
Classification of Histopathological Images of Penile Cancer using
DenseNet and Transfer Learning
Marcos Gabriel Mendes Lauande
1
, Amanda Mara Teles
2
, Leandro Lima da Silva
2
,
Caio Eduardo Falc
˜
ao Matos
1
, Geraldo Braz J
´
unior
1
, Anselmo Cardoso de Paiva
1
,
Jo
˜
ao Dallyson Sousa de Almeida
1
, Rui Miguel Gil da Costa Oliveira
2,3
, Haissa Oliveira Brito
2
,
Ana Gis
´
elia Nascimento
4
, Ana Clea Feitosa Pestana
2,4
, Ana Paula Silva Azevedo dos Santos
2
and Fernanda Ferreira Lopes
2
1
Computer Applied Group (NCA), Federal University of Maranh
˜
ao (UFMA), S
˜
ao Lu
´
ıs - MA, Brazil
2
Graduate Program in Adult Health - PPGSAD, Federal University of Maranh
˜
ao (UFMA) S
˜
ao Lu
´
ıs - MA, Brazil
3
Centre for Research and Technology of Agro-Environmental and Biological Sciences (CITAB), Inov4Agro,
University of Tr
´
as-os-Montes and Alto Douro (UTAD), Vila Real, Portugal
4
Department of Pathology, Presidente Dutra University Hospital - Federal University of Maranh
˜
ao (UFMA),
S
˜
ao Lu
´
ıs - MA, Brazil
marcos.lauande, geraldo.braz, anselmo.paiva, joao.dallyson, rui.costa, haissa.brito, ana.giselia, ana.pestana, ana.azevedo,
Keywords:
Histopathology, Penile Cancer, Deep Learning, Deep Features, Convolutional Neural Network, Transfer
Learning, Data Augmentation, Contrast Limited Adaptative Histogram Equalization.
Abstract:
Penile cancer is a rare tumor that accounts for 2% of cancer cases in men in Brazil. Histopathological analyzes
are commonly used in its diagnosis, making it possible to assess the degree of the disease, its evolution, and its
nature. About a decade ago, scientific works in the field of deep learning were developed to help pathologists
make decisions quickly and reliably, opening up possibilities for new contributions to improve such a complex
and time-consuming activity for these professionals. In this work, we present the development of a method
that uses a DenseNet to diagnose penile cancer in histopathological images, and the construction of a dataset
(via the Legal Amazon Penis Cancer Project) used to validate this method. In the experiments performed, an
F1-Score of up to 97.39% and a sensitivity of up to 98.33% were achieved in this binary classification problem
(normal or squamous cell carcinoma).
1 INTRODUCTION
Cancer is related to disordered cell growth. Depend-
ing on the degree of malignancy, it can invade adja-
cent tissues or organs, leading the patient to death if
there is no adequate early treatment. Penile cancer
is a rare tumor and represents 0.4% to 0.6% of all
cancers in Europe and North America but is consid-
erably more common in developing countries in Latin
America, Africa, and Asia (Douglawi and Masterson,
2017). According (INCA, 2021), this disease has a
higher incidence in men aged 50 years and over in
Brazil, being more common in the North and North-
east regions of the country and corresponding to 2%
of cancer cases in men. A report from the state of
Maranh
˜
ao, Brazil, indicates an age-standardized inci-
dence rate of 6.15 per 100,000 (Coelho et al., 2018),
which is very worrying. This type of tumor is linked
to some factors, such as lack of hygiene, human papil-
lomavirus (HPV) infection, the presence of phimosis,
and risky sexual behavior (Vieira et al., 2020).
According to (ACS, 2021), and (Thomas et al.,
2021), one of the exams that can be indicated for the
diagnosis of this disease is the histopathological anal-
ysis of tissues collected through biopsy, which con-
sists of the microscopic evaluation of very fine tis-
sues extracted from the region of interest. Before be-
ing taken for analysis, these tissues are stained using
Eosin and Hematoxylin, then placed on glass slides
(Neto, 2012). The pathologist verifies the structure
of tissue cells understanding the evolution, subtype,
and extent of the disease, making it possible to make
safer decisions about the type of treatment or surgery
to be prescribed. However, according to (Melo et al.,
2020), this activity tends to be very complex and time-
976
Lauande, M., Teles, A., Lima da Silva, L., Matos, C., Braz Júnior, G., Cardoso de Paiva, A., Sousa de Almeida, J., Oliveira, R., Brito, H., Nascimento, A., Pestana, A., Santos, A. and Lopes, F.
Classification of Histopathological Images of Penile Cancer using DenseNet and Transfer Learning.
DOI: 10.5220/0010893500003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
976-983
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

consuming because the process is detailed and has
a subjective conclusion, depending a lot on the pro-
fessional’s expertise. Because of this, software-based
solutions can help these professionals, bringing more
reliability, enabling a faster diagnosis, and the sooner
the disease is detected and together with adequate
treatment, the greater the probability of cure.
