
Distributed Deep Learning for Multi-Label Chest Radiography
Classification
Maram Mahmoud A. Monshi
1,2 a
, Josiah Poon
1 b
and Vera Chung
1 c
1
School of Computer Science, The University of Sydney, Camperdown, NSW, 2006, Australia
2
Department of Information Technology, Taif University, Taif, 26571, Saudi Arabia
Keywords:
Distributed Deep Learning, Chest x-ray, Multi-Label Classification.
Abstract:
Chest radiography supports the clinical diagnosis and treatment for a series of thoracic diseases, such as
cardiomegaly, pneumonia, and lung lesion. With the revolution of deep learning and the availability of large
chest radiography datasets, binary chest radiography classifiers have been widely proposed in the literature.
However, these automatic classifiers neglect label co-occurrence and inter-dependency in chest radiography
and fail to make full use of accelerators, resulting in inefficient and computationally expensive models. This
paper first studies the effect of chest radiography image format, variations of Dense Convolutional Network
(DenseNet-121) architecture, and parallel training on chest radiography multi-label classification task. Then,
we propose Xclassifier, an efficient multi-label classifier that trains an enhanced DenseNet-121 with a blur
pooling framework to classify chest radiography based on fourteen predefined labels. Xclassifier accomplishes
an ideal memory utilization and GPU computation and achieves 84.10% AUC on the MIMIC-CXR dataset and
83.89% AUC on the CheXpert dataset. The code used to generate the experiment results mentioned in this
paper can be found here: https://github.com/MaramMonshi/Xclassifier.
1 INTRODUCTION
Chest x-rays are of great importance for clinical di-
agnosis as they contain rich relationship informa-
tion among pathologies such as label co-occurrence
of multiple observations (Pham et al., 2021). The
availability of large public chest radiography datasets
(Wang et al., 2017) (Bustos et al., 2020) (Irvin et al.,
2019) (Johnson et al., 2019a) and the revolution of
deep learning offer an optimal solution for the multi-
label chest radiography classification problem. Con-
sequently, many recent models have been proposed
in the applications of classifying chest radiographs
(Rajpurkar et al., 2017) (Wang et al., 2018) (Monshi
et al., 2019) (Yarnall, 2020). However, these methods
did not capture the label dependencies in chest radio-
graphs, and effectively accomplishing this task is still
a challenge(Chen et al., 2020).
On the computation side, the computation power
grows tremendously with the introduction of a state-
of-the-art Graphics Processing Unit (GPU) such as
NVIDIA A100 (NVIDIA, 2020) and NVIDIA V100
a
https://orcid.org/0000-0001-5622-1601
b
https://orcid.org/0000-0003-3371-8628
c
https://orcid.org/0000-0002-3158-9650
(NVIDIA, 2018), but on-device memory is often con-
strained. NVIDIA A100 is the new generation of ac-
celerator GPUs but is still not supported on all plat-
forms. Parallel training on the other hand is perform-
ing multi-processes on devices of single/multiple ma-
chines. As public chest radiography datasets and the
number of deep learning layers get bigger, one GPU
quickly becomes insufficient to accelerate neural net-
work training. However, evaluating these techniques
in real-world applications such as classifying chest x-
rays is limited.
Further, existing chest radiography classifiers’
performance can be improved by leveraging label co-
occurrence (Chen et al., 2020), selecting the optimal
radiographs format (Sabottke and Spieler, 2020) and
training with an efficient approach. By studying pre-
vious methods on these issues, it is noted that existing
literature rarely discusses the efficiency of the chest
radiography classifiers.
Our contribution can be outlined as follows. Re-
garding the multi-label chest x-ray classification task,
we quantify the value of the optimal image format,
study parallels deep learning in accelerating neural
network training, and compare the performance of
variations of Dense Convolution Network (DenseNet-
Monshi, M., Poon, J. and Chung, V.
Distributed Deep Learning for Multi-Label Chest Radiography Classification.
