
Rendering Medical Images using WebAssembly
Sébastien Jodogne
Institute of Information and Communication Technologies, Electronics and Applied Mathematics,
Louvain School of Engineering, Louvain-la-Neuve, Belgium
Keywords:
Medical Imaging, DICOM, WebAssembly, Data Visualization, Teleradiology.
Abstract:
The rendering of medical images is a critical step in a variety of medical applications from diagnosis to
therapy. Specialties such as radiotherapy and nuclear medicine must display complex images that are the
fusion of several layers. Furthermore, the rise of artificial intelligence applied to medical imaging calls for
viewers that can be used in research environments and that can be adapted by scientists. However, desktop
viewers are often developed using technologies that are totally different from those used for Web viewers,
which results in a lack of code reuse and shared expertise between development teams. In this paper, we show
how the emerging WebAssembly standard can be used to tackle these issues by sharing the same code base
between heavyweight viewers and zero-footprint viewers. Moreover, we propose a full Web viewer developed
using WebAssembly that can be used in research projects or in teleradiology applications. The source code of
the developed Web viewer is available as free and open-source software.
1 INTRODUCTION
The volume of medical images that are generated
worldwide is in constant growth. This can be ex-
plained by the fact that the treatment of more and
more chronic diseases requires a longitudinal and
multimodal exploitation of medical images. For a
highly effective, personalized treatment, it is indeed
often important to consider images that have been ac-
quired throughout the entire life of the patient and to
combine multiple cues produced by different types of
imaging modalities.
The DICOM standard (National Electrical Man-
ufacturers Association, 2021) specifies how medical
images are encoded and exchanged. Fortunately, DI-
COM is adopted by any hospital dealing with digital
medical images. Thanks to the fact that DICOM is
an open standard, a large ecosystem of free and open-
source software exists to handle medical images. For
instance, when it comes to the need of storing, com-
municating and archiving DICOM images in full in-
teroperability with the information systems of an hos-
pital, free and open-source DICOM servers such as
dcm4chee (Warnock et al., 2007), Dicoogle (Costa
et al., 2011) or Orthanc (Jodogne, 2018) can be used.
Obviously, once stored inside a DICOM server,
the medical images must still be displayed. Various
use cases exist for viewers of medical images: clinical
review integrated within the electronic health records,
specialized tools dedicated to one pathology or treat-
ment, external access for the patient, quality control
of modalities, second medical opinion, telemedicine,
clinical trials. . . Moreover, the huge interest in artifi-
cial intelligence applied to medical imaging requires
the development of viewers that can be used to an-
notate regions of interest for the training of the algo-
rithms, and to display the outputs of such algorithms
(e.g. segmentation masks) (Cheng et al., 2021).
Depending on the clinical or pre-clinical scenario,
one might either need a desktop software, a mobile
application or a Web platform to display the medical
images. This led many community projects or com-
mercial products to focus only on one of those three
categories of platforms, or to have separate develop-
ment teams working on distinct viewers for the dif-
ferent types of platforms. In the latter case, develop-
ing similar features in different languages depending
on the target platform (JavaScript, C++, Java. . . ) ev-
idently comes at a large cost because of non-shared
source code. Furthermore, distinct code bases might
suffer from different issues, or conversely might fea-
ture optimizations that are not available for the other
platforms. Expertise is also less shared between de-
velopment teams who are focused on different plat-
forms and languages.
Two major open-source software libraries are cur-
rently used within software displaying medical im-
ages. The first one is VTK (Schroeder et al., 2006),
Jodogne, S.
Rendering Medical Images using WebAssembly.