With the evolution of machine learning techniques
and image processing applied to medical images, sev-
eral related scientific research based on diagnosis
using histopathological images has been developed
(Srinidhi et al., 2021), such as for breast cancer (Cruz-
Roa et al., 2014) , prostate (Linkon et al., 2021), liver
(Kiani et al., 2020), colon (Sarwinda et al., 2021) and
lung (Wei et al., 2019). Thus, deep learning archi-
tectures, specifically convolutional neural networks
(CNNs), ended up becoming quite popular for this
type of problem. This popularity was due to its perfor-
mance, in certain cases, superior to traditional meth-
ods of feature extraction and classifiers not based on
neural networks; this can be seen in the results ob-
tained in (Spanhol et al., 2016a), for example. How-
ever, concerning cancer diagnosis based on images
at cellular levels of the penile tissue, the lack of a
dataset that can support the researchers’ experiments
becomes an impediment to the development of spe-
cialized methods.
In order to fill this gap, this article proposes an au-
tomated penile cancer diagnosis method that analyzes
the histopathological images and determines whether
there is cancer or not. This method is based on
transfer learning that uses a pre-trained DenseNet-201
type convolutional network for the ImageNet chal-
lenge with an image preprocessing step using an algo-
rithm known as CLAHE (Contrast Limited Adaptive
Histogram Equalization). In addition, the developed
method was validated in a dataset built through the
Legal Amazon Penis Cancer Project, comprising 194
histopathological images classified as normal or can-
cer (squamous cell carcinoma).
Our contributions are listed below: (a) the propo-
sition of a new method for automated diagnosis of pe-
nile cancer based on deep learning using microscope
images; (b) the construction of a new dataset of penile
cancer histopathological images that served to vali-
date the developed method; and (c) analysis and com-
parison of the proposed method with another one built
by the same author based on deep features and tradi-
tional classifiers.
This paper’s remainder is organized as follows:
Section II presents the related works that served as
the basis for the research that resulted in this paper,
Section III explains how the histopathological im-
ages of penile cancer were acquired and details about
the proposed method based on deep learning, Sec-
tion IV, which presents an analysis of the results ob-
tained through the method and other experiments car-
ried out; and, finally, Section V, which concludes the
work with some final considerations.
2 RELATED WORKS
Computer recognition of medical images, specifically
cancer histopathological images, is a widely explored
research topic, with many works available to obtain
theoretical foundations that served as the basis for
the entire development of a methodology and experi-
ments.
In (Filipczuk et al., 2013), the Hough Transform
was used to detect cell nuclei in histopathological
images, and these had their characteristics extracted
and used as input for an SVM (Support Vector Ma-
chine) classifier. In work described in (Spanhol et al.,
2016b), feature extraction techniques such as Lo-
cal Binary Patterns, Gray-Level Co-Occurrence Ma-
trices, and Local Phase Quantization were used in
several experiments with traditional classifiers in a
dataset consisting of 7,909 breast cancer images.
Over time, traditional feature extraction tech-
niques were replaced by techniques based on deep
learning for this type of problem, with better results.
In (Sharma et al., 2017), a CNN architecture was
constructed and applied for cancer classification and
necrosis detection using a gastric cancer histopatho-
logical image dataset. This work showed superior
performance compared to the Random Forest classi-
fier.
The use of preprocessing techniques is essential
to improve image classification task performance. A
work developed by (Sarwinda et al., 2021) demon-
strated the use of the CLAHE algorithm together with
the ResNet-18 and ResNet-50 neural networks for a
binary classification problem (malignant or benign).
In this case, a database of large intestine tissue im-
ages had its samples converted to grayscale and pre-
processed using the CLAHE algorithm, substantially
improving the classification results.
The transfer of learning technique in the classifi-
cation of histopathological images has become very
promising and popular, especially in cases where ex-
periments are restricted to less powerful computers
and datasets with few samples. This procedure con-
sists of using pre-trained networks in a given do-
main in a similar or different one. The application of
this technique can be seen in (Spanhol et al., 2017),
(Boumaraf et al., 2021) and (Choudhary et al., 2021),
and in this last work cited, the pre-trained network
Classification of Histopathological Images of Penile Cancer using DenseNet and Transfer Learning
977

had its less important weights removed, improving the
overall result of the model. In addition, data augmen-
tation can be used to circumvent the sample quan-
tity limitation, the class imbalance, and the overfit-
ting problem, which can considerably affect the per-
formance of deep learning techniques, as they per-
form better on image bases with thousands or mil-
lions of samples, such as ImageNet (Russakovsky
et al., 2015). This technique causes the images to
undergo some kind of transformation, thus produc-
ing new samples, works such as those reported in
(Rakhlin et al., 2018) and (Tellez et al., 2019) showed
very promising results when using it.