DOI: 10.5220/0010849400003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 4: VISAPP, pages
949-956
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
949
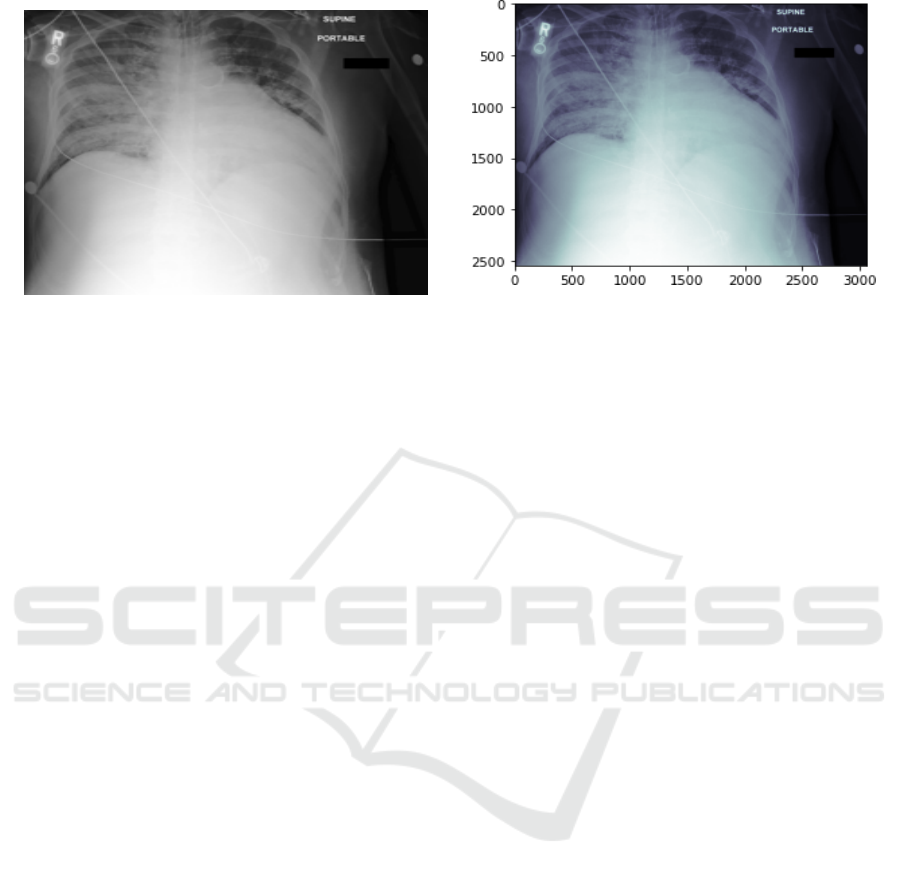
(a) Joint Photographic Experts Group (JPEG).
(b) Digital Imaging and Communications in Medicine (DI-
COM).
Figure 1: Chest X-Ray Image Format.
121). Then, we propose the Xclassifier, an efficient
and accurate multi-label chest x-ray classifier, based
on an enhanced DenseNet-121 framework with an-
tialiasing blur pooling and parallel training.
2 RELATED WORK
2.1 Chest Radiography Classification
The simplest method to solve the multi-label chest ra-
diography classification problem is using binary clas-
sification with Convolution Neural Network (CNN).
For instance, CheXNet (Rajpurkar et al., 2017),
TieNet (Wang et al., 2018), MultiViewModel (Mon-
shi et al., 2019), and VGG16-based model (Yarnall,
2020) trains independent binary classifiers for each
label with CNNs. CheXNet achieved benchmark per-
formance on detecting pneumonia using a modified
DenseNet. To improve the classification accuracy,
TieNet added text embedding information and the
MultiViewModel utilized various views of the chest
x-rays. Recently, Yarnall (Yarnall, 2020) studied
the effect of various CNN architectures with differ-
ent hyperparameters on classification accuracy. The
study used Visual Geometry Group (VGG-16) (Si-
monyan and Zisserman, 2014) with the ReLU acti-
vation function, resulting in an accuracy that ranged
from 62.23% to 83.52% for each label. However,
these single label classifiers did not consider any
pathology correlation and ignored the relationship in-
formation among labels.
From a practical perspective, some of the chest x-
rays labels might be closely linked and their inter-
dependency is very important for final diagnostics.
For example, infiltration is often associated with at-
electasis (Wang et al., 2017) and cardiomegaly tends
to be linked with pulmonary edema (Yao et al.,
2017). To examine multiple labels simultaneously,
latent-space self-ensemble model employees stacked
semi-supervised learning, using unsupervised disen-
tangled representation learning (Gyawali et al., 2019).
This model achieved a 66.97% AUC on CheXpert
(Irvin et al., 2019). Recently, the Visual-Semantic
Embedded - Graph Convolutional Networks (VSE-
GCN) model fed joint features of label embed-
dings and visual features into a GCN to model the
correlations among chest x-ray labels (Hou et al.,
2021). Differently, CheXclusion investigates fairness
gaps in deep-learning-based chest x-ray classifiers to
evaluate the true positive rates disparity for public
datasets (Seyyed-Kalantari et al., 2020). VSE-GCN
and CheXclusion achieved 72.10% and 83.40% on
MIMIC-CXR (Johnson et al., 2019a), respectively.
We extended this wave of multi-label classification re-
search using more efficient training methods.
The most common file format used to store med-
ical imaging data for patient medical scans such as
chest x-ray, CT and MRI is Digital Imaging and Com-
munications in Medicine (DICOM) (Sahu and Verma,
2011). However, most existing deep learning models
in medical image prediction utilize the Joint Photo-
graphic Experts Group (JPEG) format due to the lim-
itations of compute engine machines. Fig. 1 shows an
example of DICOM and JPEG chest x-ray. Recently,
researchers started to extract image categories from
DICOM metadata (i.e., study and image description)
and mapped them to the World Health Organization
(WHO) manual of diagnostic imaging (Dratsch et al.,
2021). However, to the best of our knowledge, there
has not been any comparison between DICOM and
JPEG formats on the performance of multi-label clas-
sifiers for chest radiographs using deep learning.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
950

2.2 Parallel Training
Training a deep learning model in parallel trains a
model across multiple GPUs to speed up neural net-
work training. This training approach is essential for
training the large public chest X-rays that have been
recently introduced one after another. For example,
ChestX-Ray14 (Wang et al., 2017), PadChest (Bus-
tos et al., 2020), CheXpert (Irvin et al., 2019), and
MIMIC-CXR (Johnson et al., 2019a) have 112,120,
160,868, 224,316, and 473,057 images, respectively.