DOI: 10.5220/0010833300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 43-51
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
43

that is mature and mainly written in the C++ lan-
guage. VTK is used by many heavyweight desktop
viewers, including the 3D Slicer platform (Kikinis
et al., 2014), the well-known OsiriX radiology appli-
cation (Rosset et al., 2004) and its fork Horos. The
second library is Cornerstone (Urban et al., 2017),
that is more recent and that is written in the JavaScript
language so as to be used as a building block for
higher-level Web applications. As it takes its roots in
modern Web technologies, Cornerstone is at the core
of many zero-footprint, lightweight Web viewers, no-
tably OHIF, and is often encountered within cloud-
based proprietary solutions. The scope of Corner-
stone is mostly focused on the hardware-accelerated
rendering of 2D images by Web browsers, whereas
VTK is feature-rich, makes active use of GPU for 3D
rendering, and can be used for scientific visualization
beyond medical imaging.
The emerging WebAssembly standard might be a
game changer in this divided landscape. WebAssem-
bly is an open standard that defines a bytecode for
Web applications (Haas et al., 2017). The core spec-
ification of WebAssembly was released as an offi-
cial W3C recommendation in December 2019. We-
bAssembly is actively backed by the industry, and is
now supported by the major Web browsers. Thanks
to the Emscripten toolchain, C++ source code can be
compiled to WebAssembly bytecode, which can then
be executed by Web browsers. This opens the path to
the creation of C++ libraries that can be used both
by native software (using a standard compiler) and
by Web platforms (using the Emscripten toolchain).
Such C++ libraries could also be used by mobile
applications using Android NDK or an Objective-C
wrapper. WebAssembly has thus a huge potential to
improve code reuse in multi-platform developments,
in particular in the field of medical imaging.
In this paper, we firstly propose a novel, free and
open-source C++ library named “Stone of Orthanc”,
whose internal architecture was driven by the con-
straints of Web development while providing compat-
ibility with native compiler toolchains. The footprint
of Stone of Orthanc is as small as possible, and is de-
signed to be as portable and versatile as possible
1
.
Historically, the Stone of Orthanc project started in
2016 by leveraging the Google’s PNaCl and Mozilla’s
asm.js technologies that are now superseded by We-
bAssembly. Secondly, different applications for med-
ical imaging that were built using the Stone of Or-
1
This intimacy with the lightweight architecture of the
Orthanc project for medical imaging (Jodogne, 2018) ex-
plains the name of the Stone of Orthanc library, the latter
being a tribute to the palantír that resides inside the tower
of Orthanc in The Lord of the Rings by J. R. R. Tolkien.
thanc library are presented. In particular, the “Stone
Web viewer” is introduced as a fully functional Web
viewer for medical images that is integrated as a plu-
gin within the Orthanc DICOM server and that can be
used by end users.
2 Stone of Orthanc
As explained in the Introduction, Stone of Orthanc
is a C++ library to render medical images, with the
goal of being compatible with WebAssembly. This
objective puts strong constraints on the architecture
of the library. Indeed, traditional C++ applications are
multi-threaded and sequential, whereas Web applica-
tions are single-threaded and driven by asynchronous
callbacks (closures).
Besides this constrained architecture, the accurate
rendering of medical images requires a careful map-
ping between pixel coordinates and 2D or 3D physi-
cal coordinates. The library must also be extensible
so as to display the various kinds of overlays (such as
segmentation masks or annotations) that might have
been produced by artificial intelligence algorithms or
manually added by physicians. Furthermore, medical
specialties such as nuclear medicine, radiotherapy or
protontherapy necessitate to display fusions of differ-
ent layers (for instance, a dose over a CT-scan, or a
contour over a PET signal). To fulfill such needs, the
architecture of Stone of Orthanc integrates a portable
2D rendering engine for vectorized graphics with sup-
port of overlays.
In the current section, we review how the high-
level architecture of Stone of Orthanc has been engi-
neered. Figure 1 provides a summary of this architec-
ture. Because the goal of this paper is not to document
the internal components of Stone of Orthanc, only the
main concepts that drove the design of Stone of Or-
thanc are presented. The interested reader is kindly
invited to dive into the source code of the project.
