
3D MRI Image Segmentation using 3D UNet Architectures: Technical
Review
Vijaya Kamble
1a
and Rohin Daruwala
2b
1
Research Scholar Department of Electronics Engineering, Veermata Jijabai Technological Institute (VJTI),
Mumbai, Maharashtra, India
2
Department of Electronics Engineering, Veermata Jijabai Technological Institute (VJTI), Mumbai, Maharashtra, India
Keywords: 3D UNet, MRI
Images, Segmentation, Brain Tumors
.
Abstract: From last few decades machine learning & deep convolutional neural networks (CNNs) used extensively and
have shown remarkable performance in almost all fields including medical diagnostics. It is used in medical
domain for automatic tissue, lesion detection, segmentation, anatomical or structure segmentation classifica-
tion & survival predictions. In this paper we presented an extensive technical literature review on 3D CNN
U-Net architectures applied for 3D brain magnetic resonance imaging (MRI) analysis. We mainly focused on
the architectures, its modifications, pre-processing techniques, types datasets, data preparation, methodology,
GPU, tumor disease types and per architectures evaluation measures in this works. Our primary goal for this
extensive technical review is to report how different 3D U-Net architectures or CNN architectures have been
used to differentiate between state-of-the-art strategies, compare their results obtained using public/clinical
datasets and examine their effectiveness. This paper is intended to present detailed reference for further re-
search activity or plan of strategy to use 3D U-Nets for brain MRI automated tumor diseases detection, seg-
mentation & survival prediction analysis. Finally, we are presenting a novel perspective to assist research
directions on the future of CNNs & 3D U-Net architectures to explore in subsequent years to help doctors &
radiologist.
1 INTRODUCTION
Over last few decades the use of machine learning and
deep learning techniques revolutionized medical
imaging field for tumor or disease segmentation,
detection & survival predication. It is helping
physicians to diagnose brain cancers quickly to boost
prognosis. A patient’s MRI is the three-dimensional
brain anatomy (Oday Ali Hassen, et al., 2021). MRI
images of different modalities such as T1-weighted,
T2-weighted, T1c, and Flair as T1c has precise data
such as tumor form, location, and scale. Different
MRI modalities are used in brain tumor extraction
and segmentation. Among all types of brain tumors
Gliomas are most common tumors in brain cancer
with high mortality rate. These brain tumors
originating from the glial cells in the central nervous
system. Gliomas are 70% of all brain tumors. The
survival duration of patients with high grade gliomas
a
https://orcid.org/0000-0002-6469-8837
b
https://orcid.org/0000-0002-3267-6270
(HGG) lead less than 2 years if prognosis is poor.
Compared with HGG, prognosis of low grade
gliomas (LGG) are more effective (Chandan
Yogananda, et al., 2020).
Different architectures of CNNs used in medical
imaging and other applications from year 1990s.
Medical Image data is sensitive patients data and not
available easily. Earlier limitations on on
performance of CNN networks years as less labeled
medical data available. But now large annotated
medical public & clinical data sets available online &
on demand and more powerful graphics processing
units (GPUs) available for data processing so this is
enabling researchers to continue working in the area
to help doctors (Chandan Yogananda, et al., 2020).
Automated or semi automated segmentation
methods saving physicians time and provide an
accurate reproducible solutions for 3D brain tumor
analysis and patient monitoring. Convolutional neural
Kamble, V. and Daruwala, R.
3D MRI Image Segmentation using 3D UNet Architectures: Technical Review.
DOI: 10.5220/0010851300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 141-146
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
141
networks (CNN) able to learn from examples so they
demonstrate state-of-the-art segmentation accuracy
both in 2D natural images (Andriy Myronenko, 2019)
and in 3D medical image modalities. Its difficult to
differentiate brain tumors from normal tissues
because tumor boundaries are ambiguous and there is
a high degree of variability in the shape, location,
intensity in homogeneity, or different intensity ranges
between the same sequences and acquisition scanners
and extent of the patient (Li Sun, et al., 2019). This
can influence the segmentation accuracy and correct
detection of tumor. Different hospitals shows
different gray-scale values for the same tumorous
cells may when they are scanned differently.