Parallel training can be achieved by Data Paral-
lel (DP) or Distributed Data Parallel (DDP) (Li et al.,
2020) techniques. DP is performing one process (i.e.,
training a deep learning model) on multiple devices
(i.e., multi-GPU) of a single machine by distributing
batches of the data on the available GPUs. Although
in DP, a batch size can be large, the processing time is
long due to the limitation of one process. Differently,
DDP enables each device to independently conduct
one process on a portion of the training dataset (Li
et al., 2020).
3 METHOD AND DATASET
3.1 Dataset
MIMIC-CXR and CheXpert were used in this study
with more than half a million chest radiographs. Each
radiography was labeled with 14 observations: atelec-
tasis, cardiomegaly, consolidation, edema, enlarged
cardiomediastinum, fracture, lung lesion, lung opac-
ity, no finding, pleural effusion, pleural other, pneu-
monia, pneumothorax, and support devices. The la-
bels contained positive, negative, uncertain, and miss-
ing values. Tables 1 and 2 show the dependencies be-
tween labels in each dataset and emphasize the impor-
tance of labeling the datasets in a multi-label method
rather than a single label method.
MIMIC-CXR is the largest publicly available
dataset with 377,110 chest x-rays and the associated
reports. There are two releases of this dataset in-
cluding, the DICOM version (Johnson et al., 2019a)
and the JPEG version (Johnson et al., 2019b), where
the latter was generated by converting DICOM files
into a more accessible format. Further, MIMIC-
CXR were labeled by two automatic labelers: namely,
NegBio labeler (Peng et al., 2018) and CheXpert
labeler (Irvin et al., 2019). Then, a board of ex-
perienced radiologists validated the generated labels
against 687 reports and concluded that CheXpert out-
performed NegBio. We utilized 356,225 chest x-rays
from MIMIC-CXR with the CheXpert labels. We ex-
plicitly examined the dependencies between labels on
the MIMIC-CXR dataset in Table 1. It illustrates, for
instance, that 37% of the cardiomegaly labeled chest
x-rays are also pleural effusion.
CheXpert contains 224,316 chest radiographs.
There are two variations of this dataset: a high-
resolution dataset and a down-sampled resolution. We
utilized 212,498 of the low-resolution images. Table
2 represents label co-occurrence in this dataset. For
instance, 43% of the atelectasis labeled chest x-rays
are also lung opacity. Note that a CheXpert compe-
tition is organized by the Stanford Machine Learning
Group, which maintains private testing data for final
evaluation of the AUC score on detecting five chosen
diseases, including atelectasis, cardiomegaly, edema,
consolidation, and pleural effusion. However, the task
of this paper is to detect 14 observations simultane-
ously.
We converted uncertain and missing values to neg-
ative in both datasets, following the U-Zeros model
(Irvin et al., 2019). We ensured that each chest x-
ray had at least one positive label because a positive
“no finding” label presents the absence of all patholo-
gies. In addition, we randomly shuffled the chest x-
rays into three splits: 80% for training, 10% for vali-
dation, and 10% for testing, using a fixed random seed
of 42.
3.2 Xclassifier Model
Data Augmentation: For data augmentation, we
squished each CXR to 224x224 pixels (i.e., resizing
each CXR by squishing it on the horizontal axis), ro-
tated it by 20°, zoomed in by 1.2 scale, warped it by
0.2 magnitude, en-lighted it by 0.3 scale, and normal-
ize it. These data augmentation parameters increased
the accuracy of detecting abnormalities from chest
x-rays based on extensive experiment results(Monshi
et al., 2021). Importantly, we have only applied data
augmentation on the training set, where the validation
and test sets always get the original images.
CNN Architecture: Xclassifier is based on DenseNet
(Huang et al., 2017) due to the success of this ar-
chitecture in recent classification models using x-ray
datasets (Rajpurkar et al., 2017)(Yao et al., 2017)(Mo
and Cai, 2019)(Chen et al., 2020)(Bressem et al.,
2020). DenseNet utilizes dense blocks to connect all
layers directly with each other by matching feature-
map sizes. As demonstrated in Fig. 2, each layer in
this CNN passed on its own feature-maps to all suc-
cessive layers and collected additional inputs from all
prior layers to maintain the feed-forward nature.