2.1 Loaders and Oracle
The Stone of Orthanc library provides developers
with many primitives that are necessary to create
medical imaging applications. In particular, the li-
brary offers facilities to download images from a DI-
COM server (notably using the DICOMweb standard,
but a custom REST API can be used as well). In this
way, Stone of Orthanc features C++ classes that con-
tain 2D or 3D medical images, and that are filled by
C++ classes that are referred to as “Loaders”. In the
case of 3D dense volumes, the associated Loaders au-
tomatically sort the individual 2D slices of the volume
BIOIMAGING 2022 - 9th International Conference on Bioimaging
44
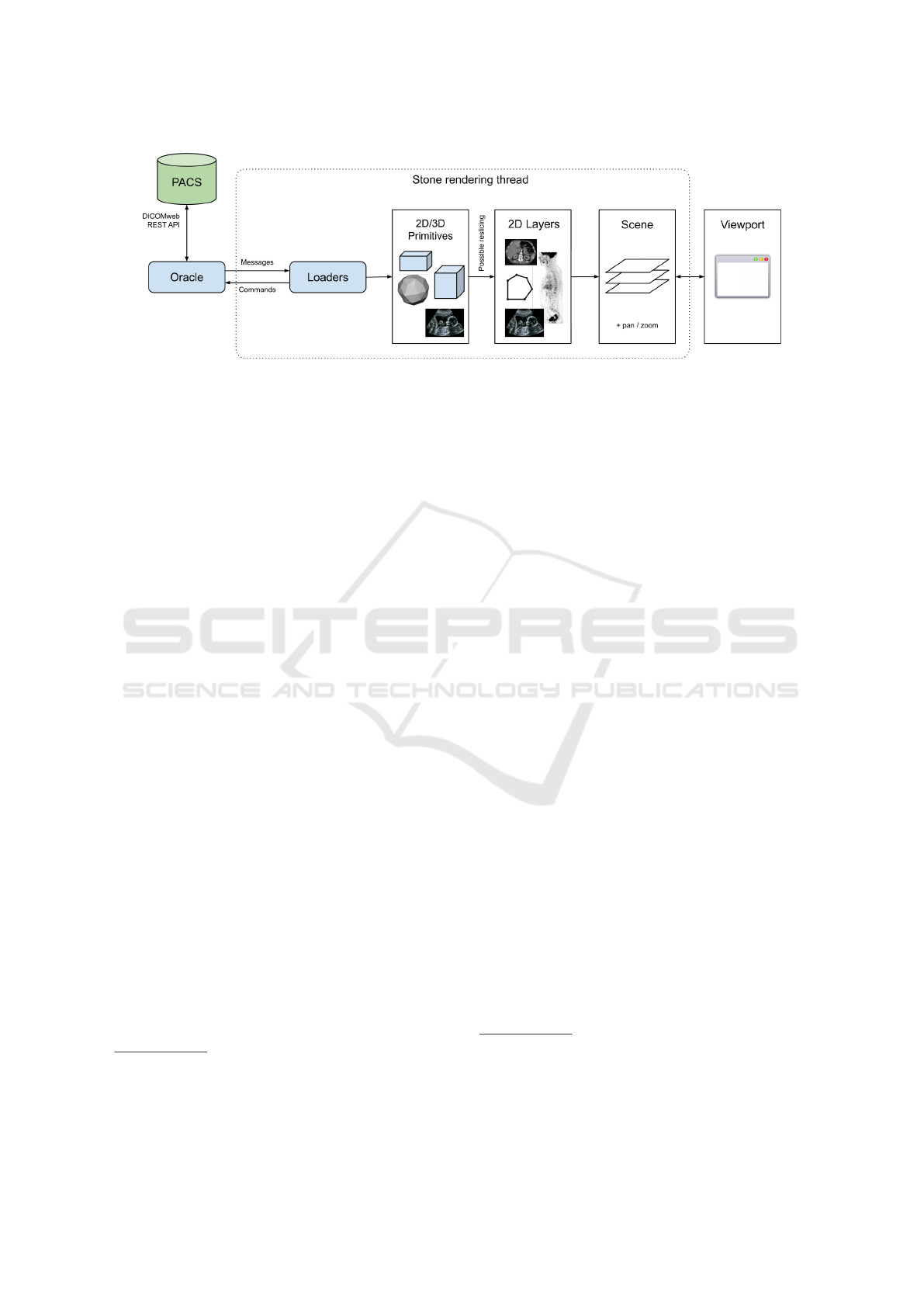
Figure 1: High-level view of the components of the Stone of Orthanc library. The Oracle abstracts the runtime environment.