Although advance automatic algorithms used for
brain tumor segmentation, the problem is still remains
a challenging task.
To address issues in this research area we have
done extensive comparative review of most cited
research papers based on 3D U-Net architectures &
3D medical imaging modalities, with different
processing techniques use of powerful GPUs
different software's with various high grade tumors
classification segmentation & survival prediction of
patients.
Summary of this extensive most cited research is
mentioned in table no 1 with reference to paper, few
prominent U-Net model parameter & methodology
discussed in short with figure. Different imaging
modalities, preprocessing techniques datasets,
evaluation parameters advantages & limitations also
mentioned. Most of the reviewed content got dice
scores above 0.75 to 0.89 range for whole tumor core
tumors & enhancing tumors. Some of the papers got
excellent accuracy, sensitivity & specificity.
2 CNN ARCHITECTURES
CNN architectures used in medical imaging for
segmentation detection & predictions of disease
diagnosis prognosis. CNN architectures can be
grouped around five sub types:
I) Based on interconnected operating modules,
II) Selection of types of input MRI modalities,
III) Selection of input patch dimension,
IV) Number of Predictions at a time
V) Based on implicit and explicit contextual
information.
In this summary of the literature review methods dis-
tinguish with the different CNN architectures mostly
on types of U-Nets, pre processing, post-processing
and target of the segmentation & tumor types.
2.1 UNet Architectures Literature
Survey
In medical imaging for brain tumor disease diagnosis
prognosis for image semantic segmentation and
classifications mostly U-Net ResNet architectures are
used.
The U-Net is one of the most popular convolu-
tional neural network end-to-end architectures in the
field of semantic segmentation.a that is designed for
fast and precise segmentation of images. In several
challenges U-Net has performed extremely well.
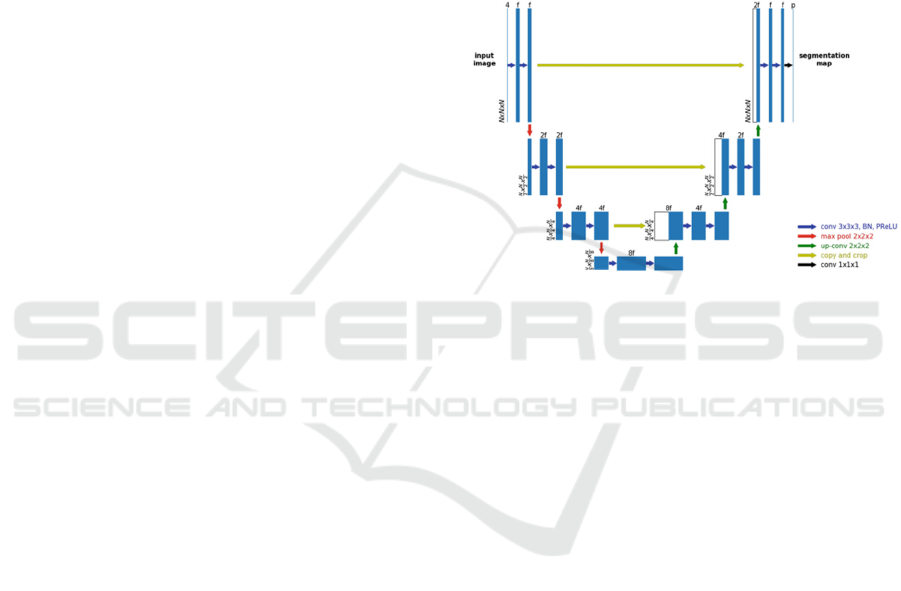
Figure 1: UNet Architecture (Xue Feng et al.).
