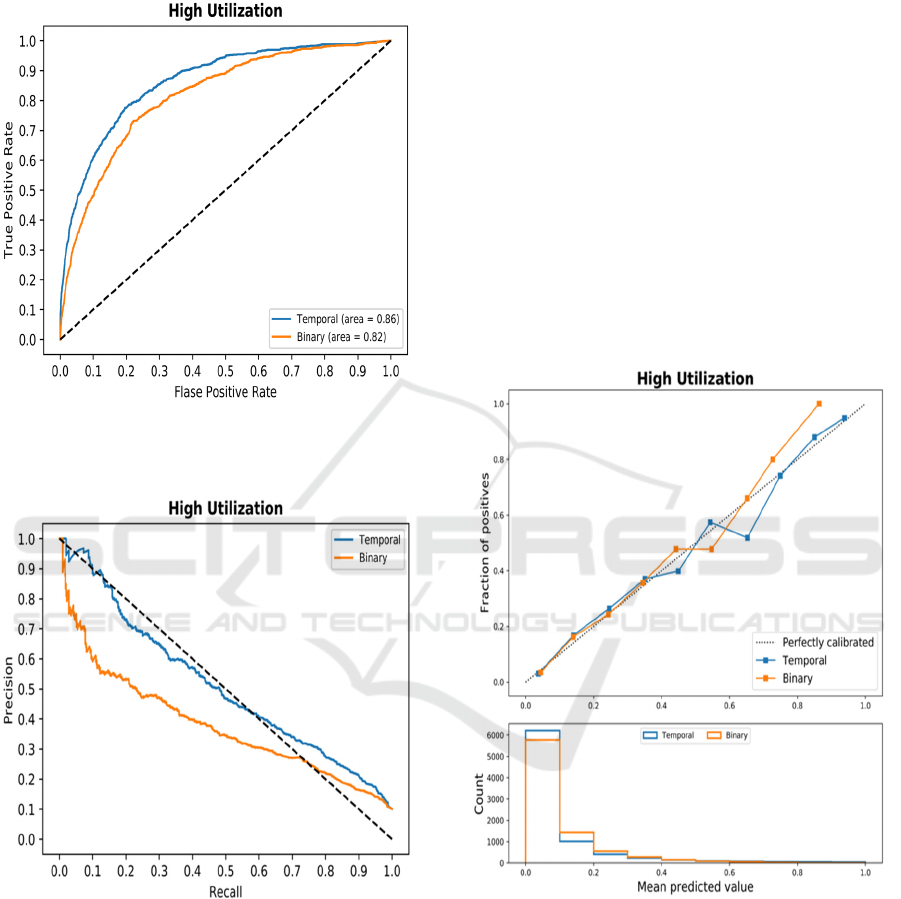
representation. Here, it is clear that the temporal
representation is superior at every possible
classification threshold.
Figure 3: Receiver Operator Curves comparing two
representation spaces in predicting high utilization of
medical services.
Figure 4: Precision-Recall Curves comparing two
representation spaces in predicting high utilization of
medical services.
Some authors argue the benefit of using Precision-
Recall Curves (PRC) and AUCPR instead of AROC.
An example of such curves is shown in Fig. 4 in
predicting high utilization (Problem 2) using GB.
Precision and recall (and F1-score) are calculated
for one specific point on the PRC, which corresponds
to a threshold that is selected by the application.
Interestingly, few works report precision and recall
for classification thresholds other than 0.5, which
makes the direct comparison of models often
impossible based on the two measures. One model
may have higher precision, while other has higher
recall. While F1-score solves the problem by
calculating a single number, it is an
oversimplification. Thus, it is suggested to fix value
of one of the measures and report the other. For
example, precision can be reported at recall of 90%
of both models, or conversely recall can be reported
at precision of 90%.
Calibration (Fig. 5) refers to the property of
models that compare their output probability to the
actual probabilities as defined by frequencies of
positive and negative examples. Numerically,
calibration is quantified with Briar score, that is mean
squared error between scores and probabilities (Flach,
2019).
Figure 5: Calibration plots for two models constructed
using Temporal Min-Max representation and binary
representation.