The Loaders are responsible for loading the graphical primitives that have to be displayed. These primitives are converted into
a set of 2D Layers (in particular, primitives corresponding to 3D volumes can be sliced along a cutting plane). The Layers are
then assembled as a Scene that is finally composed onto the rendering surface of the Viewport. The user can interact with the
Viewport to update the affine transform (pan/zoom) that is associated with the Scene.
according to their 3D location
2
.
However, as written above, the JavaScript and
WebAssembly engines are inherently single-threaded.
As a consequence, in order to be compatible with the
Web runtimes, the core of the Stone of Orthanc li-
brary is not allowed to launch threads or to maintain
a pool of threads for the Loaders to download the im-
ages, which contrasts with what would be done in a
traditional C++ application. The solution adopted by
Stone of Orthanc is to introduce a so-called “Oracle”
singleton object that abstracts the way asynchronous
operations such as downloads are actually processed.
In the case of WebAssembly, the Oracle essentially
corresponds to a wrapper that abstracts the primitives
offered the Web browser itself. In the case of native
applications, the Oracle abstracts the system SDK as-
sociated with the target platform: For instance, in or-
der to handle downloads, the Oracle internally man-
ages a set of worker threads connected to a shared
queue of pending HTTP requests. Besides down-
loading DICOM instances, other examples of asyn-
chronous operations include reading files from the
filesystem, or sleeping for a certain amount of time.
According to this discussion, the Oracle receives
a stream of “Command” objects that specify the asyn-
chronous operations that must be carried on. Follow-
ing the observer design pattern (Gamma et al., 1994),
each Command to be processed is associated with an
“Observer” object that will receive a “Message” ob-
ject that contains the result of the Command once it
has been processed. For instance, in the case of the
download of a DICOM instance from some REST
API, the Command would contain the URL to down-
2
The 3D location is determined by the “image posi-
tion patient” (0020,0032) and “image orientation patient”
(0020,0037) DICOM tags if available, or by the “instance
number” (0020,0013) DICOM tag.
load, the Observer would correspond to the Loader
object that populates the target medical image, and the
Message would contain the raw bytes of the DICOM
instance. Sending a Command to the Oracle is a non-
blocking operation, which reflects the asynchronous
nature of the Oracle. The Observer is specified as a
C++ “pointer to member function”, with a slight use
of C++ templates as syntactic sugar, and the Oracle
invokes this member function upon completion of the
Command. The Oracle is the key component that en-
ables the writing of source code that can be run indif-
ferently by WebAssembly and by native applications.
The Oracle is part of the global context of the
Stone of Orthanc library, and each class that must ex-
ecute asynchronous operations keeps a reference to
the Oracle. In order to map the single-threaded as-
pect of WebAssembly into native environments sup-
porting multi-threading, it is assumed that the global
context is only accessed from one distinguished user
thread that is responsible for the rendering. A mutex
is included in the global context to prevent the Mes-
sages emitted by the Oracle from interfering with the
rendering thread. Reference counting using shared
pointers ensures that the Observer objects are not de-
structed before all the scheduled Commands they are
receivers of have been processed
3
.