U-Net Architecure split the network into two parts:
Encoder: The encoder path is the backbone. The
encoder captures features at different scales of the
images by using a traditional stack of convolutional
and max pooling layers. A block in the encoder
consists of the repeated use of two convolutional
layers (k=3, s=1), each followed by a non-linearity
layer, and a max-pooling layer (k=2, s=2). For every
convolution block and its associated max pooling
operation, the number of feature maps is doubled to
ensure that the network can learn the complex
structures effectively.
Decoder: The decoder path is a symmetric expanding
counterpart that uses transposed convolutions. This
type of convolutional layer is an up-sampling method
with trainable parameters and performs the reverse of
(down)pooling layers such as the max pool. Similar to
the encoder, each convolution block is followed by
such an up-convolutional layer. The number of feature
maps is halved in every block. Because recreating a
segmentation mask from a small feature map is a
rather difficult task for the network, the output after
every up-convolutional layer is appended by the
feature maps of the corresponding encoder block. The
feature maps of the encoder layer are cropped if the
BIOIMAGING 2022 - 9th International Conference on Bioimaging
142

dimensions exceed the one of the corresponding
decoder layers.
In the end, the output passes another convolution
layer (k=1, s=1) with the number of feature maps
being equal to the number of defined labels. The
result is a u-shaped convolutional network that offers
an elegant solution for good localization and use of
context. Let’s take a look at the code. max pooling
operations (in each dimension) are the most
appropriate.
In these section the from most cited literature
review best architecture discussed. Researchers
proposed common U-nets, cascaded U-Nets,
modified type of Unet architectures for brain tumor
detection & survival predictions.
Xue Feng et al. explained generic 3D U-Net
structure with different hyper-parameters, deployment
of each model is for full volume prediction and final
ensemble modeling. Model fitting done for the survival
task feature extraction (Xue Feng, et al., 2019).
Figure 2: Cascaded Unet Architecture (Yan Hu, et al.).
Yan Hu et al proposed algorithm for intra-tumor
structure segmentation using three cascaded U-Net
models as shown in figure 2. They are concatenated
and further processed by two convolutional layers to
detect tumor region. The feature maps generated by
three cascaded U-Net models using T1, T1c, T2 and
FLAIR modalities.Patches are cropped within tumor
region detected for classification model (Yan Hu , et
al., 2018).
2.2 Pre-processing
In computer vision or image processing domain pre-
processing of image is preliminary but important task.
Brain MRI volumes acquired from scanners, these
volumes are with nonbrain tissues, parts of the head
or skull, eyes, fat, spinal cord. From acquired MRI
volumes extracting the brain tissue from non-brain
image is the pre processing primary task. This is
known as skull strippings. This is an essential step for
subsequent segmentation task. To achieve a good
performance in training supervised models such as
CNNs, or Unets the input training data hugely
influences the performance of the model, so having
preprocessed and well-annotated data is a crucial step
in MRI image processing. This step is very important
as it has direct impact on the performance of auto-
mated segmentation methods. Inclusion of skull or
eyes as brain tissue in MRI analysis may lead to
unexpected results missed classification or tumor
detection. In this review context every researcher
used some preprocessing methods & post processing
method for correct segmentation, predictions results.
In MRI image preprocessing there are few
common but fixed steps or algorithms stated as below:
i) Intensity Normalization-as there are different
image modalities,
ii) Bias field Corrections,
iii) Skull stripping,
iv) Image registration.
After Image preprocessing step there is data
preparation phase in CNN algorithms in that data
augmentation, 2D, 3D patch extraction before
segmentation & classification task.
2.3 Input Modalities
There are various types of MRI images based on their
scanning techniques, acquisition modes, intensities.
Basically in MRI four modalities are popular among
research community T1,T2 T2c Flair. In the literature
strategies of selection of modality for processing can
also be grouped according to the number of
modalities that are processed at the same time. The
major categories are two: single- and multi-modality.
