Separation of Concerns in Extended Epidemiological
Compartmental Models
A Yvan Guifo Fodjo
1,6,7 a
, Mikal Ziane
2,1 b
, Serge Stinckwich
3 c
, Bui Thi Mai Anh
4
and Samuel Bowong
5,6
1
CNRS, UMR 7606, LIP6, Sorbonne Universit
´
e, Paris, France
2
Universit
´
e de Paris, Paris, France
3
United Nations University Institute in Macau, Macau SAR, China
4
School of Information and Communication Technology, Laboratory of Intelligent Software Engineering,
Hanoi University of Science and Technology, Hanoi, Vietnam
5
D
´
epartement de Math
´
ematiques, Universit
´
e de Douala, Douala, Cameroon
6
IRD, UMI 209, UMMISCO, Bondy, France
7
URIFIA, Universit
´
e de Dschang, Dschang, Cameroon
Keywords:
Separation of Concerns, Compartmental Models, Contact Network, Epidemiology Modeling Tool.
Abstract:
Epidemiological models become more and more complex as new concerns are taken into account (age, sex,
spatial heterogeneity, containment or vaccination policies, etc.). This is problematic because these aspects are
typically intertwined which makes models difficult to extend, change or reuse. The Kendrick approach has
shown promising results to separate epidemiological concerns but is restricted to homogeneous compartmental
models. In this paper, we report on an attempt to generalize the Kendrick approach to support some aspects
of contact networks, thereby improving the predictive quality of models with significant heterogeneity in the
structure of contacts, while keeping the simplicity of compartmental models. This approach has been validated
on two different techniques to generalize compartmental models.
1 INTRODUCTION
Modeling and simulation have been heavily used in
epidemiology, for instance to inform control strate-
gies (Levin and Durrett, 1996). Epidemiological
models largely rely on the compartmental framework
where the individuals of a population are grouped
by their epidemiological status (Keeling and Rohani,
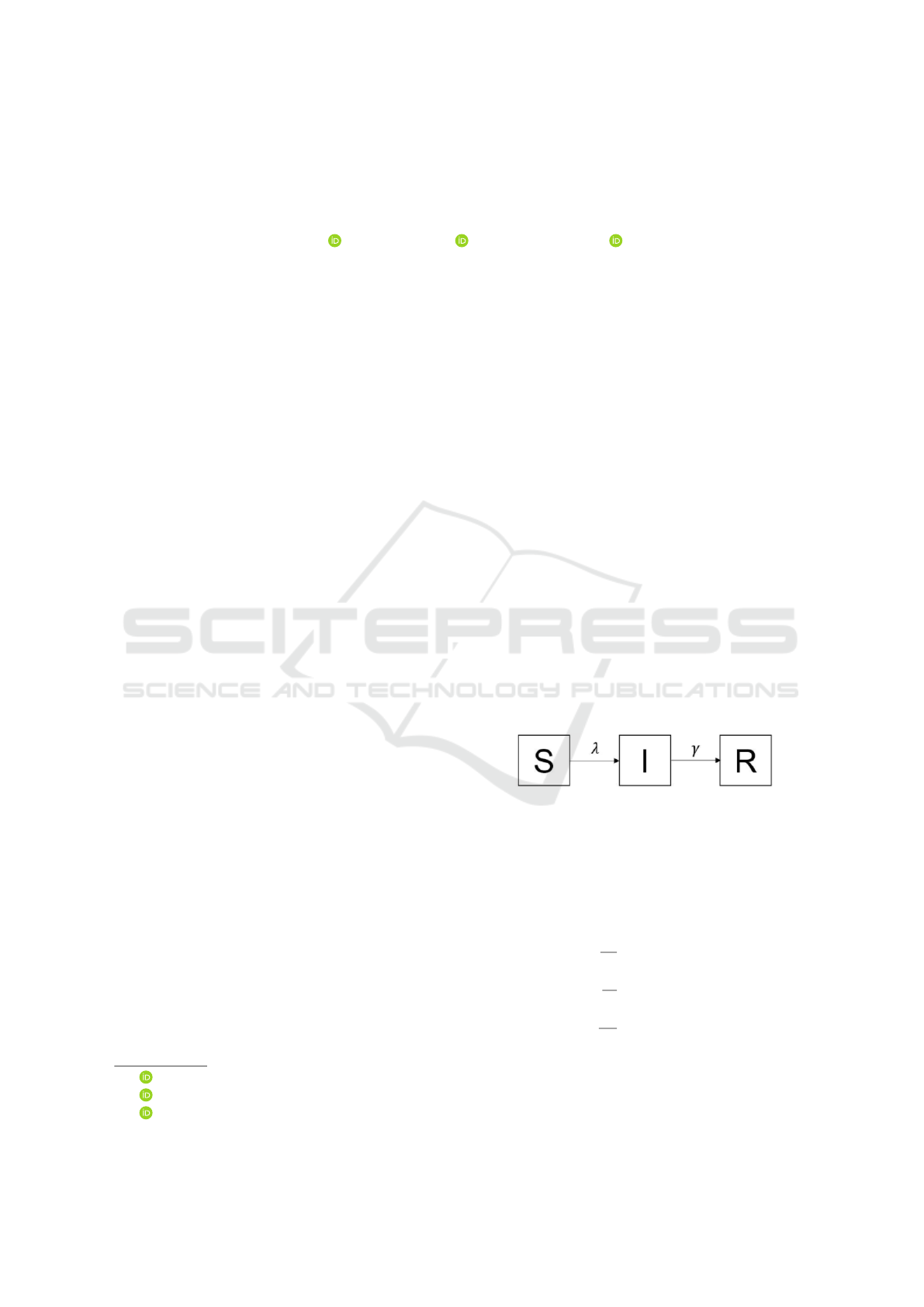
2011). Those Susceptible to the pathogen (state S)
can be infected with rate λ, the Infectious ones (state
I) can transmit the disease or become immune a.k.a
Recovered (state R) with rate γ (See Figure 1).