Finally, these works are a sample of the growing
diversity of software-based studies with the purpose
of automating the activity of analyzing histopatholog-
ical images that represent state of the art in this field
of research, being the foundation for the application
of a specialized diagnostic method in a type of cancer
image not explored in the literature.
3 MATERIALS AND METHOD
This section details the construction process of the
dataset of penile cancer histopathological images
used and the proposed method (Figure 1) that has
its following steps: image acquisition; image prepro-
cessing using the CLAHE (Contrast Limited Adaptive
Histogram Equalization) algorithm; transfer learn-
ing and classification using an ImageNet pre-trained
model based on the DenseNet convolutional network
architecture; evaluation of the results on the subset of
test image.
3.1 Histopathological Images Dataset
Construction
The dataset of images provided by the Legal Ama-
zon Penis Cancer Project, which served to validate the
proposed method, consists of 194 RGB images with
a resolution of 2048x1536 pixels. These files were
grouped by magnification and pathological classifica-
tion according to Table 1.
Table 1: Distribution of images according to magnification
and pathological classification.
Category/Magnification 40X 100X
Normal 40 40
Cancer (squamous cell carcinoma) 57 57
Total 97 97
The image capture process was carried out in
2021 using penile tissue samples representing tumors
and adjacent non-tumor areas, stained with hema-
toxylin and eosin stored at the Maranh
˜
ao Tumour and
DNA Biobank. Two graduate students photographed
the samples using a high-definition camera (Leica
ICC50 HD) coupled to a brightfield microscope (Le-
ica DM500). With the aid of specific software (Leica
Aperio ImageScope), the images were analyzed and
classified by two pathologists as penile cancer or non-
tumor tissue, according to the international classifica-
tion of penile tumors (Epstein et al., 2020). Some
examples of these images are shown in Figure 2.
3.2 Preprocessing
Before training or testing the model based on convo-
lutional neural networks, we verified that the images
had differences in light distribution. To minimize the
effect, we propose a preprocessing via CLAHE (Con-
trast Limited Adaptive Histogram Equalization) algo-
rithm presented in (Zuiderveld, 1994).
According to (Kumar and Shaik, 2015) CLAHE
is an evolution of Adaptive Histogram Equalization
(AHE) and its most basic precursor, Histogram Equal-
ization (HE). Created for medical images, this algo-
rithm is easy to implement and has good results when
applied to microscopic images that are most often af-
fected by low lighting effects at the time of acquisition
(Kumar and Shaik, 2015). The main difference com-
pared to its predecessors is in the way the histogram
equalization is applied, not being on the whole image
at once but on regions of adjustable size. The image
is divided into blocks that are adaptively equalized,
then combined and treated to avoid edge effects using
bilinear interpolation.
In this work, initially, the images are resized to
224x224 pixels and converted from RGB to YUV.
This transformation is necessary because all channels
in RGB space carry color information. Therefore, un-
desirable effects on image colors would be noticeable
after the equalization of these channels. Thus, the
CLAHE algorithm was applied to the Y channel of
the YUV color space (Vill
´
an, 2019). This channel
represents the intensity information that needs to be
improved. After this operation, the equalized images
were converted to RGB. An example of this technique
can be seen in Figure 3, where it is possible to see how
evident the cellular structures of the tissue are after
the operation.
3.3 Transfer Learning with
Convolutional Networks
Convolutional Neural Networks are special architec-
tures inspired by the biological mechanisms of vi-
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
978

Figure 1: Proposed method.
Figure 2: Examples of histopathological images of penile cancer by category and magnification.
Figure 3: This example demonstrates the application of the
CLAHE enhancement algorithm on a histopathological im-
age of penile cancer.
sion of living beings, being used in classification,
segmentation and object recognition tasks, for exam-
ple. What makes them different from other neural
network-based architectures are convolution opera-
tions in some of their layers. Still, in a complementary
way, their structure is usually composed of other lay-
ers, such as pooling and fully connected layers. The
first publication of this type of neural network was
made by (Lecun et al., 1998). Still, they only became
popular through the work of (Krizhevsky et al., 2012)
for the 2012 ImageNet challenge as commented on in
(Zhang et al., 2021), after that, many other new archi-
tectures emerged, being widely applied in the context
of medical image analysis.
The complete training of a CNN started with ran-
dom weights depends on an extremely large amount
of images and many computational resources to pro-
cess them. The quantity of samples tends to be a
problem in the case of medical images, as there is
an entire technical procedure to acquire this data in a
correct and controlled manner to produce quality arti-
facts. Furthermore, there is another factor which is the
availability of patients in a particular medical condi-
tion (Morid et al., 2021). To get around this problem,
the use of transfer learning technique is the most indi-
cated, in this approach the weights learned by a neu-
ral network in a given problem domain can be reused
in another different domain, but as the classification
layer is specific for the original problem, it is neces-
sary to change the last layer to adapt this CNN archi-
tecture to a new problem. According to (Tajbakhsh
et al., 2016), the initial layers of convolutional net-
works learn low-level characteristics that are gener-
ally applicable to most computer vision tasks. In con-
trast, those from deeper layers learn high-level char-
acteristics specific to the problem domain by which
they are being applied. Based on these statements, an
auxiliary technique can be used together with trans-
fer learning to improve training results, called fine-
tuning, which consists of training the aggregated lay-
ers and those from the pre-trained CNN.