3 EVALUATION
MEASUREMENTS
Final Deep CNN models performance majorly
depend on the types of dataset, modalities,
types of
tumors regions, sub-regions & model parameters. In
this extensive research survey of 3D UNet & MRI
imaging following evaluation metrics were used for
segmentation & classification of tumors:
i) Global accuracy,
ii) Dice coefficient,
iii) Recall,
iv) Precision and
v) Housedraf distance measure.
3D MRI Image Segmentation using 3D UNet Architectures: Technical Review
143

Following are the 6 Equations for evaluation
parameters with the well known terms False Positives
(FP), False Negatives (FN), True Negative(TN):
𝐺𝑙𝑜𝑏𝑎𝑙𝐴𝑐𝑐𝑢𝑟𝑎𝑐𝑦
𝑇𝑃 𝑇𝑁
𝑇𝑃
𝑇𝑁
𝐹𝑃
𝐹𝑁
(1)
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛
𝑇𝑃
𝑇𝑃
𝐹𝑃
(2)
𝑅𝑒𝑐𝑎𝑙𝑙
𝑇𝑃
𝑇𝑃
𝐹𝑁
(3)
𝐹1 2
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 ∗ 𝑅𝑒𝑐𝑎𝑙𝑙
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 𝑅𝑒𝑐𝑎𝑙𝑙
(4)
𝐷𝑖𝑐𝑒
2𝑇𝑃
2𝑇𝑃
𝐹𝑃
𝐹𝑁
(5)
𝑆𝑝𝑒𝑐𝑖𝑓𝑖𝑐𝑖𝑡𝑦
𝑇𝑁
𝑇𝑁
𝐹𝑃
(6)
The main evaluation measures for the challenges
mentioned pre-viously are DSC, specificity,
sensitivity, positive predictive value (pre-cision),
average surface distance (ASD), average volumetric
difference (AVD) and modified Hausdorff distance
(MHD).
Table 1 summarizes the types of architectures,
databases, numbers of samples, MRI modalities
considered, tumor diseases types, GPU types,
Software's used, evaluation measurements applied
and corresponding results reported of the extensive
technical surveyed work (Chandan Yogananda, et al.,
2020 - http://www.tomography.org/).
4 MEDICAL CNN-BASED
SOFTWARES
Now a days most of researchers release their winning
competitions or challenges source codes to the public
it helps for research in the medical & other fields.
There are few free deep learning libraries for MRI
segmentation as listed below:
i) Tensorflow,
ii) Theano,
iii) Caffe,
iv) Keras
v) PyTorch.
There are few CNN open-source frameworks
namely NiftyNet 17 and DLTK.
Researchers work on clinical & publically
available datasets depending on their application.
5 CONCLUSION AND FUTURE
SCOPE
Automatic brain tumor segmentation for cancer
diagnosis & prediction is challenging task. Most recent
advancements in medical diagnostic research using
Deep Convolution Neural network 3D Unet
architectures discussed in this technical most cited
literature review paper. This 3D Unet architectures &
modified frameworks indicate significant potential to
segment classify &predict the brain tumors lesions
from the 3D MRI images. Even though MRI images
are of different modalities intensities and categories
still complex features from these MRI images can be
automatically extracted from 3D Unet architectures it
also segment tumor with subregions. There is always
chance of improvements and modifications in CNN
architectures, Unet architectures to improve the
efficiency of segmentation, detection & predictions of
cancerous brain tumors.
With this deep technical review we observed and
analyzed that most of the proposed methods are based
on specific 3D MRI modalities for high grade tumor
segmentation so they have computational
complexities as well as memory constraints & in need
of specific GPU speed for software's. In most of
papers deep learning software libraries are used to
implement layers of deep CNNs. They are arranged
either parallel or distributed or cascaded frameworks,
which help researchers to train their models in multi-
core architectures or GPUs. Mostly Nvidia GPUs &
Intel GPUs used for training and implementation of 3
D Unet CNN models. It is observed from evaluation
measures that the training and validation for brain
image analysis is significantly affected by the data
imbalance problem .Lesions are smaller than the
entire volume so it affects generalization & robust
model. We observed that the full capacity of 3D Unet
CNN architectures has not yet been fully leveraged in
brain MRI analysis. More sophisticated dedicated
softwares are available for Medical imaging or Brain
MRI analysis. But there is always a challenge for
domain adaptation techniques, more research in this
sense is needed for permanent solutions for high
grade and low grade tumors for correct diagnosis
without experts interventions.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
144

Table 1: Comparison with different U-Net models.