Compartmental models are typically first defined
as ordinary differential equations (ODEs), such as
Equation 1 below, which can be studied analytically
and/or simulated using algorithms such as Runge-
Kutta. It is however considered more realistic to adopt
a
https://orcid.org/0000-0002-0714-6737
b
https://orcid.org/0000-0003-1860-485X
c
https://orcid.org/0000-0002-8755-9848
Figure 1: Flow diagram of the SIR (Susceptible, Infected,
Recovered) mathematical model.
a stochastic viewpoint on these models, typically con-
sidering them as Continuous-Time Markov Chains
(CTMCs), that can be derived from the ODEs modulo
some widely-accepted, albeit simplifying, probabilis-
tic assumptions.
dS
dt
= −λS
dI
dt
= λS −γI
dR
dt
= γI
N = S +I + R
(1)
Aside from this core epidemiological concern,
other concerns may have to be taken into account
such as the age structure, the social or sexual mix-
152
Guifo Fodjo, A., Ziane, M., Stinckwich, S., Anh, B. and Bowong, S.
Separation of Concerns in Extended Epidemiological Compartmental Models.
DOI: 10.5220/0010881900003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 3: BIOINFORMATICS, pages 152-159
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

ing and the spatial heterogeneity of the transmission
that may be caused by various considerations. Each of
these additional aspects may lead to further partition-
ing the population. Unfortunately, concerns are typ-
ically intertwined in epidemiological models in both
executable code and mathematical equations.
At first sight at least, it seems difficult to sepa-
rate these concerns since, when a concern is added
to a model, it is precisely because it interferes with
the core epidemiological concern to introduce some
kind of heterogeneity. For example, in a spatial SIR
model, the spatial concern impacts the epidemic con-
cern at each region which assigns a specific value to
an epidemic parameter. If the epidemic parameters
had the same value on the whole space, there would
be little point in introducing a spatial concern in the
first place.
The Kendrick approach(Bui et al., 2016) consists
in defining concerns (age, sex, spatial heterogene-
ity, ...) as independent, possibly incomplete, mod-
els that are then combined into Stochastic Automata
Networks (SANs) (Plateau and Stewart, 2000) using a
tensor-sum operator. The stochastic dependencies be-
tween concerns are then introduced in a second phase
so that the independent concerns can be reused and
combined in other models much more easily.
A typical assumption of compartmental models is
that individuals mix (meet others individual with po-
tential disease transmission) uniformly, at least in the
same compartments, i.e. with the same probability.
This assumption is getting more and more criticized
as unrealistic as it has been observed that, in many
outbreaks, a few super-spreaders initially infect other
individuals more quickly than homogeneous models
predict. Later in an epidemic, on the contrary, when
a lot of individuals are infected, the reverse has been
observed: homogeneous models predict more infec-
tions than what happens, because the remaining sus-
ceptible individuals are typically the most isolated
ones.
It has thus been proposed to define, study or simu-
late contact-network models in which nodes denote
individuals or groups of individuals and edges de-
note potential contacts. These models, however, are
more involved to define and study than compartmen-
tal models. It is thus natural to try and extend com-
partmental models to include some contact hetero-
geneity.
Several adaptations have been published to im-
prove the homogeneous compartmental approach for
at least some classes of contact networks. Bansal et
al. (Bansal et al., 2007) have then proposed a general
framework to capture such adaptations as changes to
the parameters that define the force of infection, and
have redefined two of these adaptations, that they
deemed typical enough, using their proposal: the
Stroud (Stroud et al., 2006) and the Aparicio (Apari-
cio and Pascual, 2007) adaptations.
In this paper, we report on our attempt to integrate
Bansal’s framework in the Kendrick approach to sep-
aration of concerns(Bui et al., 2016). The challenge
consists in expressing Bansal’s framework as well as
any specialization of it such as Stroud’s or Aparicio’s
adaptation as separate Kendrick concerns. The bene-
fit of our proposal is that adaptations to the homoge-
neous compartmental model may now take advantage
of separation of concerns and be combined with other
kind of concerns that are not directly related to the
contact network.