It is worth noticing that the Oracle mechanism can
be used as an accelerator for certain operations that
can be time-intensive. For instance, Stone of Orthanc
introduces a Command to download then parse DI-
COM instances as a single operation: In the case of a
multi-threaded environment, this allows to offload the
3
The observer design pattern and reference counting are
used elsewhere in Stone of Orthanc for sending messages
from one object to another, and for broadcasting messages
from one object to a set of Observers using a subscription
mechanism.
Rendering Medical Images using WebAssembly
45

DICOM parsing to a thread without locking the mutex
of Stone of Orthanc, which brings more concurrency
in native environments
4
. This also allows to maintain
a cache of parsed DICOM files. As another example,
a new type of Command could be defined to delegate
some expensive computation to the CPU (such as pro-
jecting a DICOM RT-STRUCT along the sagittal or
coronal planes) or to the GPU (such as generating a
maximum intensity projection – MIP – rendering of a
3D volume).
2.2 Viewports and Layers
When it comes to the client-side rendering of med-
ical images, the 2D operations of panning, zooming
and changing windowing must be as fast as possi-
ble. This calls for taking advantage of the 2D hard-
ware acceleration that is provided by any GPU. To
this end, both native applications and WebAssembly
applications have access to the OpenGL API, the for-
mer through development libraries such as Qt or SDL,
the latter through WebGL.
On the other hand, in order to reduce the network
bandwidth in the presence of large images (typically
3D volumes), the developers of viewers might want to
use streaming: In this scenario, a central server gen-
erates a stream of “screenshots” of projections of the
volumes it stores in its RAM, and those renderings are
then sent to a thin client in charge of their actual dis-
play to the user. In the case of streaming, GPU is not
necessarily available on the server, as multiple users
might be connected to the same server. GPU might
also be unavailable if printing an image, if generat-
ing a secondary capture image, or if the viewer runs
on a low-power or embedded computer (such as the
popular Raspberry Pi).
To accommodate with all such situations in a
cross-platform and lightweight way, the architecture
of Stone of Orthanc is designed to support both hard-
ware acceleration (using OpenGL) and software ren-
dering (using the widespread cairo drawing library).
Furthermore, in order to maximize portability, the
Stone of Orthanc library implements its own OpenGL
shaders for the GPU-accelerated drawing of lines,
texts and bitmaps.
Internally, each rendering surface (the application
widget, the HTML5 canvas or the memory buffer) is
associated with a so-called “Viewport” abstract class
that is responsible for the rendering of a 2D “Scene”.
The Scene is an object that encodes a superposition
of “Layer” objects, each Layer being a 2D vectorized
graphics with an alpha layer to allow for transparency.
4
In the future, similar optimizations could be offered in
the WebAssembly environment by leveraging Web workers.
Each Scene and Layer is agnostic of the actual render-
ing surface. In a nuclear medicine application for in-
stance, a Scene would consist of three superimposed
Layers, the foremost being the contours (as encoded
in a DICOM RT-STRUCT instance), the second be-
ing the PET-scan signal (with a heat colormap hav-
ing transparency), and the background being the CT-
scan slice. The Scene also contains a 2D-to-2D affine
transform that maps the coordinates of Layers to the
Viewport coordinates. Altering this transform is used
to pan and zoom the Scene onto the Viewport.
Depending on the type of its rendering surface,
the Viewport will use a different algorithm to ren-
der a Scene, following the strategy design pattern.
This separation of concerns between Viewports and
Scenes, is similar to that between the Oracle and
Commands. If a Layer containing a bitmap must
be rendered, the Viewport is also responsible for the
proper conversion of the pixel values (that typically
correspond to floating-point numbers or 16 bit inte-
gers) to a bitmap that is suitable for rendering onto
the surface (in general, red/green/blue/alpha values in
8 bits per pixel). This conversion must take into ac-
count the windowing parameters of the Layers, if any.
Note that a single application is allowed to manage
multiple Viewports in order to show different Scenes.