DenseNets were presented in (Huang et al., 2017),
their main objectives are: to reduce the so-called van-
ishing gradient effect that often occurs in deep neural
networks, in addition to strengthening the propaga-
tion of features and significantly reducing the number
of parameters to be learned. The main characteris-
tic of this type of CNN is how data is sent between
the layers, each of which has connections with the
other layers later, something very similar to ResNets.
A DenseNet network is composed of several dense
Classification of Histopathological Images of Penile Cancer using DenseNet and Transfer Learning
979

blocks that are separated by a transition layer that
aims to reduce the size of the generated feature maps
that will be sent to the next layers. There are several
versions of DenseNets from the original work (Huang
et al., 2017), numbered by the number of layers; in
this case, the architecture used in this work has ex-
actly 201 layers by default.
The proposed method uses a pre-trained model
through the DenseNet-201 architecture in the Ima-
geNet image base available in the Keras library writ-
ten in Python. The use of this CNN type is jus-
tified because it had a superior performance in ex-
periments performed concerning other architectures
such as Xception and InceptionResnetV2, for exam-
ple. This model was then trained using an Nvidia
Geforce 3060 RTX in the penile cancer histopatholog-
ical image dataset for both types of magnification in
accordance with the transfer-learning technique. The
classification layers were removed. The other layers
of the network remained frozen. Three additional lay-
ers were added: an average pooling layer, a dense
layer that uses the Relu activation function with 256
neurons, and an output layer that uses the Softmax al-
gorithm. Furthermore, a dropout with an empirically
defined probability of 0.35 has been added between
the last two layers. Figure 4 shows the entire CNN ar-
chitecture with the dense blocks, its connections be-
tween the transition layers, and the layers added by
the author that are suitable for this binary classifica-
tion problem.
Each training was performed in 35 epochs using
the Adam optimizer configured with a learning rate
of 0.0001. The batch size was adjusted to 32, and
some experiments used data augmentation (horizontal
and vertical flip and random rotation up to 90º). Af-
ter each training, the resulting model was fine-tuned
to improve the results. All layers of this network
were unfrozen, and the model was retrained with eight
epochs using a learning rate equal to 0.00001.
4 RESULTS AND DISCUSSION
After preprocessing the color images, the model was
trained using k-fold cross-validation to assess the gen-
eralizability of the model. Each experiment was con-
figured for five folds in a stratified way, and for each
of the five rounds, a different fold of images is se-
lected for testing; the others are used to compose the
training and validation partitions, respectively 80%
and 20% also in a stratified way. Table 2 presents
the experiments performed that demonstrate the in-
fluence of preprocessing and data augmentation on
the results. In order to evaluate the proposed method,
the following metrics were used: accuracy, sensitiv-
ity, specificity, precision, and F1-Score, with the latter
indicator being the criterion used to indicate the best
model.
Table 2: List of experiments performed to demonstrate the
contribution of preprocessing with CLAHE and data aug-
mentation to method results.
Experiments
No. Mag Preprocessing Augmentation
1 40X Raw No
2 100X Raw No
3 40X CLAHE No
4 100X CLAHE No
5 40X Raw Yes
6 100X Raw Yes
7 40X CLAHE Yes
8 100X CLAHE Yes
As shown in Table 3, the application of the
CLAHE contributed to the method obtaining a good
result for the 40X magnification based on the F1-
Score metric, which is the harmonic mean between
precision and recall; therefore, for experiment 3, the
result was 97.39%(+/-2.13). For the 100X magnifi-
cation, it is verified that the data augmentation con-
tributed together with the CLAHE algorithm in exper-
iment 8, which obtained 97.31 (+/-3.62) of F1-Score.
In addition to good F1-Scores, these trained mod-
els had significant results regarding recall. This met-
ric reports the proportion of images that have cancer
and that were rated positively. Therefore, experiment
3, carried out on images of 40X magnification, had
the result 98.33%(+/-3.33), being more stable with
a smaller standard deviation than other experiments;
for the 100X magnification, experiment 8 resulted in
98.18%(+/-3.64).
In addition to the experiments carried out to ver-
ify the performance of the method proposed in this
dataset of histopathological images, the author exper-
imented with another similar method based on the
deep features technique. DenseNet was used to ex-
tract features from the resized images (224x244 pix-
els) that were highlighted by the CLAHE algorithm.