2 INTEGRATING BANSAL’S
FRAMEWORK IN THE
KENDRICK APPROACH
Seen from a software engineer’s viewpoint, our idea
is somehow similar to applying the Template Method
design pattern (Gamma et al., 1995) to epidemic
model equations
1
: it consists in expressing λ as a
function of 3 parameters, α, i
t
and τ.
α is the average number of individuals with which
a susceptible individual has contact or the average de-
gree of nodes in a contact network, i
t
is the proportion
of contacts that are infectious
2
and τ is the per-contact
rate at which the disease is transmitted between an in-
fectious and a susceptible individual.
Following Bansal et al. we have renamed these
3 usual parameters to α
gen
, it
gen
and τ
gen
to insist
on their special meaning when applied to a model
based on a contact-network and to signal that they are
generic hot-spots, i.e. varying points. The generic
definition for the force of infection λ is given by
Equation 2. We have, however, for the sake of sim-
plicity and of readability, often kept the original sim-
ple names without the ”gen” subscript, especially
when they have their usual homogeneous meaning,
typically before a model is made generic using the
Kendrick approach. We have used names with ”gen”
in final generic definitions of models and in Kendrick
code.
λ = α
gen
τ
gen
it
gen
(2)
1
The most obvious difference with the Template pattern
is that there is no object-oriented inheritance involved here
but there was of course no such inheritance either in the
original patterns from Alexander.
2
Caveat: this means the proportion of infectious individ-
uals among the individuals a susceptible individual meets.
Separation of Concerns in Extended Epidemiological Compartmental Models
153

In traditional (homogeneous-compartmental)
models i
t
is typically defined this way:
i
t
=
I
N
(3)
(Bansal et al., 2007) reports on two proposals,
Stroud et al. (Stroud et al., 2006) and Aparicio et
al. (Aparicio and Pascual, 2007), to generalize com-
partmental models to capture some aspects of contact-
network models. Both proposals were captured by
giving (α
gen
, it
gen
and τ
gen
) specific values. In order to
explain these approaches in the limited allowed space
for this paper, we have chosen to simplify the rational
and hypotheses behind them and more generally the
epidemiological questions in this paper. The inter-
ested reader will need to refer to the original papers
to get the full explanations of these models.
Consider Figures 5, 6 and 7. The curves are the
daily incidence
3
predicted by different models assum-
ing a Poisson, exponential and scale-free contact net-
work respectively . The red line denotes a traditional
(homogeneous) compartmental model. By definition,
it does not depend on the network but then, in some
cases, it suffers from two problems: the height of the
peak is wrong and it happens too late.
The blue and green curves are run using our
Kendrick compartmental tool but using the approach
from Stroud et al. and from Aparicio et al., respec-
tively, to approximate some aspects of contact net-
works. The former approach improves (lowers) the
height of the peak on some classes of networks. The
latter approach improves the timing of the peak on
some classes of networks although sometimes exag-
gerating its height. Ideally, it should be easy to try
one or several of these approaches to decide which
approach best matches the observed data.
In this paper, we thus want to check if both these
approaches can be captured as Kendrick concerns to
keep the benefits of separation of concerns. This way
these approaches can be changed more easily than in
monolithic models where all concerns are mixed to-
gether (Bui et al., 2016).
In case of success, our middle-term objective, out-
side of the scope of this paper, would be to investigate
what class of extensions can be defined as Kendrick
concerns so that compartmental models can more eas-
ily be enriched with some of the properties of contact
networks.
3
Incidence is the number of new infections per day,
sometimes normalized by dividing by some value. (Bansal
et al., 2007) do not give which value they chose for the de-
nominator we thus did not normalize incidence.
2.1 Stroud et al.’s Extension
In traditional homogeneous-mixing compartmental
modeling, the expected number of new infections per
day per infectious person is assumed to be propor-
tional to the fraction of the population that is suscep-
tible. In real social structures, however, some suscep-
tible individuals have a greater chance to receive and
transmit the disease than others. (Stroud et al., 2006)
claim that ”epidemic models can practically incorpo-
rate inhomogeneous mixing by taking the number of
new infections per day per infectious person to scale
as a power (greater than one) of the fraction of the
population that is susceptible”.
Introducing this power law can be done in two
steps. First, combining Equations 1, 2 and 3 leads
to:
dS
dt
= −ατI
S
N
(4)
Then, this equation is generalized by introducing a
constant, ν, greater than 1, that is postulated to be a
power of S/N. This constant must be ”fitted” from real
data i.e. estimated to minimize errors. Intuitively,
a higher value for ν represents a higher degree of
heterogeneity, especially (although not necessarily) in
the number of contacts among individuals or similarly
in the degree of nodes in a contact network.