According to this discussion, the rendering engine
of Stone of Orthanc is inherently oriented toward 2D
vectorized graphics. This choice is similar to that of
the Cornerstone library, but contrasts with the VTK
library, that is more 3D-oriented. The advantage of
focusing on 2D rendering is to make the rendering of
multi-layer scenes more natural to the developers than
if 3D must also be taken into account. Importantly,
just like the Oracle/Commands/Observers architec-
ture, this Viewport/Scene/Layers architecture is com-
patible with both native applications and WebAssem-
bly applications. It is worth noticing that Stone of
Orthanc could be used as a rendering engine for vec-
torized graphics in native, mobile or Web applications
out of the field of medical imaging.
Evidently, viewers for medical imaging need to
display 3D volumes, not just 2D images. Section 2.1
explained how Loaders are used to download 3D im-
ages and store them into RAM. To render 3D vol-
umes, Stone of Orthanc introduces the concept of
“VolumeReslicer” that is an abstract class taking as
its inputs one 3D image and one cutting 3D plane,
and producing as its output a 2D slice. This slice
can then be injected as a Layer into the Scene to be
rendered. Volume reslicers are currently implemented
for multiplanar reconstruction (MPR, cf. Section 3.3)
and for reslicing along an arbitrary cutting plane (cf.
Section 3.4). As discussed above, more reslicers will
BIOIMAGING 2022 - 9th International Conference on Bioimaging
46
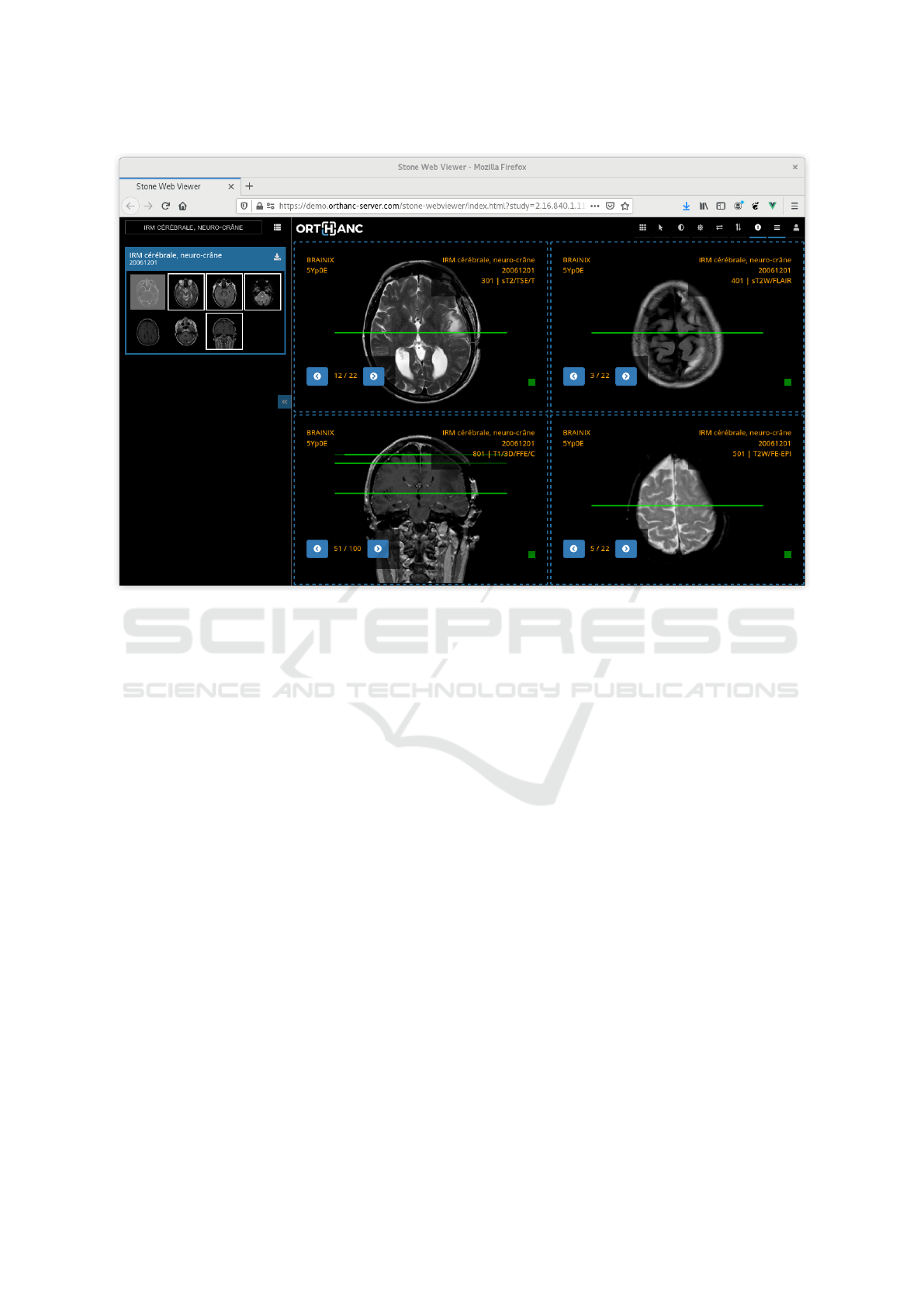
Figure 2: Screenshot of the Stone Web viewer.
be implemented in the future for MIP rendering, pos-
sibly using the Oracle as an accelerator for the com-
putations.
3 APPLICATIONS
The core features of the Stone of Orthanc library have
been explained in the previous sections. In this sec-
tion, several higher-level applications that were built
using Stone of Orthanc are reviewed.
3.1 Simple Desktop Viewer
The source code of Stone of Orthanc comes with a
basic viewer that can be used to display one 2D frame
of a DICOM instance downloaded from an Orthanc
server. This viewer is a standalone desktop applica-