Finally, it is important to compare models’
performance for specific sub-populations. ROC and
PRCs as well as calibration can be investigated. The
model performance analysis on sup-population has
been popularized with the growing focus on fairness
in Artificial Intelligence (AI). In short, one needs to
check if model accuracy is comparable for sub-
HEALTHINF 2022 - 15th International Conference on Health Informatics
714
populations, especially those representing minorities
and vulnerable populations.
4 OUTPUT COMPARISON
Comparing calibration of models gives good insight
about their outputs and allows for some comparison
at different output levels. However, neither
calibration analysis nor any of the other statistical
methods described in Section 2 provide case-by-case
insights into models’ performance.
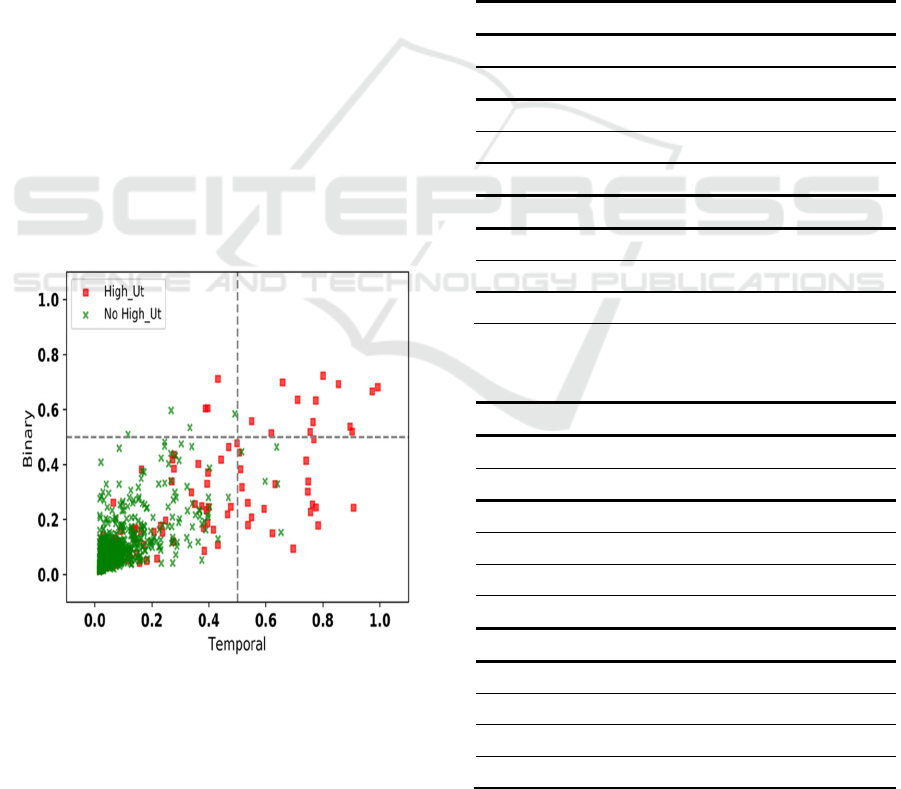
Model Correlation Plots (MCP) allow for visual
case-by-case comparison of model outputs
(Wojtusiak et al., 2017). The MCPs are scatterplots
with axis corresponding to outputs of two models, and
points representing individual cases (i.e., patients) for
which predictions are made. If two models are
identical, all points are located at the diagonal and
M1(x) = M2(x) for every x in dataset (training or
testing). Further, MCPs encode true class by color or
symbol as shown in Fig. 6, which in this case red
points represent high utilizers and green non-high
utilizers, and may include regression lines.
The corresponding aggregated data are presented
in Table 1 for both mortality and high utilization
prediction problems. Wilcoxon signed-rank test was
used to compare the results and significance in
differences is indicated in Table 1.
Figure 6: Model Correlation Plot that compares two
Gradient Boost models based on Binary and Temporal Min-
Max representations in predicting high utilization of
medical services.
When observing values in Fig 6, it is clear that
both sets are nonempty with large number of cases. It
can be also observed that the Temporal Min-Max-
based model tends to produce overall higher scores
for positive test cases. The average output probability
among cases with positive labels was significantly
higher across Temporal Min-Max representation-
based models (also shown in Table 1). Overall, higher
probabilities of positive labels and lower probability
of negative labels in Temporal representation suggest
that this method is generally more likely to correctly
classify both classes. Also, higher recall in Temporal
representation of diagnoses suggests that the method
allows the algorithms to select more positive cases
that are missed in Binary representation method, thus
leading to overall higher recall (Table 2).