The generalized equation becomes:
dS
dt
= −α
gen
τ
gen
I(
S
N
)
ν
(5)
which leads to:
it
gen
=
I
S
(
S
N
)
ν
(6)
The parameters α
gen
and τ
gen
are constants whose val-
ues are best set outside of models themselves, in so-
called simulation scenarios. Capturing this idea in
Kendrick is quite simple and merely consists in as-
signing the above value to it
gen
which will implicitly
declare an additional parameter ν to be assigned in
simulation scenarios. The Kendrick code of this con-
cern is given in Section 3. Remember that the point
was to try and keep the implementation of this idea
separate from the rest of the model, here from the SIR
concern.
2.2 Aparicio et al.’s Extension
Homogeneous models often fail to predict when the
peek of an epidemic will occur. Early in epidemics,
the most likely individuals to be infected are typically
”hubs” that have a lot contacts and thus are also likely
to secondary infect more individuals than an average
individual would. Late in epidemics, on the contrary,
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
154

the new infected individuals are typically more iso-
lated and induce less secondary infections. On Fig-
ures 5, 6 and 7, the peek of the red curve comes too
late.
In (Aparicio and Pascual, 2007), the authors pro-
pose an SIYR model to better account for secondary
infections. Their idea is to split the usual I compart-
ment into I and Y, where their I is individuals that
are infected and infectious and Y individuals that are
infected but not infectious, meaning that they do not
produce secondary infections.
Their model is given by 7 where γ
e
= τ + γ, is the
constant rate at which infectious nodes become in-
active while γ is the constant rate at which infected
nodes become recovered, g is the constant rate at
which inactive nodes recover, and R
0
, the basic re-
production rate, depends on the network.
dS
dt
= −γ
e
R
0
S
N
I
dI
dt
= γ
e
R
0
S
N
I − γ
e
I
dY
dt
= γ
e
I − gY
dR
dt
= gY
N = S +I +Y + R
(7)
Note that there is here a potential confusion be-
tween the usual τ parameter and the generic τ
gen
which in this specific model is not equal to τ. From
the above formulas we deduce: α
gen
= γ
e
; it
gen
=
I
N
and τ
gen
= R
0
. (Bansal et al., 2007) (supplementary
material) gives R
0
= T
hk
2
i−hki
hki
where hki is the mean
degree of the network (set to 10 in our examples),
while hk
2
i is the mean square degree and T =
τ
γ
e
is
the probability of transmission of the pathogen.
For a Poisson random network hk
2
i = hki(hki +1)
(Barab
´
asi et al., 2016) but for more general networks
it is an issue to estimate hk
2
i without an actual net-
work. (Bansal et al., 2007) relies on computations on
the random networks they used.
For the exponential network, we have generated a
sequence of degrees with an exponential distribution
and have computed hk
2
i.
For the scale-free network, in order to calculate
the mean square degree, we proceed in a similar way
as in the exponential case, but we use the (Barab
´
asi
and Albert, 1999) algorithm to generate the graph.
3 VALIDATION AND
DISCUSSION
To validate our approach, we have replicated the ex-
periments of (Bansal et al., 2007) comparing the
simulation results from homogeneous compartmental
models with the models of Stroud et al. and of Apari-
cio et al. with those obtained by (Bansal et al., 2007)
for Poisson, exponential and scale-free networks.
4
.
The homogeneous model is a special case of Stroud
et al.’s model with ν = 1.
All models were defined using the Kendrick
tool(Bui et al., 2019) even though we used the
low-level Kendrick language rather than its domain-
specific language which is currently under revision.
The parameters α
gen
, it
gen
and τ
gen
are noted in
Kendrick as alpha gen, it gen and tau gen.
Our implementation of Stroud et al. is shown Fig-
ure 2. The code is divided into three parts: defining
the concerns (SIR and Stroud), initializing the param-
eters, running the simulation. The code for the Apari-
cio et al. case is divided in a similar way (See Figure
4).
The results of the simulations can be seen on Fig-
ures 5, 6 and 7 are similar to those of Figures 6 (b),
6 (d) and 6 (f) in (Bansal et al., 2007), taking into
account that used random networks and that differ-
ent scale-free or exponential networks may have dif-
ferent values for hk
2
i. This suggests that integrating
Bansal’s idea was successful.
Both adaptations alter the homogeneous case in
very different ways. As heterogeneity in the degree
of nodes grows from the Poisson to the exponential
to the scale-free case the Stroud adaptation further
shrinks the peek of daily incidence but does not fix its
dynamics: it still comes too late. On the contrary, the
Aparicio adaptation shifts this peak to the left (earlier
in the outbreak) but outside of the case of their pub-
lication (Poisson networks) the height of the peek is
deemed exaggerated by (Bansal et al., 2007).
The challenge was to check if the Kendrick ap-
proach to separation of concerns could capture ap-
proaches such as those from Stroud et al. or Apari-
cio et al. while keeping the familiar compartmental
framework.