SrNo Article Dataset Number of scans Model GPU Softwares Segmentation
tasks
Evaluation Measure
1 Yue Zhao
et al.
BraTS 2018 75 low grade and 210 high
grade gliomas
3D recurrent
multi-fiber network
NVIDIA Tesla
V100 32 GB GPU.
Pytorch MS lesion
Whole brain,
tissue and sub-
cortical
structure
Dice scores of WT 89.62%,
TC 83.65% and ET 78.72%
2 Chandan
Ganesh
Bangalore
Yogananda
et al
BraTS2017,
BraTS2018,
Oslo data set 52
LGG and HGG
(age > 18 years)
scanned from
2003 to 2012
(200 cases 150 HGG and 50
LGG), validation (65 48 HGG
and 17 LGG) and 10% (20 12
HGG and 8 LGG)
3D-Dense-Unets
Combination of WT-net,
TC net, ET net
Tesla P100, P40 or
K80 NVIDIA-
GPUs
Tensorflow,
Keras,
python
p
ackage, and
Pycharm
IDEs with
(Adam)
MS lesion
Whole brain,
tissue and sub-
cortical
structure,
Stroke
Dice-scores for WT 0.90, TC
0.84, and ET 0.80
3 Oday Ali
Hassen et al
BRATS 2019
and BRATS
2017. At
BRATS 2019
3D-MRI of 336 heterogeneous
gliomas patients, 259 HGG and
76 Low-Grade Gliomas LGG
Population-based Artificial
Bee Colony Clustering (P-
ABCC) methodology, K-
means
Intel (R), Core
(TM) i3 CPU, 8.00
GB RAM
MATLAB
R2018a
Brain Tumour Entire Tumor (WT), Tumor
Center (TC), Improved (ET) by
0.03%, 0.03%, and 0.01%
respectively. At BRATS 2017,
an increase in precision for WT
was reached by 5.27%.
4
Jing Huang
and Minhua
Zheng et al
BRATS 2017
285 patients, 210 HGG images,
75 LGG images
NVIDIA RTX
TITAN 24GB GPU
Brain Tumour
Dice scores Similarity
WT 0.9089, TC 0.7165, and
ET 0.8398
5 Parvez
Ahmad et
al
BRATS
2018
228 training images 57 testing
images out of data set is 285.
Residual 3D U-net, Dense
inception-like architecture
with multiple dilated
convolutional layers
Keras Brain Tumour Dice Similarity
WT 87.16, ET 84.81, 80.20,
Whole 86.42, Sensitivity
Core, Enhancing 82.15,80.01
6 Hassan A.
Khalil et al
BRATS 2017 Clustering technique
integrates k-means and the
dragonfly algorithm
Intel, Core i3 CPU
with 8.00 GB of
RAM
MATLAB
software
R2018a
Brain Tumour Accuracy 98.20, Recall 95.13,
Precision 93.21
7 Xue Feng
et al
CBICA’s Image
Processing
Portal
163 training subjects, 285
training subjects, 66 subjects
were provided as validation
An ensemble of 3D U-Nets
with different hyper-
parameters for brain tumor
segmentation
Nvidia Titan Xp
GPU with 12 Gb
Tensorflow
framework
was used
with Adam
optimizer
Brain Tumour Accuracy was 0.321, MSE
was 99115.86, median SE
was 77757.86, std SE was
104291.596 and Spearman
Coefficient was 0.264
8
Andriy
Myronenko
et al
BraTS 2018
285 Training cases validation
(66 cases) and the testing sets
(191 cases)
Encoder-decoder based
CNN architecture
asymmetrically larger
encoder to smaller decoder
NVIDIA Tesla,
V100 32 GB GPU
Tensorflow Brain Tumour
Dice Similarity ET 0.7664,
WT 0.8839 and TC 0.8154
9
Wei Chen,
Boqiang
Liu et al.