Table 1: Comparison of output probability for Temporal
Min-Max (Tem) vs. Binary (Bin) Representations. *
indicates significance (p<0.05).
Problem 1 - Mortality
Positive Label Negative Label
Alg Temp Bin Temp Bin
RF .1859
*
.1459 .0758
*
.0765
GB .1883
*
.1452 .0637
*
.0674
LR .1579
*
.1487 .0665
*
.0673
Problem 2 – High Utilization
RF .3040
*
.2300 .0879
*
.0946
GB .3282
*
.2385 .0748
*
.0850
LR .2680
*
.2414 .0817
*
.0848
Table 2: Comparison of the number correct classified cases
for Temporal Min-Max vs. Binary representations.
Problem 1 - Mortality
Positive Label Negative Label
Alg Temp Bin Temp Bin
RF 143 8 16 75
GB 465 33 67 348
LR 99 28 78 159
DT 980 715 5244 4767
Problem 2 – High Utilization
RF 1115 118 174 417
GB 1443 196 418 811
LR 524 167 240 439
DT 1902 1114 6133 5031
Let’s consider two models 𝑀
and 𝑀
. Let 𝑇𝑆
be
Discussion on Comparing Machine Learning Models for Health Outcome Prediction
715

a set of examples from the test set TS such that
⋁
𝑀
(𝑥) > 𝑀
(𝑥) + 𝜀
∈
. Similarly, one can
define 𝑇𝑆
as
⋁
𝑀
(𝑥) < 𝑀
(𝑥) − 𝜀
∈
. The sets
𝑇𝑆
and 𝑇𝑆
define points for which the models
produce higher scores retrospectively. Instead of
counting correct/incorrect cases or calculating
precision and recall, one can compare examples in
sets TS
1
and TS
2
. This analysis can be further stratified
by patient demographics and plots constructed
separately for genders, races, age groups, or different
diagnoses. On aggregate, this is equivalent to
calculating Mean Squared Error or Mean Absolute
Error from the 0/1 classification methods.
5 INPUT COMPARISON
Comparing model outputs allows for visually or
statistically inspecting differences between models
on individual cases. However, one needs to
understand if there are any patterns within input
values that correspond to differences in outputs of the
compared models. In other words, are there patterns
in input values that correspond to outputs visualized
in model correlation plots? The patterns can be
described in terms of attributes present in the data or
derived from them.
Let 𝑇𝑆
be a set of positive cases in testing set TS
and 𝑇𝑆
be a set of negative cases in TS. Let’s
consider now four subsets of the testing set.
𝐶𝑃𝑀
={𝑥∈𝑇𝑆
: 𝑀
(𝑥) ≥ 𝜏 ∧ 𝑀
(𝑥) < 𝜏}
𝐶𝑁𝑀
={𝑥∈𝑇𝑆
: 𝑀
(𝑥) < 𝜏 ∧ 𝑀
(𝑥) ≥ 𝜏}
𝑆𝑃𝑀
={𝑥∈𝑇𝑆
: 𝑀
(𝑥) ≥ 𝑀
(𝑥) + 𝜀}
𝑆𝑁𝑀
={𝑥∈𝑇𝑆
:𝑀
(
𝑥
)
<𝑀
(
𝑥
)
−𝜀}
𝐶𝑃𝑀
and 𝐶𝑁𝑀
are respectively positive and
negative cases correctly classified by model 𝑀
but
not 𝑀
. 𝑆𝑃𝑀
and 𝑆𝑁𝑀
are positive and negative
cases better classified by model 𝑀
. These four sets
can be compared in terms of values of input attributes.
The sets 𝐶𝑃𝑀
, 𝐶𝑁𝑀
, 𝑆𝑃𝑀
, and 𝑆𝑃𝑀
are defined
analogously for results superior by model 𝑀
.
For example, let’s consider again the two
problems of predicting mortality and high utilization
for medical services. Let 𝑀
be model based on
Temporal Min-Max representation and 𝑀
model
based on standard Binary representation. One simple
way to assess patient risk for both problems is a
simple count of underlying medical conditions
(present diagnosis codes). For all cases correctly
classified by one model but incorrectly by another,
what are the counts of present medical conditions.