Then the resulting feature vectors served as input
to a classifier in the training and testing steps. To
find a good model among several classifiers (Deci-
sion Tree, Random Forest, and K-Nearest Neighbors),
we used the GridSearch algorithm with stratified 5x5
folds nested cross-validation. The results by magnifi-
cation and classifier can be seen in table 4. The re-
sults, in this case, were very promising, especially
for the selected model trained in the KNN classi-
fier for 40X magnification, which had its indicators
slightly higher than the others, obtaining an F1-Score
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
980
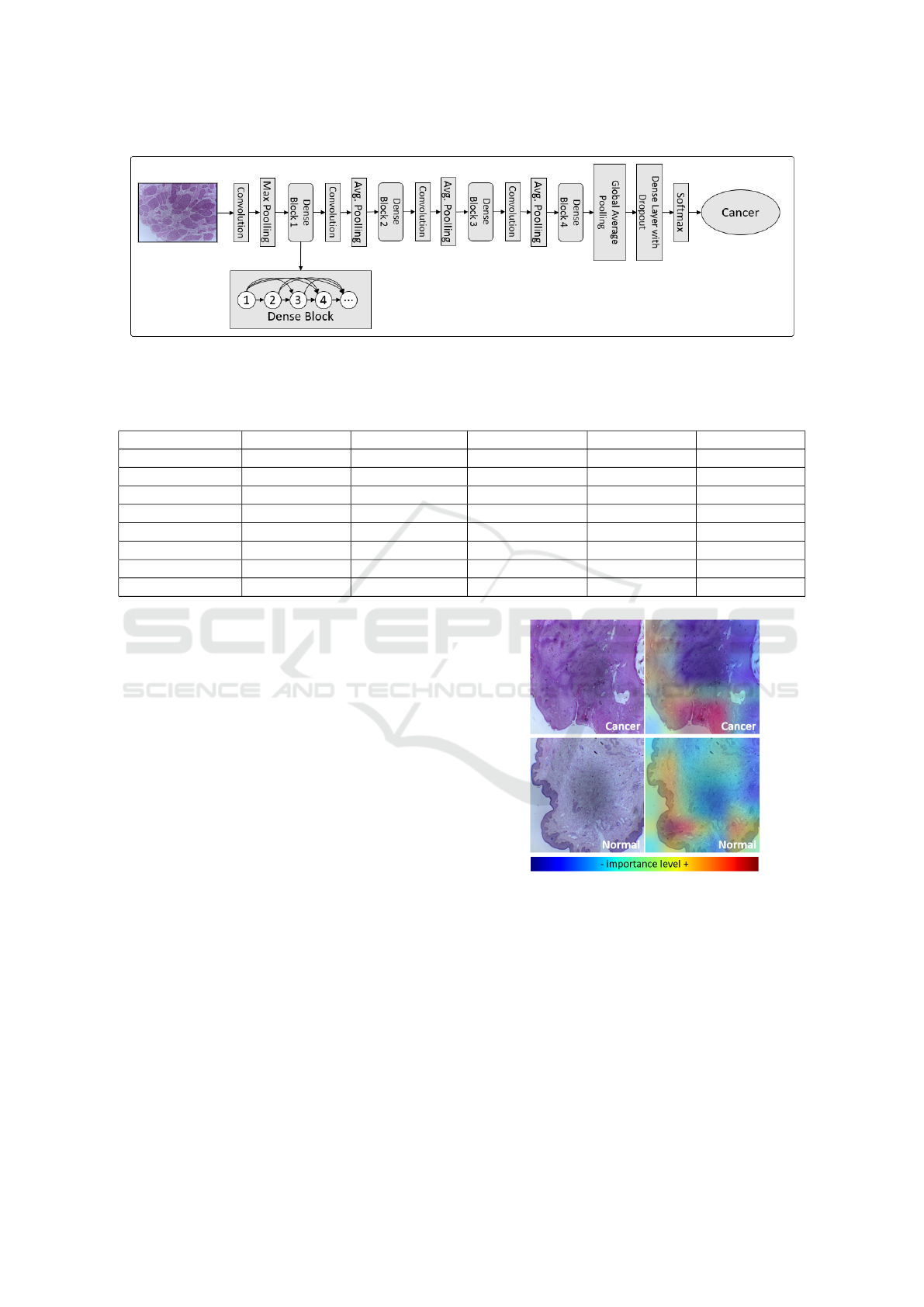
Figure 4: DenseNet-201 architecture along with the layers added by the author: shortcut connections are pertinent to each
dense block, and the operations between blocks are the transition layers; finally, additional layers were included in order to
improve and adapt the network to the binary classification problem.
Table 3: Results of the proposed experiments.