Distributed Deep Learning for Multi-Label Chest Radiography Classification
951
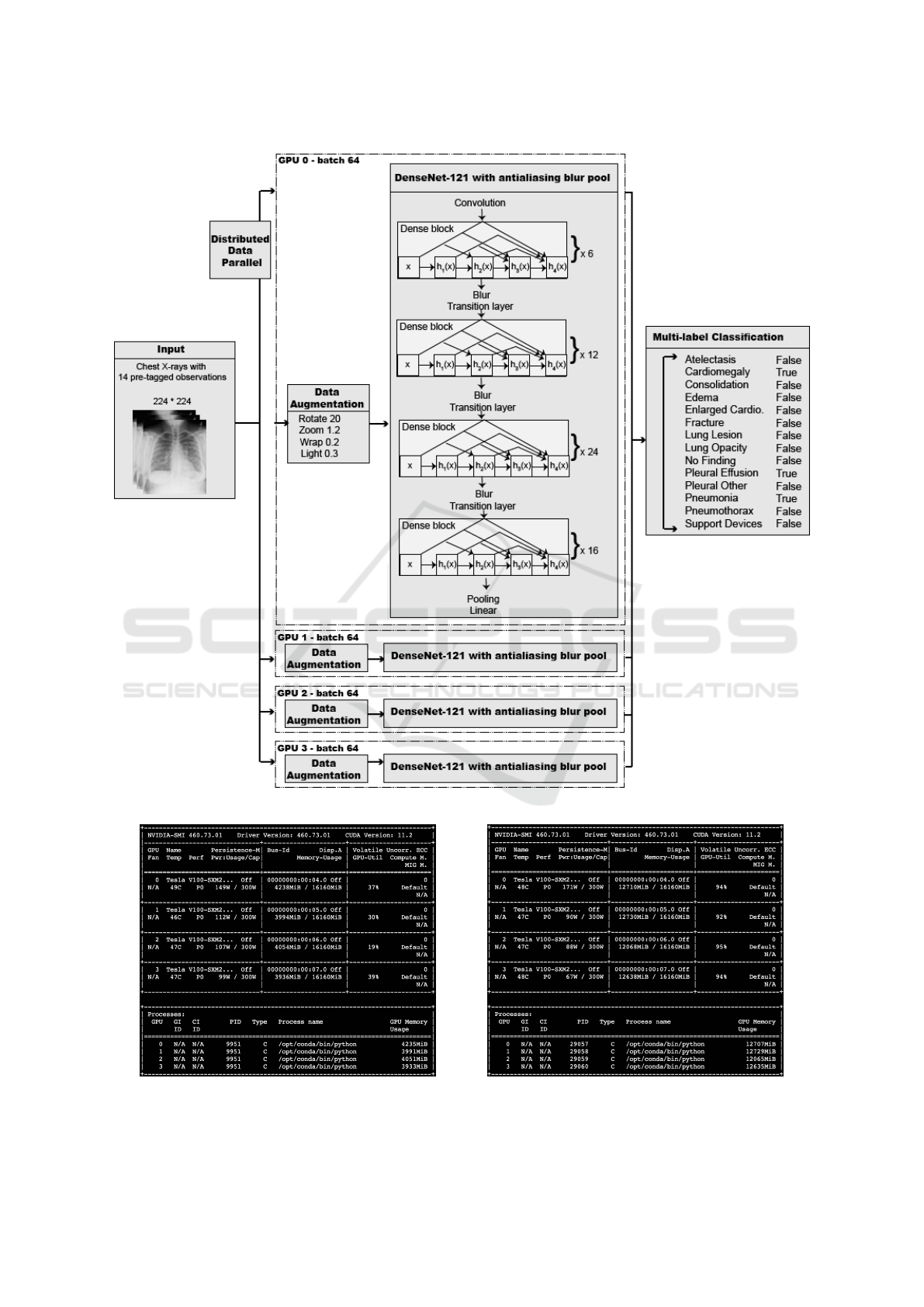
Figure 2: Xclassifier Structure.
(a) Data Parallel (DP).
(b) Distributed Data Parallel (DDP).
Figure 3: Visualizing Parallel Training Approaches. We used four Tesla V100 GPUs and trained DenseNetblur-121d for
multi-label classification tasks.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
952

Table 1: Positive Label Co-occurrence of the MIMIC-CXR.