These numbers are reported in Table 3. Note the
extremely small value 1.1 for Binary representation
in random forest classifier. That number is due to very
small number of cases correctly classified when using
Binary representation and incorrectly with Temporal
Min-Max representation. Similarly, Table 4 presents
values for cases better predicted by one of the models.
Table 3: Comparison of the number of present codes for
Temporal Min-Max vs. Binary representations for correct
predictions.
Problem 1 - Mortality
Positive Label
TS
+
Negative Label
TS
-
Alg
Temp
𝐶𝑃𝑀
Bin
𝐶𝑃𝑀
Temp
𝐶𝑁𝑀
Bin
𝐶𝑁𝑀
RF 83.0
*
1.1 10.5
*
77.3
GB 77.9 87.0 85.8
*
78.5
LR 80.0 86.5 84.9
*
78.8
DT 68.9
*
65.0 57.8
*
59.5
Problem 2 – High Utilization
RF 85.3
*
103.7 101.5
*
88.9
GB 79.7
*
97.0 96.8
*
80.8
LR 85.8
*
99.7 98.1
*
85.6
DT 76.9
*
78.6 63.6
*
64.0
Table 4: Comparison of the number of present codes for
Temporal Min-Max vs. Binary representations for superior
predictions.
Problem 1 - Mortality
Positive Label
TS
+
Negative Label
TS
-
Alg
Temp
𝑆𝑃𝑀
Bin
𝑆𝑃𝑀
Temp
𝑆𝑁𝑀
Bin
𝑆𝑁𝑀
RF 60.68
*
52.52 46.25
*
50.13
GB 60.52
*
52.92 47.71
*
46.23
LR 57.35 57.08 49.19
*
45.13
DT 68.46
*
64.24 56.89
*
58.49
Problem 2 – High Utilization
RF 71.74
*
70.50 47.24
*
50.83
GB 70.70 70.99 49.22
*
39.08
LR 69.72
*
72.65 44.79
*
45.71
DT 76.84
*
78.54 62.85 63.19
Diving deeper into comparison of the models
constructed with Binary and Temporal Min-Max
representations, it is possible to compare the actual
HEALTHINF 2022 - 15th International Conference on Health Informatics
716

values within cases based on these representations.
Temporal Min-Max representation has more
information than binary representation, and more
specifically the numbers of days. Thus, a reasonable
comparison is one that looks at numbers of days from
diagnoses for cases correctly classified by either of
the models (but not both) as shown in Table 5, or
better predicted by either of the models as shown in
Table 6. The structure of these tables is analogous to
the Tables 3 and 4, but values are average numbers of
days between diagnosis and prediction.
Depending on the actual types of models
constructed and the representation spaces used, one
needs to design appropriate comparisons on input
values. These depend on the nature of data, such as
static or temporal, types of input attributes, numbers
of attributes, and types of models used.
Table 5: Comparison of the average number of days for
Temporal Min-Max vs. Binary representations for correct
predictions.
Problem 1 - Mortality
Positive Label
TS
+
Negative Label
TS
-
Alg
Temp
𝐶𝑃𝑀
Bin
𝐶𝑃𝑀
Temp
𝐶𝑁𝑀
Bin
𝐶𝑁𝑀
RF 1259.6* 2599.1 2405.3
*
1243.4
GB 1353.6 1413.3 1898.0
*
1380.5
LR 1181.7
*
1906.9 2608.1
*
1193.3
DT 1595.1
*
1680.8 1981.4
*
1838.6
Problem 2 – High Utilization
RF 1432.5
*
1913.5 2070.2
*
1555.0
GB 1468.2
*
1754.1 1834.7
*
1525.5
LR 1184.2
*
2195.7 2362.4
*
1213.2
DT 1579.9
*
1669.6 1944.5
*
1754.2
Another possible way to compare the models is to
construct a model C
M1,M2
that predicts when one
models is significantly better than the other, that is to
classify 𝑆𝑃𝑀
and 𝑆𝑁𝑀
against 𝑆𝑃𝑀
and 𝑆𝑁𝑀
.
Alternatively, 𝑆𝑃𝑀
against 𝑆𝑃𝑀
can be built if one
is concerned only with positive cases. AROC > 0.5
for such a model indicates a pattern on cases for
which one model dominates the other. Note such a
model does not tell us if either of the models 𝑀
or
𝑀
are correct, but rather which is more likely to
produce a better result for a given case. Typically, one
should prefer CM1,M2 to be an easy to interpret
model such as logistic regression or decision tree so
that conclusions can be drawn from the discovered
patterns.