Experiment No. Accuracy(%) Recall(%) Especificity(%) Precision(%) F1-Score(%)
1 92.74(+/-4.26) 96.52(+/-4.27) 87.50(+/-7.91) 91.79(+/-5.28) 93.98(+/-3.52)
2 92.89(+/-5.06) 100.00(+/-0.00) 82.50(+/-12.75) 89.81(+/-6.69) 94.50(+/-3.72)
3 96.89(+/-2.54) 98.33(+/-3.33) 95.00(+/-6.12) 96.67(+/-4.08) 97.39(+/-2.13)
4 94.89(+/-3.16) 100.00(+/-0.00) 87.50(+/-7.91) 92.27(+/-4.55) 95.92(+/-2.44)
5 91.68(+/-6.37) 92.88(+/-6.77) 90.00(+/-9.35) 93.16(+/-6.71) 92.87(+/-5.53)
6 93.89(+/-3.72) 98.33(+/-3.33) 87.50(+/-7.91) 92.14(+/-4.56) 95.06(+/-2.97)
7 92.74(+/-4.26) 94.55(+/-7.27) 90.00(+/-5.00) 93.26(+/-3.48) 93.72(+/-4.01)
8 96.84(+/-4.21) 98.18(+/-3.64) 95.00(+/-6.12) 96.52(+/-4.27) 97.31(+/-3.62)
of 95.47%(+/-5.23). For the 100X magnification, the
model trained by the RF classifier obtained an F1-
Score of 95.71%(+/-2.89). Despite these results, the
proposed method did better according to the compar-
ison presented in table 5.
Complementary, some experiments were carried
out on the test images to verify which regions the
convolutional neural network took into account to
make a given classification decision. This verifi-
cation was performed based on the results of the
Gradient-weighted Class Activation Mapping algo-
rithm, known as Grad-CAM (Selvaraju et al., 2019),
which is based on the gradient data of the last convo-
lutional layer of a CNN. This approach allowed a heat
map type image superimposed on each image clas-
sified by the DenseNet network. Examples of these
experiments can be seen in Figure 5 for the 40X mag-
nification images and in Figure 6 for the 100X mag-
nification images. In this case, we conclude that the
neural network considered the tissue edge regions in
all cases as the most important for classification.
5 CONCLUSION
In this work, a specialized method based on deep
learning and image enhancement for the problem of
binary classification of penile cancer histopatholog-
Figure 5: Grad-CAM algorithm result when overlaying the
heat maps with the evaluated 40X magnification images.
ical images was presented. As discussed above, it
achieved promising results with an F1-Score of up
to 97.39%(+/-2.13) on an imaging database used for
the first time in experiments to automate image-based
medical diagnoses.
As future work, we suggest to use techniques
that can better take advantage of the characteristics
of high-resolution images, such as patch extraction
methods and the use of weakly supervised learning
through multiple instance learning techniques, for ex-
ample. We pretend, furthermore, to update the dataset
Classification of Histopathological Images of Penile Cancer using DenseNet and Transfer Learning
981

Table 4: Additional results for ablation experiments based on deep features and on the use of GridSearch for the selection of
models. The classifiers were represented by the following abbreviations: DT = Decision Tree; RF = Randon Forest; and KNN
= K-Nearest Neighbors.
Mag. Classifier Accuracy(%) Recall(%) Especificity(%) Precision(%) F1-Score(%)
40X DT 87.68(+/-3.99) 91.36(+/-9.14) 82.50(+/-15.00) 89.59(+/-8.63) 89.69(+/-3.42)
40X RF 93.84(+/-1.92) 98.33(+/-3.33) 87.50(+/-7.91) 92.14(+/-4.56) 94.98(+/-1.34)
40X KNN 94.84(+/-5.77) 94.70(+/-7.20) 95.00(+/-6.12) 96.46(+/-4.39) 95.47(+/-5.23)
100X DT 78.47(+/-5.48) 82.73(+/-8.87) 72.50(+/-9.35) 81.44(+/-4.89) 81.75(+/-5.10)
100X RF 94.84(+/-3.33) 98.18(+/-3.64) 90.00(+/-5.00) 93.44(+/-3.32) 95.71(+/-2.89)
100X KNN 91.84(+/-8.27) 94.70(+/-7.20) 87.50(+/-13.69) 92.18(+/-7.90) 93.27(+/-6.78)
Table 5: Comparison between the proposed method (line 1
and 2) and another one created by the same author based on
deep features (line 3 and 4).
Mag. Feat. Ext. Classifier F1-Score(%)
40X DenseNet DenseNet 97.39(+/-2.13)
100X DenseNet DenseNet 97.31(+/-3.62)
40X DenseNet KNN 95.47(+/-5.23)
100X DenseNet RF 95.71(+/-2.89)
Figure 6: Grad-CAM algorithm result when overlaying the
heat maps with the evaluated 100X magnification images.
with new images and information that will make it
possible to evolve the method presented in order to
classify images with cancer by the presence of HPV
and histological grade.