tion that is started from the command line by provid-
ing the URL of the Orthanc REST API, and the iden-
tifier of the DICOM instance to be displayed.
This viewer uses SDL to manage its window, mak-
ing it eminently cross-platform. A screenshot of this
application can be found in Figure 3. Zooming, pan-
ning and windowing are supported, as well as taking
measures. The mouse interactions are handled by a
dedicated abstract class called “ViewportInteractor”
that is responsible for tracking the mouse events, and
that is part of the Stone of Orthanc library. Interest-
ingly, because the DICOM image is downloaded from
the Orthanc server, it is decoded server-side, which
frees the viewer from handling the various transfer
syntaxes of the DICOM standard, hereby simplifying
the development.
3.2 Stone Web Viewer
The so-called “Stone Web viewer” is currently the
main application that leverages the Stone of Orthanc
library. The Stone Web viewer is a fully functional
teleradiology solution, as depicted in Figure 2. It can
display multiple DICOM series, and implements fea-
tures such as measurements, synchronized browsing,
3D cross-hair, and 3D reference lines. The Stone
Web viewer communicates with a DICOM server
using the DICOMweb protocol, so it is in theory
compatible with any PACS environment equipped
with DICOMweb. Contrarily to the simple desk-
top viewer described in Section 3.1, the Stone Web
viewer doesn’t rely on the remote server to decode
the DICOM files, and embeds the DCMTK library in
the WebAssembly bytecode to this end.
Very importantly, to the best of our knowledge,
the Stone Web viewer is the first free and open-source
Rendering Medical Images using WebAssembly
47

Figure 3: Simple 2D viewer displaying a radiography im-
age. In this case, the Viewport is a SDL window, with one
background Layer containing a 16bpp bitmap. The win-
dowing can be changed using the mouse. The viewer can be
used to take measures of lines, circles and angles in phys-
ical coordinates. These measures are geometric primitives
stored in the foreground Layers of the Scene.
viewer of medical images that uses WebAssembly.