BraTS 2018
285 subjects, of which 210 are
GBM/HGG and 75 are LGG
Separable 3D U-Net
GeForce GTX
1080Ti GPU
PyTorch
toolbox,
Adam
Brain Tumor
Dice scores of ET 0. 68946,
WT 0. 83893 and TC 0.
78347
10 Xiaojun Hu
et al.
BRATS 2015,
ISLES 2017
database
3D Brain SegNet 4 Titan Xp
GPUs,8G memory
for each GPU
Pytorch Brain Tumor Dice Score 0.30± 0.22,
0.35±0.27, 0.43±0.27
11 Li Sun et
al.
BraTS 2018 210 HGG and 75 LGG Three different 3D CNN
architectures (CA-CNN,
DFKZ Net, 3D U-Net,
Wnet, Tnet, Enet)
Brain Tumor,
survival
prediction
61.0% accuracy
12 Dmitry
Lachinov et
al
BraTS 2018 285 MRIs for training (210
high grade and 75 low grade
glioma images), 67 validation
and 192 testing MRIs.
Multiple Encoders Unet,
Cascaded UNet
GTX 1080TI MXNet
framework
Brain Tumor,
survival
prediction
Dice score of ET 0.720, WT
0.878, TC 0.785
13
Ping Liu et
al
BraTS 2017
285 samples with manually
annotated and confirmed
ground truth labels
Deep supervised 3D
Squeeze-and-Excitation V-
Net (DSSE-V-Net)
4 NVIDIA Titan
1080 TI 11GB
GPUs
Pytorch
Brain Tumor,
survival
prediction
Dices of WT and TC of DS-U-
Net increased to 0.8953 and
0.7828 from 0.8799 and 07693
of 3D U-Net, respectively
14
Pawel
Mlynarski
et al
BRATS 2017
285 scans (210 high grade
gliomas and 75 low grade
gliomas
CNN-based model,short-
range 3D context and the
long-range 2D context
Keras,
Tensor
Flow
Brain Tumor,
survival
prediction
Dice scores of WT 0.918, TC
0.883 ET 0.854
15 Suting
Peng et al
BraTS 2015 220 HGG and 54 LGG Multi-Scale 3D U-Nets
architecture
NVIDIA GeForce
GTX 1080Ti GPU
Brain Tumor,
survival
prediction
Dice similarity
WT 0.85, ET 0.72, WC 0.61
16 Mina
Ghaffari et
al
BraTS 2018 230 cases for training, and the
remaining 55 cases were
reserved for testing.
Modified version of the
well-known U-Net
architecture
4 x
NVIDIA Tesla
Pascal P100
Brain Tumor Dice similarity WT,0.87, ET
0.79 WC0.66
17 Parvez
Ahmad et
al
BraTS 2018 80% of subjects for training and
20% for validation
3D Dense Dilated
Hierarchical Architecture
Brain Tumor, Dice similarity
WT 0.8480, TC 0.8574 CT
0.8219
18
Shangfeng
Lu et al
Brats2019
259 high-grade gliomas (HGG)
and 76 low-grade gliomas
(LGG)
Multipath feature extraction
3D CNN
NVIDIA 1080ti
GPU with 11G
RAM
pytorch Brain Tumour
Dice similarity
WT 0.881, TC 0.837, ET
0.815
19
Saqib
Qamar et al
BraTS 2018
210 patients, to train and test
our model
3D Hyper-dense Connected
Convolutional Neural
Network
Brain Tumour
Dice similarity
WT 0.87, ET 0.81, CT 0.84
0 Yan Hu et
al
BraTS 2017 285 training subjects, 46
validation subjects and 146 test
bjects.