We first defined a classical SIR model (line 1-14)
in a usual way except for the definition of lambda
(lines 9-10). Note however that this definition of
lambda is general and not, by any means, restricted
to trying and capture some aspects of contact net-
works. The SIR model defined this way can thus be
4
Python and Kendrick code of our experiments are
available online: https://github.com/KendrickOrg/
BIOINFORMATICS22-code
Separation of Concerns in Extended Epidemiological Compartmental Models
155

Figure 2: Definition of SIR and Stroud concerns.
Figure 3: Definition of the scenario’s parameters.
used quite generally.
The Stroud concern is then quite simple: it merely
gives a value to the it
gen
parameter (lines 16-18). This
concern is separate from the SIR in the sense that it is
a distinct syntactic structure. The SIR concern can be
reused without the Stroud concern.
One issue is the fact that the SIR concern was not
reused in the SIYR one for the implementation of
Aparicio et al’s approach. Factoring out commonal-
ities between the core epidemic concerns (SIR, SEIR,
SIYR...) that define the epidemic status is a bit in-
volved and does not always simplify models.
It might be tempting to introduce some kind of in-
heritance between models (using the Kendrick DSL)
but the benefits are not impressive as far as core con-
cerns are considered. In low-level Kendrick it is pos-
sible to copy SIR into SIYR, add a Y status, redefine
the transitions and so on, but this is not clearly simpler
than redefining SYIR from scratch as we did. There
are only a few kinds of core concerns so that redefin-
Figure 4: Definition of an Aparicio concern.
Figure 5: Daily incidence for the homogeneous, Stroud and
Aparicio approaches on a Poisson network of 10000 nodes
with a mean degree of 10.
ing them from scratch is not a heavy burden.
Other concerns, on the contrary, are quite varied
and are the focus of the Kendrick approach. Because
it relies on a specific core concern (SIYR), imple-
menting the Aparicio et al. extension is more involved
than with the Stroud et al. one. It is however still pos-
sible to benefit from the Kendrick approach and com-
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
156
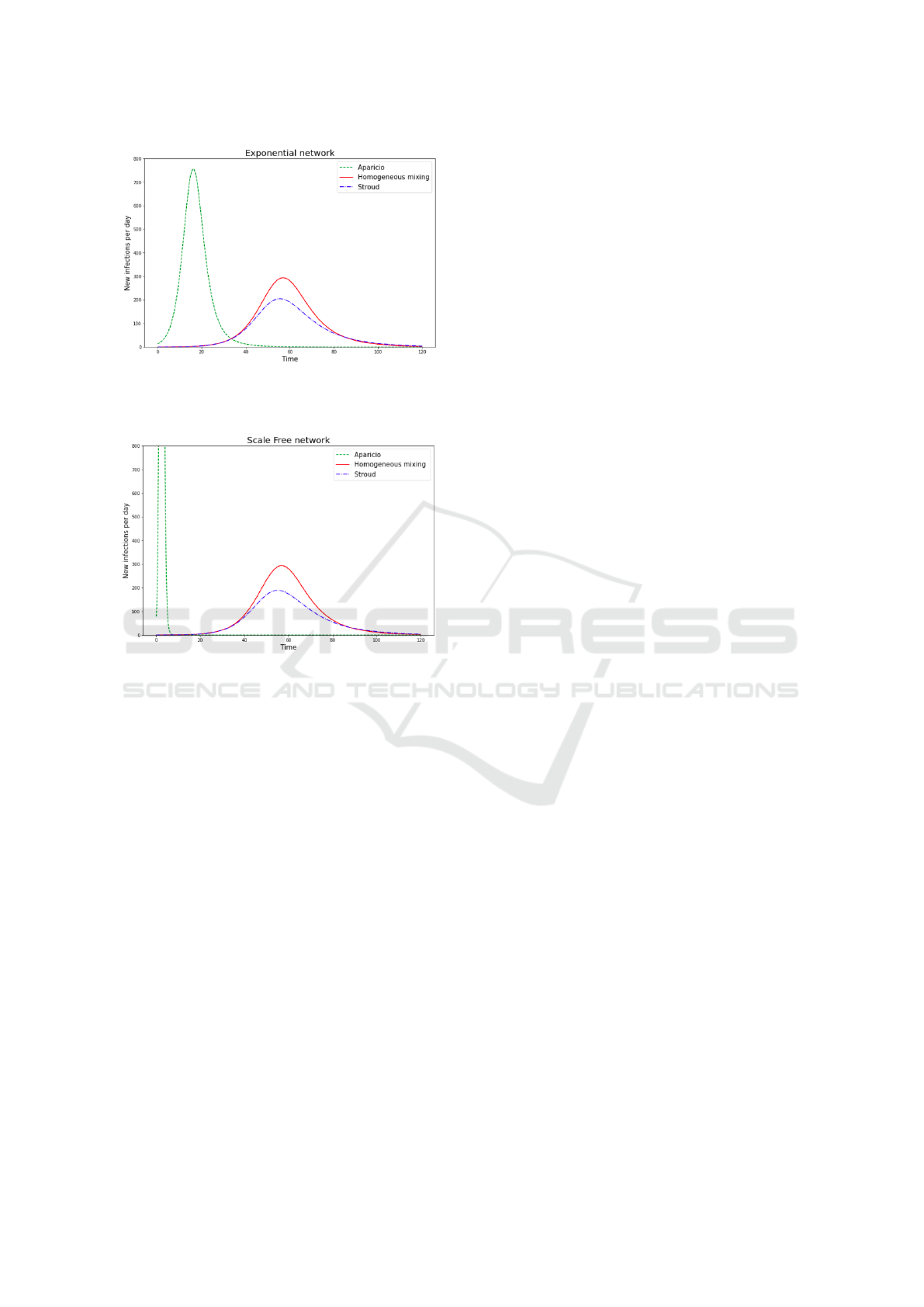
Figure 6: Daily incidence for the homogeneous, Stroud and
Aparicio approaches on an exponential network of 10000
nodes with a mean degree of 10.