Label
% of all % of label co-occurrence
data At Ca Co Ed EC Fr LL LO NF PE PO Pa Px SD
Atelectasis (At) 18 100 29 5 13 5 2 3 31 0 48 1 8 6 39
Cardiomegaly (Ca) 18 28 100 5 23 4 2 2 25 0 37 1 8 4 41
Consolidation (Co) 4 22 23 100 21 5 2 6 27 0 50 1 22 4 44
Edema (Ed) 10 24 40 8 100 4 1 2 29 0 51 1 11 2 37
Enlarged Cardiom. (EC) 3 32 23 7 14 100 3 6 33 0 36 2 7 8 45
Fract (Fr) 2 21 19 2 6 4 100 3 19 0 21 3 4 9 23
Lung Lesion (LL) 3 18 13 8 6 5 2 100 46 0 26 3 11 4 18
Lung Opacity (LO) 21 27 21 5 14 4 2 7 100 0 32 2 17 4 31
No Finnding (NF) 40 0 0 0 0 0 0 0 0 100 0 0 0 0 10
Pleural Effusion (PE) 22 41 31 10 24 5 2 4 31 0 100 1 9 6 41
Pleural Other (PO) 1 15 25 4 9 6 7 8 39 0 26 100 10 5 25
Pneumonia (Pa) 7 20 18 12 15 3 1 5 48 0 26 1 100 1 21
Pneumothorax (Px) 4 28 17 5 6 6 5 3 21 0 33 1 3 100 54
Support Devices (SD) 24 31 31 8 16 5 2 2 28 16 37 1 7 9 100
Table 2: Positive Label Co-occurrence of the CheXpert.
Label
% of all % of label co-occurrence
data At Ca Co Ed EC Fr LL LO NF PE PO Pa Px SD
Atelectasis (At) 16 100 12 6 27 5 4 3 43 0 49 1 2 9 60
Cardiomegaly (Ca) 13 14 100 5 43 7 3 2 48 0 44 1 2 3 58
Consolidation (Co) 7 14 10 100 21 4 3 5 38 0 50 2 7 5 52
Edema (Ed) 25 17 22 6 100 4 2 2 53 0 51 1 2 3 64
Enlarged Cardiom. (EC) 14 18 6 20 20 100 6 5 48 0 36 2 1 7 52
Fract (Fr) 4 14 9 4 11 7 100 4 40 0 27 3 2 12 40
Lung Lesion (LL) 4 11 7 8 9 6 4 100 58 0 36 3 5 9 35
Lung Opacity (LO) 50 13 12 5 26 5 3 5 100 0 49 2 4 9 58
No Finnding (NF) 11 0 0 0 0 0 0 0 0 100 0 0 0 0 39
Pleural Effusion (PE) 41 19 14 9 31 5 3 4 61 0 100 1 2 8 61
Pleural Other (PO) 2 11 9 9 9 5 8 9 53 0 26 100 4 7 39
Pneumonia (Pa) 3 10 8 17 20 3 2 8 67 0 29 2 100 2 29
Pneumothorax (Px) 9 16 4 4 8 4 5 4 47 0 34 1 1 100 60
Support Devices (SD) 55 17 13 7 29 5 3 3 53 8 46 1 2 10 100
Equation (1) represents the dense connectivity,
where [x
0
, x
1
, x
2
..] donates concatenation of the fea-
ture maps produced by [0, 1, ..L
t
h] layers. Each
DenseNet architecture consisted of four dense blocks
with a varying number of layers. Xclassifier had
[6,12,24,16] layers in the four dense blocks as in
DenseNet-121. We did not use the deeper architec-
tures of DenseNet (i.e., 161, 169, 201, and 264) be-
cause increasing the number of DenseNet hidden lay-
ers would not improve chest x-ray classification per-
formance (Yarnall, 2020).
X
l
= H
l
([x
0
, x
1
, ..., x
l−1
]) (1)
Antialiasing and Subsampling: Before each down-
sampling step in DenseNet, we inserted a blur kernel
m ×m as an antialiasing filter. We found that this mi-
nor modification increased the chest x-ray classifica-
tion accuracy as illustrated in Table 3. Besides, pre-
vious research showed that modifying the backbone
of several CNN architectures, by adding a blur ker-
nel, can increase the accuracy of ImageNet classifi-
cation (Zhang, 2019). We applied the antialiasing, as
depicted in Eq. (2) at stride 2 of DenseNet. Note
that BlurPool
m,s
donates the image processing func-
tion that combines blurring and subsampling, where k
is the kernel and s is the stride.
Relu ◦Conv
k,s
→ BlurPool
m,s
◦ Relu ◦Conv
k,1
(2)
Fine-tuning: To fine-tune Xclassifier, we adopted
the one-cycle policy (Smith, 2018), and the discrimi-
native learning rates (Howard and Ruder, 2018). This
policy of cyclical learning rates worked as a regular-
ization technique to converge faster and better train-
ing and hence kept the network from overfitting.
Distributed Data Parallel (DDP): With the DDP
technique (Li et al., 2020), we could use a large batch
size of 64 images for each of the 4 GPUs to accel-
erate the convergence. In every training iteration,
the one-device memory is frequently above 91% dur-
ing backward propagation, where each GPU indepen-
Distributed Deep Learning for Multi-Label Chest Radiography Classification
953

Table 3: DenseNet-121 variations models and training performance. We used the full MIMIC-CXR dataset and trained for 10
epochs.
Model Description Accuracy AUC
DenseNet-121 Single 7x7 convolution layer with no antialiasing layer 90.69 81.34
DenseNet-121d Three 3x3 convolution layers with no antialiasing layer 90.73 81.28
DenseNetblur-121d Three 3x3 convolution layers with antialiasing blur pool 90.80 81.96
Table 4: Image formats for chest x-rays and training performance. We used 10% of the MIMIC-CXR and trained ResNet18
for 10 epochs.