3D Deep neural network Intel Xeon 2.10
GHz CPU, NVIDIA
GTX 1080 Ti GPU,
32 GB RAM
Brain Tumour Dice similarity
WT 0.81, CT 0.69 and ET 0.55
3D MRI Image Segmentation using 3D UNet Architectures: Technical Review
145

REFERENCES
Chandan Ganesh Bangalore Yogananda, Bhavya R. Shah,
Maryam Vejdani-Jahromi, Sahil S. Nalawade,
Gowtham K. Murugesan, Frank F. Yu, Marco C. Pinho,
Benjamin C. Wagner, Kyrre E. Emblem, Atle Bjørne-
rud, Baowei Fei, Ananth J. Madhuranthakam, and Jo-
seph A. Maldjian “A Fully Automated Deep Learning
Network for Brain Tumor Segmentation” tomogra-
phy.org, volume 6, number 2, June 2020, ISSN 2379-
1381 https://doi.org/10.18383/j.tom.20 19.00026
Oday Ali Hassen, Sarmad Omar Abter, Ansam A. Ab-
dulhussein, Saad M. Darwish, Yasmine M. Ibrahim 4
and Walaa Sheta, “Nature-Inspired Level Set Segmen-
tation Model for 3D-MRI Brain Tumor Detection”,
2021.
Yue Zhao, Xiaoqiang Ren, Kun Hou, Wentao Li, “Recur-
rent Multi-Fiber Network for 3D MRI Brain Tumor
Segmentation”, Symmetry 2021, 13, 320.
https://doi.org/10.3390/sym13020320,https://www.md
pi.com/journal/symmetry
Parvez Ahmad, Hai Jin, Saqib Qamar, Ran Zheng, and
Wenbin Jiang, “Combined 3D CNN for Brain Tumor
Segmentation”, 2020 IEEE Conference on Multimedia
Information Processing and Retrieval (MIPR), 978-1-
7281-4272-2/20/, DOI 10.1109/MIPR49039.2020.0
0029,
Hassan A. Khalil, Saad Darwish, Yasmine M. Ibrahim and
Osama F. Hassan, “3D-MRI Brain Tumor Detection
Model Using Modified Version of Level Set Segmen-
tation Based on Dragonfly Algorithm”, Symmetry
2020, 12, 1256; doi:10.3390/sym12081256,
www.mdpi.com/journal/symmetry,
Xue Feng, Nicholas Tustison, Craig Meyer, “Brain Tumor
Segmentation Using an Ensemble of 3D U-Nets and
Overall Survival Prediction Using Radiomic Features”,
Springer Nature Switzerland AG 2019. A. Crimi et al.
(Eds.): BrainLes 2018, LNCS 11384, pp. 279–288,
2019. https://doi.org/10.1007/978-3-030-11726-9_25,
Andriy Myronenko, “3D MRI Brain Tumor Segmentation
Using Autoencoder Regularization, “Springer Nature
Switzerland AG 2019 A. Crimi et al. (Eds.): BrainLes
2018, LNCS 11384, pp. 311–320, 2019.
https://doi.org/10.1007/978-3-030-11726-9 _ 28.
Wei Chen, Boqiang Liu, Suting Peng, Jiawei Sun, and Xu
Qiao, “S3D-UNet: Separable 3D U-Net for Brain
Tumor Segmentation”, Springer Nature Switzerland
AG 2019, A. Crimi et al. (Eds.): BrainLes 2018, LNCS
11384, pp. 358–368, 2019. https://doi.org/10.1007/
978-3-030-11726-9_32.
Xiaojun Hu, Weijian Luo, Jiliang Hu, Sheng Guo, Weilin
Huang, Matthew R. Scott, Roland Wiest, Michael
Dahlweid and Mauricio Reyes, “Brain SegNet: 3D
local refinement network for brain lesion
segmentation”, Hu et al. BMC Medical Imaging, (2020)
20:17, https://doi.org/s12880-020-0409-2.