Figure 7: Daily incidence for the homogeneous, Stroud
and Aparicio approaches on an scale-free network of 10000
nodes with a mean degree of 10.
bine the SIYR and the Aparicio concerns with other
concerns such as the age or sex structure. For this
reason we consider that this was also a success.
Another issue is the relationship between the
Stroud concern and SIR and more generally of non-
core concerns with the core concern of the model.
Non-core concerns are expected not to structurally
depend (use names introduced in other concerns) on
each other even though this may possibly happen in
huge models (e.g. a global pandemic model) that
are quite beyond the scope of this paper. Non-core
concerns may however depend on the core one. In-
deed, the Stroud concern gives value to a parameter
introduced in SIR and uses names that are defined
in SIR. In low-level Kendrick there is little support,
aside from the host system complaining at run time,
to avoid pitfalls such as misspelling a name or avoid-
ing name conflicts across concerns. This is obviously
something the Kendrick DSL must address e.g. by
prefixing names by that of their concern, importing
the names of concern, ...
The reader may wonder if it would be possible to
combine the approaches of Stroud et al. and from
Aparicio et al. The Kendrick approach would support
it but the respective domains of validity, or at least of
reasonable quality of prediction, of both approaches
do not necessarily intersect so that neither (Bansal
et al., 2007) nor we, have considered this possibility.
Finally, in (Bansal et al., 2007) the idea to
adapt compartmental models, that was transposed to
Kendrick in this paper, was further developed to dis-
tinguish the edges of Susceptible individuals from
those of Infected individuals. Including other param-
eters to concerns is not difficult but Bansal et al. con-
sider that obtaining values (by fitting on simulations
run on a contact network) or formulas (by analyti-
cal methods) for these parameters may become more
cumbersome than using network models. We think
this suggests that it may be worth in future work to try
and capture more aspects of contact networks in con-
cerns and check if this helps combining or switching
between the compartmental approach and the contact-
network one in models.
4 RELATED WORK
Separation of concerns (H
¨
ursch and Lopes, 1995) is a
major goal of software engineering and more gener-
ally of any discipline where models or artifacts may
be too complex to grasp, change or reuse easily when
they are not decomposed into separate parts whose de-
pendencies on each other are kept minimal.
The Kendrick approach, language and tool (Bui
et al., 2016; Bui et al., 2019) achieve separation
of concerns in compartmental epidemic models by
defining each concern (age, sex, spatial heterogene-
ity, containment or vaccination policies, etc.), i.e.
each potential source of heterogeneity, as a sepa-
rate stochastic automaton and by deferring combining
concerns until a composition phase where they are put
together into a SAN using a tensor sum operator.
Process algebras and SANs have similar objec-
tives and have both been used to define compartmen-
tal epidemic models, although not, as far as we know,
to support a general approach to separation of con-
cerns that introduce heterogeneity in epidemic models
(Mccaig et al., 2009). Moreover, process algebras are
not initially meant to model epidemic models which
makes them awkward.
Bio-PEPA, for instance, is an extension of PEPA
(Performance Evaluation Progress Algebra) (Gilmore
and Hillston, 1994) with some features for biologi-
cal system modeling. Some PEPA models however
cannot be translated into ODEs which requires check-
ing some conditions to do it (Benkirane, 2011). Also,
Separation of Concerns in Extended Epidemiological Compartmental Models
157

defining compartments is tedious and PEPA’s syntax
”might look daunting” (Benkirane, 2011). A subse-
quent adaptation of Bio-PEPA was thus introduced to
better support compartmental epidemic models (Cioc-
chetta and Hillston, 2010). We are not aware of any
extensions to Bio-PEPA to support contact networks.