Chest x-ray format Accuracy AUC Avg. time per epoch (min)
DICOM 89.40 80.02 111
JPEG 89.58 81.57 6
Table 5: Training approaches and training performance. We used the NVIDIA V100 GPU.
Training Approach Dataset Accuracy AUC Avg. time per epoch (min)
Single GPU (1 x GPU) CheXpert 88.09 78.55 16
Data parallel (4 x GPUs) CheXpert 88.36 79.25 14
Distributed data parallel (4 x GPUs) CheXpert 88.33 80.10 4
Data parallel (4 x GPUs) MIMIC-CXR 90.27 80.97 181
Distributed data parallel (4 x GPUs) MIMIC-CXR 90.31 81.76 54
Table 6: Comparing the Xclassifier with the benchmark.
Multi-label classifier Dataset Accuracy AUC
Latent-space self-ensemble (Gyawali et al., 2019) CheXpert 66.97
CheXclusion (Seyyed-Kalantari et al., 2020) CheXpert 80.50
Xclassifier CheXpert 89.61 83.89
VSE-GCN (Hou et al., 2021) MIMIC-CXR 72.10
CheXclusion (Seyyed-Kalantari et al., 2020) MIMIC-CXR 83.40
Xclassifier MIMIC-CXR 92.17 84.10
dently performed one copy of the training on a part
of the dataset. Fig. 3b captures a live example of the
Xclassifier training job using four Tesla V100-SXM2-
16GB GPUs. It shows the normalized GPU utilization
of both compute core and memory usage.
4 EXPERIMENT
For distributed deep learning, we used PyTorch DDP
(Li et al., 2020), Pytorch image models (timm)
(Wightman, 2021), the Fastai v2 library (Howard and
Gugger, 2020), and an n1-highmem-32 (32 vCPUs,
208 GB memory) machine with four NVIDIA Tesla
V100 GPUs. We used a batch size of 64 for each of
the 4 GPUs and trained the model for 30 epochs.
5 RESULTS AND DISCUSSION
A comparison via accuracy and areas under receiver
operator characteristic curve (AUC) values for DI-
COM vs. JPEG for the multi-label classification task
is demonstrated in Table 4. Despite that, the DICOM
format is more readily applicable than JPEG to clini-
cal practice. It did not improve automated neural net-
work accuracy. In fact, it took significantly more time
to train DICOM (i.e., 111 min per epoch) than the
JPEG counterparts (i.e., 6 min per epoch), using 10%
of the MIMIC-CXR dataset. Therefore, we decided
not to train the DICOM files any further.
A comparison via accuracy and AUC values for
DenseNet-121 vs. DenseNet-121d vs. DenseNetblur-
121d for the multi-label classification task is shown
in Table 3. DenseNet-121 with the blur pooling out-
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
954
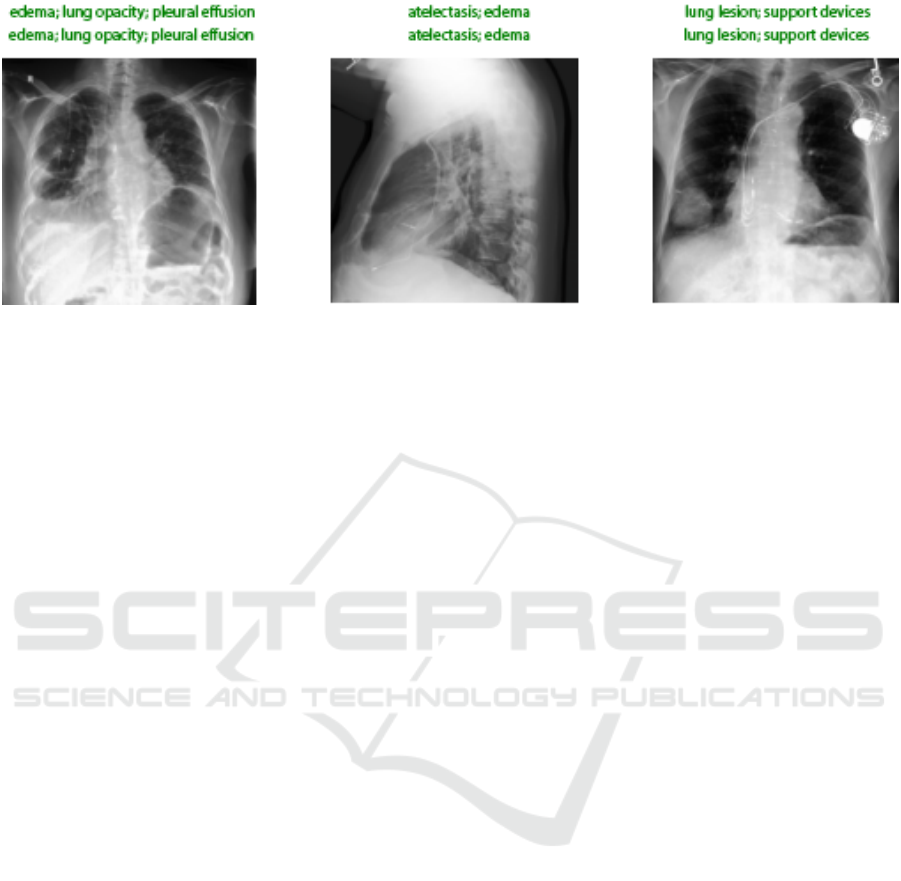
Figure 4: Correct Output Sample by the Xclassifier Model.