ACKNOWLEDGEMENTS
This work was supported by the Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
(CAPES) - Finance Code 001 and CAPES PDPG
Amaz
ˆ
onia Legal 0810/2020 - 88881.510244/2020-
01; the Fundac¸
˜
ao de Amparo
`
a Pesquisa e ao De-
senvolvimento Cient
´
ıfico e Tecnol
´
ogico do Maranh
˜
ao
(FAPEMA); and the Conselho Nacional de Desen-
volvimento Cient
´
ıfico e Tecnol
´
ogico (CNPq).
REFERENCES
ACS (2021). Tests for penile cancer - american cancer
society. https://www.cancer.org/cancer/penile-
cancer/detection-diagnosis-staging/how-
diagnosed.html. (Accessed on 10/06/2021).
Boumaraf, S., Liu, X., Zheng, Z., Ma, X., and Ferk-
ous, C. (2021). A new transfer learning based ap-
proach to magnification dependent and independent
classification of breast cancer in histopathological im-
ages. Biomedical Signal Processing and Control,
63:102192.
Choudhary, T., Mishra, V., Goswami, A., and Sarangapani,
J. (2021). A transfer learning with structured filter
pruning approach for improved breast cancer classifi-
cation on point-of-care devices. Computers in Biology
and Medicine, 134:104432.
Coelho, R., Pinho, J., Moreno, J., Garbis, D., Nascimento,
A., Larges, J., Calixto, J., Ramalho, L., Silva, A.,
Nogueira, L., Feitoza, L., and Barros-Silva, G. (2018).
Penile cancer in maranh
˜
ao, northeast brazil: The high-
est incidence globally? BMC Urology, 18.
Cruz-Roa, A., Basavanhally, A., Gonz
´
alez, F., Gilmore, H.,
Feldman, M., Ganesan, S., Shih, N., Tomaszewski, J.,
and Madabhushi, A. (2014). Automatic detection of
invasive ductal carcinoma in whole slide images with
convolutional neural networks. In Gurcan, M. N. and
Madabhushi, A., editors, Medical Imaging 2014: Dig-
ital Pathology, volume 9041, pages 1 – 15. Interna-
tional Society for Optics and Photonics, SPIE.
Douglawi, A. and Masterson, T. (2017). Updates on the epi-
demiology and risk factors for penile cancer. Transla-
tional Andrology and Urology, 6:785–790.
Epstein, J., of Pathology, A. R., Magi-Galluzzi, C., Zhou,
M., Cubilla, A., of Pathology Staff, A. R., and
of Pathology (U.S.) Staff, A. F. I. (2020). Tumors of
the Prostate Gland, Seminal Vesicles, Penis, and Scro-
tum. AFIP atlas of tumor and non-tumor pathology.
Fifth series. American Registry of Pathology.
Filipczuk, P., Fevens, T., Krzy
˙
zak, A., and Monczak, R.
(2013). Computer-aided breast cancer diagnosis based
on the analysis of cytological images of fine needle
biopsies. IEEE Transactions on Medical Imaging,
32(12):2169–2178.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In 2017 IEEE Conference on Computer Vision
and Pattern Recognition (CVPR), pages 2261–2269.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
982

INCA (2021). Types of cancer — national can-
cer institute - jos
´
e alencar gomes da silva -
inca. https://www.inca.gov.br/tipos-de-cancer/cancer-
de-penis. (Accessed on 10/06/2021).
Kiani, A., Uyumazturk, B., Rajpurkar, P., Wang, A., Gao,
R., Jones, E., Yu, Y., Langlotz, C., Ball, R., Mon-
tine, T., Martin, B., Berry, G., Ozawa, M., Hazard, F.,
Brown, R., Chen, S., Wood, M., Allard, L., Ylagan,
L., and Shen, J. (2020). Impact of a deep learning
assistant on the histopathologic classification of liver
cancer. npj Digital Medicine, 3.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neu-
ral networks. In Proceedings of the 25th Interna-
tional Conference on Neural Information Processing
Systems - Volume 1, NIPS’12, page 1097–1105, Red
Hook, NY, USA. Curran Associates Inc.
Kumar, A. and Shaik, F. (2015). Image Processing in Dia-
betic Related Causes. SpringerBriefs in Applied Sci-
ences and Technology. Springer Singapore.
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
Linkon, A. H. M., Labib, M. M., Hasan, T., Hossain,
M., and Marium-E-Jannat (2021). Deep learning
in prostate cancer diagnosis and gleason grading in
histopathology images: An extensive study. Informat-
ics in Medicine Unlocked, 24:100582.
Melo, R. C. N., Raas, M. W. D., Palazzi, C., Neves, V. H.,
Malta, K. K., and Silva, T. P. (2020). Whole slide
imaging and its applications to histopathological stud-
ies of liver disorders. Frontiers in Medicine, 6:310.