Generally, traditional mathematical models (com-
partmental models) used in epidemiology, make sim-
plifying assumptions about the interaction between
hosts. This is because they implicitly define host-
to-host contact and assume that hosts have identical
contact rates, which does not always agree with real-
world models. However, these models have the par-
ticularity of being manageable, easily implementable,
robust and predictive (Anderson and May, 1992; Mol-
lison et al., 1994). However, faced with their diffi-
culties in reflecting certain real-world models, in re-
cent years epidemiologists have largely focused on
so-called contact network models, which have the
characteristic of explicitly capturing the various mod-
els of interactions that underlie transmission of dis-
ease (Watts and Strogatz, 1998; Pastor-Satorras and
Vespignani, 2001; Shirley and Rushton, 2005). These
models also have the advantage of providing very
good prediction but have the disadvantage of requir-
ing more programming skills. In order to find an
ideal compromise between compartment-based mod-
els and contact network models, at least from the
point of view of implementation, the Kendrick (Bui
et al., 2016; Bui et al., 2016; Bui et al., 2019) ap-
proach was thus generalized in order to take into ac-
count aspects of contact network models from models
based on compartments that are known to be easily
manipulated.
Many epidemiology modeling and simulation
platforms take into account deterministic/stochastic
models based on compartments or/and contact net-
works (Muellner et al., 2018; Bui et al., 2016; Bui
et al., 2019; Picault et al., 2019; Miller and Ting,
2020; Hladish et al., 2012). Hladish et al. introduce
EpiFire (Hladish et al., 2012) - an API
5
implemented
in C++ for generating network models of epidemi-
ology. EpiFire also provides a graphical user inter-
face (GUI) which allows to fast configure the struc-
ture of different networks (i.e., random, small-world,
scale-free etc.) for SIR models. Although the au-
thors have achieved the separation between the net-
work construction and the simulation of the disease
spreading through networks, they have ignored other
epidemiological concerns, only considered the SIR
structure. The most recent work on the field of con-
tact network modeling for epidemiology is EoN of
Miller et al. (Miller and Ting, 2020). EoN provides
5
Application Programming Interface
the same features as EpiFire with the aim of model-
ing the spread of SIR and SIS models over different
networks. It is arguable that most of these tools do not
formalize the principle of separations of concerns in
epidemiology as the Kendrick (Bui et al., 2016; Bui
et al., 2019) and Emulsion (Picault et al., 2019) tools
do. Indeed, Emulsion is a platform that was built with
the aim of helping modelers to focus on the design of
models rather than on the programming aspect. It is
a domain specific language which makes it possible
to make explicit all the components of an epidemio-
logical model (structure, process, parameters, ...) in
the form of a structured text file. Even if the authors
of Emulsion do not specifically specify how this prin-
ciple of separation of concerns was formalized, they
highlight the fact that it allows modelers to design
processes (infection, demography, detection, control,
etc.) and different scales (individuals, populations,
meta-populations) independently, which would allow
the management of multiple hosts, the diversity of
pathogens, as well as realistic detection methods and
control measures.
5 CONCLUSION
In this paper, we have proposed to generalize the
Kendrick approach to separation of concerns in com-
partmental epidemic models (Bui et al., 2016) to eas-
ily capture some aspects of contact network models.
To do this we have applied an idea from (Bansal et al.,
2007) which consists in defining the usual λ parame-
ter of epidemic models as a kind of template method
with three extension points.
We have then applied this approach to 2 exten-
sions of compartmental models, Stroud et al. (Apari-
cio and Pascual, 2007) and Aparicio et al. (Stroud
et al., 2006) and we have been able to get results close
to those of (Bansal et al., 2007). Stroud et al.’s ex-
tension was defined as a very simple concern that was
separate from the core SIR model. Aparicio et al.’s ex-
tension was a bit more complex to implement because
it relies on a specific core concern, namely SIYR,
but the resulting concerns can still be combined with
other epidemic concerns in Kendrick.
Building on these promising results we aim at
generalizing our approach to express more general
aspects of contact networks as separate concerns.
(Bansal et al., 2007) pointed out that the approach we
have reported on was probably not enough to cope
with heavy heterogeneity in the contact network un-
less much more significant efforts are made to adapt
compartmental models and suggested to abandon
them in this case for a full-fledged contact-network
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
158

approach. It is thus interesting to see whether sepa-
ration of concerns can be even further generalized to
alleviate these additional efforts. This will probably
lead to developing concerns that capture more infor-
mation of contact networks to the point that the orig-
inal compartmental framework may become a mere
specific concern itself. Finally, we also plan to in-
clude this generalized approach in the Kendrick DSL
to offer better support to avoid some caveats, espe-
cially those involving name clashes or ambiguities in
the global model.
ACKNOWLEDGEMENTS
The authors would like to thank the anonymous refer-
ees for their help in improving this paper.
REFERENCES
Anderson, R. M. and May, R. M. (1992). Infectious diseases
of humans: dynamics and control. Oxford university
press.