performs its variations, so we built the Xclassifier on
top of this architecture. Due to the shift variant nature
of CNN, antialiasing filters are used to increase the
accuracy of the Xclassifier.
A comparison via the average time per epoch for
single GPU vs. DP vs. DDP for the multi-label
classification task using DenseNetblur-121d is illus-
trated in Table 5. DDP is the best training approach
for CheXpert in terms of time efficiency, providing a
4× speedup over a single GPU, and a 1.14× to 3.35×
speedup over DP.
The proposed Xclassifier improves the multi-label
classification performance by 0.70% AUC (84.10%
vs. 83.40%) on the MIMIC-CXR and by 3.39% AUC
(83.89% vs. 80.50%) on the CheXpert, refer to Ta-
ble 6. As it depends on the DDP of DenseNet blur
121, it allows CNN layers to be deeper, more accu-
rate in learning label co-occurrence, and efficient to
train. Fig. 4 represents a sample of the correct pro-
duced labels by the Xclassifier model.
6 CONCLUSIONS AND FUTURE
WORK
We introduce Xclassifier, an efficient multi-label clas-
sifier that trains an enhanced DenseNet-121 frame-
work with blur pooling to detect 14 observations from
a chest x-ray. It accomplishes an ideal memory uti-
lization, GPU computation, and high AUC on two
large chest radiography, MIMIC-CXR, and CheX-
pert. Xclassifier uses features of all complexity levels
to handle label co-occurrence training. DDP is a true
process and data parallelism. It is useful in perform-
ing multi-processes on devices of multiple machines
but also can be used on devices of just a single ma-
chine as well.
In practice, radiologists use a finer resolution of a
CXR, DICOM format and rely on additional informa-
tion, such as the patient electronic health records, to
detect multiple observations. However, in deep learn-
ing, our findings suggest that utilizing JPEG images is
more efficient than their DICOM counterparts in the
multi-label classification task. Therefore, for future
work, we plan to investigate the use of DICOM in de-
tecting diseases with small and complex structures to
offer a greater degree of understanding of our initial
findings. Further, we plan to concatenate patient data
such as age and gender to the flattened layer to im-
prove prediction.
ACKNOWLEDGEMENTS
This material is based upon work supported by the
Google Cloud research credits program.
REFERENCES
Bressem, K. K., Adams, L. C., Erxleben, C., Hamm, B.,
Niehues, S. M., and Vahldiek, J. L. (2020). Comparing
different deep learning architectures for classification
of chest radiographs. Scientific reports, 10(1):1–16.
Bustos, A., Pertusa, A., Salinas, J.-M., and de la Iglesia-
Vay
´
a, M. (2020). Padchest: A large chest x-ray image
dataset with multi-label annotated reports. Medical
image analysis, 66:101797.
Chen, B., Li, J., Lu, G., Yu, H., and Zhang, D. (2020). Label
co-occurrence learning with graph convolutional net-
works for multi-label chest x-ray image classification.
IEEE journal of biomedical and health informatics,
24(8):2292–2302.
Dratsch, T., Korenkov, M., Zopfs, D., Brodehl, S., Baessler,
B., Giese, D., Brinkmann, S., Maintz, D., and Pinto
dos Santos, D. (2021). Practical applications of deep
learning: classifying the most common categories of
Distributed Deep Learning for Multi-Label Chest Radiography Classification
955

plain radiographs in a PACS using a neural network.
European Radiology, 31(4):1812–1818.
Gyawali, P. K., Li, Z., Ghimire, S., and Wang, L. (2019).
Semi-supervised learning by disentangling and self-
ensembling over stochastic latent space. In Inter-
national Conference on Medical Image Computing
and Computer-Assisted Intervention, pages 766–774.
Springer.
Hou, D., Zhao, Z., and Hu, S. (2021). Multi-label learn-
ing with visual-semantic embedded knowledge graph
for diagnosis of radiology imaging. IEEE Access,
9:15720–15730.
Howard, J. and Gugger, S. (2020). Fastai: A layered API
for deep learning. Information, 11(2):108.
Howard, J. and Ruder, S. (2018). Universal language model
fine-tuning for text classification. arXiv preprint
arXiv:1801.06146.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 4700–
4708.
Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S.,
Chute, C., Marklund, H., Haghgoo, B., Ball, R., and
Shpanskaya, K. (2019). Chexpert: A large chest radio-
graph dataset with uncertainty labels and expert com-
parison. In Proceedings of the AAAI Conference on
Artificial Intelligence, volume 33, pages 590–597.