Li Sun, Songtao Zhang, Hang Chen, Lin Luo, “Brain Tu-
mor Segmentation and Survival Prediction Using Mul-
timodal MRI Scans With Deep Learning”, Frontiers in
Neuroscience, www.frontiersin.org, August 2019, Vol-
ume 13, Article 810.
Dmitry Lachinov, Evgeny Vasiliev, and Vadim Turlapov,
“Glioma Segmentation with Cascaded Unet”, Springer
Nature Switzerland AG 2019, A. Crimi et al. (Eds.):
BrainLes 2018, LNCS 11384, pp. 189–198, 2019,
https://doi.org/10.1007/978-3-030-11726-9, 17.
Ping Liu, Qi Dou, Qiong Wang and Pheng-Ann Heng, “An
Encoder-Decoder Neural Network With 3D Squeeze-
and-Excitation and Deep Supervisionfor Brain Tumor
Segmentation”, IEEE Access 2020, Digital Object
Identifier 10.1109/ACCESS.2020.2973707.
Pawel Mlynarski, Hervé Delingette, Antonio Criminisi, Nich-
olas Ayache, “3D convolutional neural networks for tu-
mor segmentation using long-range 2D context,
https://doi.org/10.1016/j.compmedimag.2019.02.001
0895-6111/, 2019 Elsevier Ltd.
Suting Peng, Wei Chen, Jiawei Sun, Boqiang Liu, “Multi-
Scale 3D U-Nets: An approach to automatic segmenta-
tion of brain tumor”, 2019, Wiley Periodicals, Inc.,
DOI: 10.1002/ima.22368
Mina Ghaffari, Arcot Sowmya, Ruth Oliver, Len Hamey,
“Multimodal brain tumour segmentation using densely
connected 3D convolutional neural network”, 978-1-
7281-3857-2/19/, 2019, IEEE.
Parvez Ahmad, Hai Jin, Saqib Qamar, Ran Zheng, Wenbin
Jiang, Belal Ahmad, Mohd Usama, “3D Dense Dilated
Hierarchical Architecture for Brain Tumor Segmenta-
tion”, 2019, Association for Computing Machinery,
ACM, ISBN 978-1-4503-6278-8/19/05, http://doi.org/
10.1145/3335484.3335516
Shangfeng Lu, Xutao Guo, Ting Ma, Chushu Yang, Tong
Wang, Member, Pengzheng Zhou, “Effective Multipath
Feature Extracion 3D CNN for Multimodal Brain Tu-
mor Segmentation”, 2019 International Conference on
Medical Imaging Physics and Engineering (ICMIPE).
Saqib Qamar, Hai Jin, Ran Zheng, Parvez Ahmad, “3D Hy-
per-dense Connected Convolutional Neural Network
for Brain Tumor Segmentation”, 978-1-7281-0441-
6/18/, DOI 10.1109/SKG.2018.00024.
Jing Huang and Minhua Zheng, Peter X. Liu, “Automatic
Brain Tumor Segmentation Using 3D Architecture
Based on ROI Extraction”, Proceeding of the IEEE In-
ternational Conference on Robotics and Biomimetics,
978-1-7281-6321-5/19/.
Yan Hu, Yong Xia, “3D Deep Neural Network-Based Brain
Tumor Segmentation Using Multimodality Magnetic
Resonance Sequences”, Springer International Publish-
ing AG, part of Springer Nature, 2018. A. Crimi et al.
(Eds.): BrainLes 2017, LNCS 10670, pp. 423–434,
2018. https://doi.org/10.1007/978-3-319-75238-9 _ 36.
Despotovic I., Goossens B., Philips W., MRI segmentation
of the human brain: challenges, methods, and applica-
tions. Computational and mathematical methods in
medicine, 2015.
www.tomography.org
BIOIMAGING 2022 - 9th International Conference on Bioimaging
146