Aparicio, J. P. and Pascual, M. (2007). Building epidemi-
ological models from R0: an implicit treatment of
transmission in networks. Proceedings of the Royal
Society B: Biological Sciences, 274(1609):505–512.
Bansal, S., Grenfell, B. T., and Meyers, L. A. (2007). When
individual behaviour matters: homogeneous and net-
work models in epidemiology. Journal of the Royal
Society Interface, 4(16):879–891.
Barab
´
asi, A.-L. and Albert, R. (1999). Emergence of scal-
ing in random networks. science, 286(5439):509–512.
Barab
´
asi, A.-L. et al. (2016). Network Science. Cambridge
University Press.
Benkirane, S. (2011). Process algebra for epidemiology:
evaluating and enhancing the ability of PEPA to de-
scribe biological systems. PhD thesis, University of
Stirling.
Bui, T., Papoulias, N., Stinckwich, S., Ziane, M., and
Roche, B. (2019). The Kendrick modelling platform:
language abstractions and tools for epidemiology [+
correction art. no 439, 1 p.]. BMC Bioinformatics, 20.
Bui, T. M. A., Ziane, M., Stinckwich, S., Ho, T. V., Roche,
B., and Papoulias, N. (2016). Separation of concerns
in epidemiological modelling. In Companion pro-
ceedings of the 15th international conference on mod-
ularity, pages 196–200.
Ciocchetta, F. and Hillston, J. (2010). Bio-PEPA for epi-
demiological models. Electronic Notes in Theoretical
Computer Science, 261:43–69.
Gamma, E., Helm, R., Johnson, R., Vlissides, J., and Pat-
terns, D. (1995). Elements of reusable object-oriented
software, volume 99. Addison-Wesley Reading, Mas-
sachusetts.
Gilmore, S. and Hillston, J. (1994). The PEPA workbench:
A tool to support a process algebra-based approach to
performance modelling. In Haring, G. and Kotsis, G.,
editors, Computer Performance Evaluation Modelling
Techniques and Tools, pages 353–368, Berlin, Heidel-
berg. Springer Berlin Heidelberg.
Hladish, T., Melamud, E., Barrera, L. A., Galvani, A., and
Meyers, L. A. (2012). EpiFire: An open source C++
library and application for contact network epidemiol-
ogy. BMC bioinformatics, 13(1):1–12.
H
¨
ursch, W. L. and Lopes, C. V. (1995). Separation of con-
cerns. Technical Report NU-CCC-95-03, Northeast-
ern University, Boston, USA.
Keeling, M. J. and Rohani, P. (2011). Modeling infectious
diseases in humans and animals. Princeton university
press.
Levin, S. A. and Durrett, R. (1996). From individu-
als to epidemics. Philosophical Transactions of the
Royal Society of London. Series B: Biological Sci-
ences, 351(1347):1615–1621.
Mccaig, C., Norman, R., and Shankland, C. (2009). From
individuals to populations: A symbolic process alge-
bra approach to epidemiology. Mathematics in Com-
puter Science, 2:535–556.
Miller, J. C. and Ting, T. (2020). EoN (Epidemics on Net-
works): a fast, flexible python package for simulation,
analytic approximation, and analysis of epidemics on
networks. arXiv preprint arXiv:2001.02436.
Mollison, D., Isham, V., and Grenfell, B. (1994). Epi-
demics: models and data. Journal of the Royal
Statistical Society: Series A (Statistics in Society),
157(1):115–129.
Muellner, U., Fourni
´
e, G., Muellner, P., Ahlstrom, C.,
and Pfeiffer, D. U. (2018). epidemix—an interactive
multi-model application for teaching and visualizing
infectious disease transmission. Epidemics, 23:49–54.
Pastor-Satorras, R. and Vespignani, A. (2001). Epidemic
spreading in scale-free networks. Physical review let-
ters, 86(14):3200.
Picault, S., Huang, Y.-L., Sicard, V., Arnoux, S., Beaun
´
ee,
G., and Ezanno, P. (2019). Emulsion: Transparent
and flexible multiscale stochastic models in human,
animal and plant epidemiology. PLoS computational
biology, 15(9):e1007342.
Plateau, B. and Stewart, W. J. (2000). Stochastic automata
networks. In Computational Probability, pages 113–
151. Springer.
Shirley, M. D. and Rushton, S. P. (2005). The impacts of
network topology on disease spread. Ecological Com-
plexity, 2(3):287–299.
Stroud, P. D., Sydoriak, S. J., Riese, J. M., Smith, J. P.,
Mniszewski, S. M., and Romero, P. R. (2006). Semi-
empirical power-law scaling of new infection rate to
model epidemic dynamics with inhomogeneous mix-
ing. Mathematical biosciences, 203(2):301–318.
Watts, D. J. and Strogatz, S. H. (1998). Collective dynamics
of ‘small-world’ networks. Nature, 393(6684):440–
442.
Separation of Concerns in Extended Epidemiological Compartmental Models
159