Table 6: Comparison of the average number of days for
Temporal Min-Max vs. Binary representations for superior
predictions.
Problem 1 - Mortality
Positive Label
TS
+
Negative Label
TS
-
Alg
Temp
𝑆𝑃𝑀
Bin
𝑆𝑃𝑀
Temp
𝑆𝑁𝑀
Bin
𝑆𝑁𝑀
RF 1641.26
*
1771.76 1920.35
*
1816.88
GB 1549.92
*
1896.64 2004.06
*
1699.10
LR 1510.49
*
1933.96 2103.39
*
1643.55
DT 1600.81
*
1682.96 1977.14
*
1842.69
Problem 2 – High Utilization
RF 1538.88
*
1795.15 2048.76
*
1644.65
GB 1497.31
*
1833.94 2055.30
*
1587.85
LR 1338.79
*
2082.75 2188.95
*
1459.63
DT 1579.53
*
1669.15 1942.51
*
1755.21
6 CONCLUSIONS
Model comparison is not a trivial task. Through
comparisons, one can select the best model, but also
gain useful insight into why specific models perform
differently than others. It is clear that comparing
models based on accuracy alone is not sufficient.
This position paper was not intended to present
the best models or results for specific problems, nor
to argue for the use of specific modeling methods.
The two examples were used only for informative
purposes and could be replaced with any other cases.
The paper did also not mean to provide a full set of
metrics and tasks to be performed in comparing
models. Selection of the specific metrics should be
done carefully to gain maximum insights into the
application area. Instead, this paper was intended to
contribute to an ongoing discussion on model
evaluation and comparison. With ML field being
dominated with statistical methods, researchers often
assume that statistical model evaluation and
comparison are sufficient. This cannot be further
from the truth, especially in the medical or health care
fields in which every “test case” is a patient whose
treatments and potentially life-altering decisions may
be made based on predictions.
While presented in a different order, the presented
methodology fits into a general framework used by
Discussion on Comparing Machine Learning Models for Health Outcome Prediction
717

the authors in which models are described by their
inputs, models themselves and their outputs. The
framework can be used to study model performance,
explainability, fairness, and other factors that may
ultimately lead to end users’ trust and model
adoption. It emphasizes the concept of machine
learning that makes sense, in which the application of
machine learning results in correctly constructed and
evaluated models for which inputs mimic measurable
real-world characteristics or modeled objects, and
outputs directly correspond to outcomes of interest.
REFERENCES
Baldi, P., Brunak, S., Chauvin, Y., Andersen, C. A., & Nielsen,
H. (2000). Assessing the accuracy of prediction algorithms
for classification: an overview. Bioinformatics, 16(5), 412-
424.
Batra, M., & Agrawal, R. (2018). Comparative analysis of
decision tree algorithms. In Nature inspired computing (pp.
31-36). Springer, Singapore.
Beaulieu-Jones, B. K., Yuan, W., Brat, G. A., Beam, A. L.,
Weber, G., Ruffin, M., & Kohane, I. S. (2021). Machine
learning for patient risk stratification: standing on, or looking
over, the shoulders of clinicians?. NPJ digital medicine, 4(1),
1-6.
Boyd, K., Eng, K. H., & Page, C. D. (2013, September). Area
under the precision-recall curve: point estimates and
confidence intervals. In Joint European conference on
machine learning and knowledge discovery in databases (pp.
451-466). Springer, Berlin, Heidelberg.
Breiman, L. (2001). Random forests. Machine learning, 45(1), 5-
32.
Breiman, L., Friedman, J. H., Olshen, R. A., & Stone, C. J. (2017).
Classification and regression trees. Routledge.
Castellazzi, G., Cuzzoni, M. G., Cotta Ramusino, M., Martinelli,
D., Denaro, F., Ricciardi, A., ... & Gandini Wheeler-
Kingshott, C. A. (2020). A machine learning approach for the
differential diagnosis of Alzheimer and Vascular Dementia
Fed by MRI selected features. Frontiers in
neuroinformatics, 14, 25.
Fawcett, T. (2006). An introduction to ROC analysis. Pattern
recognition letters, 27(8), 861-874.