Johnson, A. E. W., Pollard, T. J., Berkowitz, S. J., Green-
baum, N. R., Lungren, M. P., Deng, C.-y., Mark, R. G.,
and Horng, S. (2019a). MIMIC-CXR, a de-identified
publicly available database of chest radiographs with
free-text reports. Scientific data, 6(1):1–8.
Johnson, A. E. W., Pollard, T. J., Greenbaum, N. R.,
Lungren, M. P., Deng, C.-y., Peng, Y., Lu, Z.,
Mark, R. G., Berkowitz, S. J., and Horng, S.
(2019b). MIMIC-CXR-JPG, a large publicly available
database of labeled chest radiographs. arXiv preprint
arXiv:1901.07042.
Li, S., Zhao, Y., Varma, R., Salpekar, O., Noordhuis, P., Li,
T., Paszke, A., Smith, J., Vaughan, B., and Damania,
P. (2020). PyTorch Distributed: Experiences on Ac-
celerating Data Parallel Training. Proceedings of the
VLDB Endowment, 13(12).
Mo, S. and Cai, M. (2019). Deep learning based multi-
label chest x-ray classification with entropy weight-
ing loss. In 2019 12th International Symposium on
Computational Intelligence and Design (ISCID), vol-
ume 2, pages 124–127. IEEE.
Monshi, M. M. A., Poon, J., and Chung, V. (2019). Convo-
lutional neural network to detect thorax diseases from
multi-view chest x-rays. In International Conference
on Neural Information Processing, pages 148–158.
Springer.
Monshi, M. M. A., Poon, J., Chung, V., and Monshi,
F. M. (2021). CovidXrayNet: Optimizing Data Aug-
mentation and CNN Hyperparameters for Improved
COVID-19 Detection from CXR. Computers in Bi-
ology and Medicine, 133(0010-4825):104375.
NVIDIA (2018). DGX-2 : AI Servers for Solving Complex
AI Challenges — NVIDIA.
NVIDIA (2020). NVIDIA DGX A100 System Architec-
ture.
Peng, Y., Wang, X., Lu, L., Bagheri, M., Summers, R.,
and Lu, Z. (2018). Negbio: a high-performance tool
for negation and uncertainty detection in radiology re-
ports. AMIA Summits on Translational Science Pro-
ceedings, 2018:188.
Pham, H. H., Le, T. T., Tran, D. Q., Ngo, D. T., and Nguyen,
H. Q. (2021). Interpreting chest X-rays via CNNs that
exploit hierarchical disease dependencies and uncer-
tainty labels. Neurocomputing, 437:186–194.
Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan,
T., Ding, D., Bagul, A., Langlotz, C., and Shpan-
skaya, K. (2017). Chexnet: Radiologist-level pneu-
monia detection on chest x-rays with deep learning.
arXiv preprint arXiv:1711.05225.
Sabottke, C. F. and Spieler, B. M. (2020). The effect of
image resolution on deep learning in radiography. Ra-
diology: Artificial Intelligence, 2(1):e190015.
Sahu, B. K. and Verma, R. (2011). DICOM search in medi-
cal image archive solution e-Sushrut Chhavi. In 2011
3rd International Conference on Electronics Com-
puter Technology, volume 6, pages 256–260. IEEE.
Seyyed-Kalantari, L., Liu, G., McDermott, M., Chen, I. Y.,
and Ghassemi, M. (2020). CheXclusion: Fairness
gaps in deep chest X-ray classifiers. In BIOCOM-
PUTING 2021: Proceedings of the Pacific Sympo-
sium, pages 232–243. World Scientific.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Smith, L. N. (2018). A disciplined approach to neural net-
work hyper-parameters: Part 1–learning rate, batch
size, momentum, and weight decay. arXiv preprint
arXiv:1803.09820.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Sum-
mers, R. M. (2017). Chestx-ray8: Hospital-scale chest
x-ray database and benchmarks on weakly-supervised
classification and localization of common thorax dis-
eases. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2097–
2106.
Wang, X., Peng, Y., Lu, L., Lu, Z., and Summers, R. M.
(2018). Tienet: Text-image embedding network for
common thorax disease classification and reporting in
chest x-rays. In Proceedings of the IEEE conference
on computer vision and pattern recognition, pages
9049–9058.
Wightman, R. (2021). Pytorch image models. https:
//github.com/rwightman/pytorch-image-models.
Yao, L., Poblenz, E., Dagunts, D., Covington, B., Bernard,
D., and Lyman, K. (2017). Learning to diagnose
from scratch by exploiting dependencies among la-
bels. arXiv preprint arXiv:1710.10501.
Yarnall, J. (2020). X-Ray Classification Using Deep Learn-
ing and the MIMIC-CXR Dataset.
Zhang, R. (2019). Making convolutional networks shift-
invariant again. In International conference on ma-
chine learning, pages 7324–7334. PMLR.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
956