Morid, M. A., Borjali, A., and Del Fiol, G. (2021). A scop-
ing review of transfer learning research on medical
image analysis using imagenet. Computers in Biology
and Medicine, 128:104115.
Neto, A. D. (2012). Cytopathology technician: reference
book 3: Histopathology techniques — bras
´
ılia;
ministry of health of brazil ; 2012. 93 p.— ms.
https://pesquisa.bvsalud.org/bvsms/resource/pt/mis-
37140. (Accessed on 10/11/2021).
Rakhlin, A., Shvets, A., Iglovikov, V., and Kalinin, A. A.
(2018). Deep convolutional neural networks for breast
cancer histology image analysis. In Campilho, A.,
Karray, F., and ter Haar Romeny, B., editors, Im-
age Analysis and Recognition, pages 737–744, Cham.
Springer International Publishing.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S.,
Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bern-
stein, M., et al. (2015). Imagenet large scale visual
recognition challenge. International journal of com-
puter vision, 115(3):211–252.
Sarwinda, D., Paradisa, R. H., Bustamam, A., and Anggia,
P. (2021). Deep learning in image classification using
residual network (resnet) variants for detection of col-
orectal cancer. Procedia Computer Science, 179:423–
431. 5th International Conference on Computer Sci-
ence and Computational Intelligence 2020.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2019). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. International Journal of Computer Vi-
sion, 128(2):336–359.
Sharma, H., Zerbe, N., Klempert, I., Hellwich, O., and Huf-
nagl, P. (2017). Deep convolutional neural networks
for automatic classification of gastric carcinoma using
whole slide images in digital histopathology. Com-
puterized Medical Imaging and Graphics, 61:2–13.
Selected papers from the 13th European Congress on
Digital Pathology.
Spanhol, F. A., Oliveira, L. S., Cavalin, P. R., Petitjean, C.,
and Heutte, L. (2017). Deep features for breast cancer
histopathological image classification. In 2017 IEEE
International Conference on Systems, Man, and Cy-
bernetics (SMC), pages 1868–1873.
Spanhol, F. A., Oliveira, L. S., Petitjean, C., and Heutte, L.
(2016a). Breast cancer histopathological image classi-
fication using convolutional neural networks. In 2016
International Joint Conference on Neural Networks
(IJCNN), pages 2560–2567.
Spanhol, F. A., Oliveira, L. S., Petitjean, C., and Heutte, L.
(2016b). A dataset for breast cancer histopathological
image classification. IEEE Transactions on Biomedi-
cal Engineering, 63(7):1455–1462.
Srinidhi, C. L., Ciga, O., and Martel, A. L. (2021). Deep
neural network models for computational histopathol-
ogy: A survey. Medical Image Analysis, 67:101813.
Tajbakhsh, N., Shin, J. Y., Gurudu, S. R., Hurst, R. T.,
Kendall, C. B., Gotway, M. B., and Liang, J. (2016).
Convolutional neural networks for medical image
analysis: Full training or fine tuning? IEEE Trans-
actions on Medical Imaging, 35(5):1299–1312.
Tellez, D., Litjens, G., B
´
andi, P., Bulten, W., Bokhorst, J.-
M., Ciompi, F., and van der Laak, J. (2019). Quanti-
fying the effects of data augmentation and stain color
normalization in convolutional neural networks for
computational pathology. Medical Image Analysis,
58:101544.
Thomas, A., Necchi, A., Muneer, A., Tobias-Machado, M.,
Tran, A. T. H., Van Rompuy, A.-S., Spiess, P. E., and
Albersen, M. (2021). Penile cancer. Nature Reviews
Disease Primers, 7(1):1–24.
Vieira, C., Feitoza, L., Pinho, J., Teixeira-J
´
unior, A., Lages,
J., Calixto, J., Coelho, R., Nogueira, L., Cunha, I.,
Soares, F., and Barros-Silva, G. (2020). Profile of pa-
tients with penile cancer in the region with the highest
worldwide incidence. Scientific Reports, 10.
Vill
´
an, A. (2019). Mastering OpenCV 4 with Python: A
practical guide covering topics from image process-
ing, augmented reality to deep learning with OpenCV
4 and Python 3.7. Packt Publishing.
Wei, J., Tafe, L., Linnik, Y., Vaickus, L., Tomita, N., and
Hassanpour, S. (2019). Pathologist-level classifica-
tion of histologic patterns on resected lung adenocar-
cinoma slides with deep neural networks. Scientific
Reports, 9.
Zhang, A., Lipton, Z. C., Li, M., and Smola, A. J.
(2021). Dive into deep learning. arXiv preprint
arXiv:2106.11342.
Zuiderveld, K. J. (1994). Contrast limited adaptive his-
togram equalization. In Graphics Gems.
Classification of Histopathological Images of Penile Cancer using DenseNet and Transfer Learning
983