Flach, P. (2019, July). Performance evaluation in machine
learning: the good, the bad, the ugly, and the way forward. In
Proceedings of the AAAI Conference on Artificial
Intelligence (Vol. 33, No. 01, pp. 9808-9814).
Friedman, J. H. (2001). Greedy function approximation: a
gradient boosting machine. Annals of statistics, 1189-1232.
Friedman, J. H. (2002). Stochastic gradient boosting.
Computational statistics & data analysis, 38(4), 367-378.
Goutte, C., & Gaussier, E. (2005, March). A probabilistic
interpretation of precision, recall and F-score, with
implication for evaluation. In European conference on
information retrieval (pp. 345-359). Springer, Berlin,
Heidelberg.
Hanley, J. A., & McNeil, B. J. (1982). The meaning and use of
the area under a receiver operating characteristic (ROC)
curve. Radiology, 143(1), 29-36.
Hosmer Jr, D. W., Lemeshow, S., & Sturdivant, R. X. (2013).
Applied logistic regression (Vol. 398). John Wiley & Sons.
McDermott, M. B., Wang, S., Marinsek, N., Ranganath, R.,
Foschini, L., & Ghassemi, M. (2021). Reproducibility in
machine learning for health research: Still a ways to go.
Science Translational Medicine, 13(586).
McHugh, M. L. (2012). Interrater reliability: the kappa statistic.
Biochemia medica, 22(3), 276-282.
Mehrabi, N., Morstatter, F., Saxena, N., Lerman, K., & Galstyan,
A. (2021). A survey on bias and fairness in machine learning.
ACM Computing Surveys (CSUR), 54(6), 1-35.
Lee, E. A., & Sangiovanni-Vincentelli, A. (1998). A framework
for comparing models of computation. IEEE Transactions on
computer-aided design of integrated circuits and systems,
17(12), 1217-1229.
Powers, D. M. (2020). Evaluation: from precision, recall and F-
measure to ROC, informedness, markedness and correlation.
arXiv preprint arXiv:2010.16061.
Rahane, W., Dalvi, H., Magar, Y., Kalane, A., & Jondhale, S.
(2018, March). Lung cancer detection using image
processing and machine learning healthcare. In 2018
International Conference on Current Trends towards
Converging Technologies (ICCTCT) (pp. 1-5). IEEE.
Saha, P., Sadi, M. S., & Islam, M. M. (2021). EMCNet:
Automated COVID-19 diagnosis from X-ray images using
convolutional neural network and ensemble of machine
learning classifiers. Informatics in medicine unlocked, 22,
100505.
Tseng, Y. J., Wang, H. Y., Lin, T. W., Lu, J. J., Hsieh, C. H., &
Liao, C. T. (2020). Development of a machine learning
model for survival risk stratification of patients with advanced
oral cancer. JAMA network open, 3(8), e2011768-e2011768.
Vaccaro, M. G., Sarica, A., Quattrone, A., Chiriaco, C., Salsone,
M., Morelli, M., & Quattrone, A. (2021).
Neuropsychological assessment could distinguish among
different clinical phenotypes of progressive supranuclear
palsy: A Machine Learning approach. Journal of
Neuropsychology, 15(3), 301-318.
Wang, Q., Ma, Y., Zhao, K., & Tian, Y. (2020). A comprehensive
survey of loss functions in machine learning. Annals of Data
Science, 1-26.
Wojtusiak, J., Elashkar, E., & Nia, R. M. (2017, February). C-
Lace: Computational Model to Predict 30-Day Post-
Hospitalization Mortality. HEALTHINF 2017 (pp. 169-177).
Wojtusiak J. Reproducibility, Transparency and Evaluation of
Machine Learning in Health Applications. HEALTHINF
2021 (pp. 685-692).
Wojtusiak, J., Asadzaehzanjani, N., Levy, C., Alemi, F., &
Williams, A. E. (2021). Online Decision Support Tool that
Explains Temporal Prediction of Activities of Daily Living
(ADL). HEALTHINF 2021 (pp. 629-636).
Wojtusiak, J., Asadzadehzanjani, N., Levy, C., Alemi, F., &
Williams, A. E. (2021). Computational Barthel Index: an
automated tool for assessing and predicting activities of daily
living among nursing home patients. BMC medical
informatics and decision making, 21(1), 1-15.
HEALTHINF 2022 - 15th International Conference on Health Informatics
718