Most of its source code is written in C++ and com-
piled using the Emscripten toolchain. The buttons and
the HTML5 canvas are managed by a simple Vue.js
application that calls the WebAssembly bytecode us-
ing an automatically generated JavaScript wrapper.
The Stone Web viewer is a client-side application
that is entirely run by the Web browser. Its assets
only consist of a set of static resources (HTML files,
JavaScript sources and WebAssembly bytecode) that
can be served by any HTTP server (such as nginx,
Apache or Microsoft IIS). To fulfill the same-origin
security policy, it is advised to map the DICOMweb
server as a reverse proxy by the HTTP server. The
configuration file of the Stone Web viewer has an op-
tion to provide the root of the DICOMweb API.
However, this manual configuration of a HTTP
server might sound complex to users without a back-
ground in network administration. To ease the de-
ployments, a plugin for Orthanc has been developed
to directly serve the assets of the Stone Web viewer
using the embedded HTTP server of Orthanc. Thanks
to this Stone Web viewer plugin and thanks to the DI-
COMweb plugin for Orthanc, it is extremely easy to
start the Stone Web viewer. Furthermore, this plu-
gin is directly available in the Docker images, the Mi-
crosoft Windows installers and the macOS packages
provided by the Orthanc project
5
.
Figure 4 provides the big picture of the differ-
5
An online demo of the Stone Web viewer plugin is also
available at: https://demo.orthanc-server.com/
ent components that are involved in the Stone Web
viewer. The Stone Web viewer is a high-level appli-
cation that is built on the top of the Stone of Orthanc
library. As can be seen, the Stone of Orthanc library
reuses some classes from the Orthanc project that are
referred to as the “Orthanc Framework”. The Stone
Web viewer can either be served from a HTTP server,
or be run as a plugin to the main Orthanc server.
3.3 Rendering Oncology Images
In the context of radiotherapy, patients rarely have the
opportunity to view the images of their own treatment
plan from home. This is because the Web portals
used in hospitals are mostly focused on the radiology
workflow, and thus are typically unable to display ad-
vanced information such as contours or dose delivery.
Yet, viewing one’s own treatment plan might have
positive impact on the empowerment of the patient
and on the reduction of stress during the treatment.
In collaboration with the radiotherapy department
of the University Hospital of Liège, we have con-
tributed to a controlled randomized clinical trial in or-
der to evaluate the impact of a viewer of radiotherapy
plans in the context of patient education. The radio-
therapy viewer that is used in this trial is a desktop
application built using the Qt toolkit and the Stone of
Orthanc library. In this clinical trial, the patients re-
ceive a USB key containing their own images and the
viewer application, after a personal meeting with their
oncologist and with a nurse who explains how to use
the viewer. This clinical trial is still work in progress.
The user interface of the viewer is depicted in Fig-
ure 5 and is designed to be as simple as possible.
It displays the fusion of the simulation CT-scan, the
planned dose (DICOM RT-DOSE), and the different
contours (DICOM RT-STRUCT) following a MPR
(multiplanar reconstruction) layout. The user can se-
lect which contours are displayed.
Following the terminology of Section 2.2, the user
interface of this application is made of three different
Scene objects: one for the axial projection, one for
the sagittal projection, and one for the coronal projec-
tion. In turn, each of these three Scene objects con-
tains three Layer objects: The foremost Layer renders
polygons that correspond to the contours, the central
Layer renders a colorized version of the slice of the
dose (with an alpha channel for transparency), and the
backward Layer renders the CT-scan slice. The mul-
timodal fusion is thus solved by blitting the three 2D
Layer objects of each Scene onto its drawing surface.
From an algorithmic point of view, the surface
rendering of the 3D contours is done by reading the
content of one RT-STRUCT instance. In such an in-
BIOIMAGING 2022 - 9th International Conference on Bioimaging
48
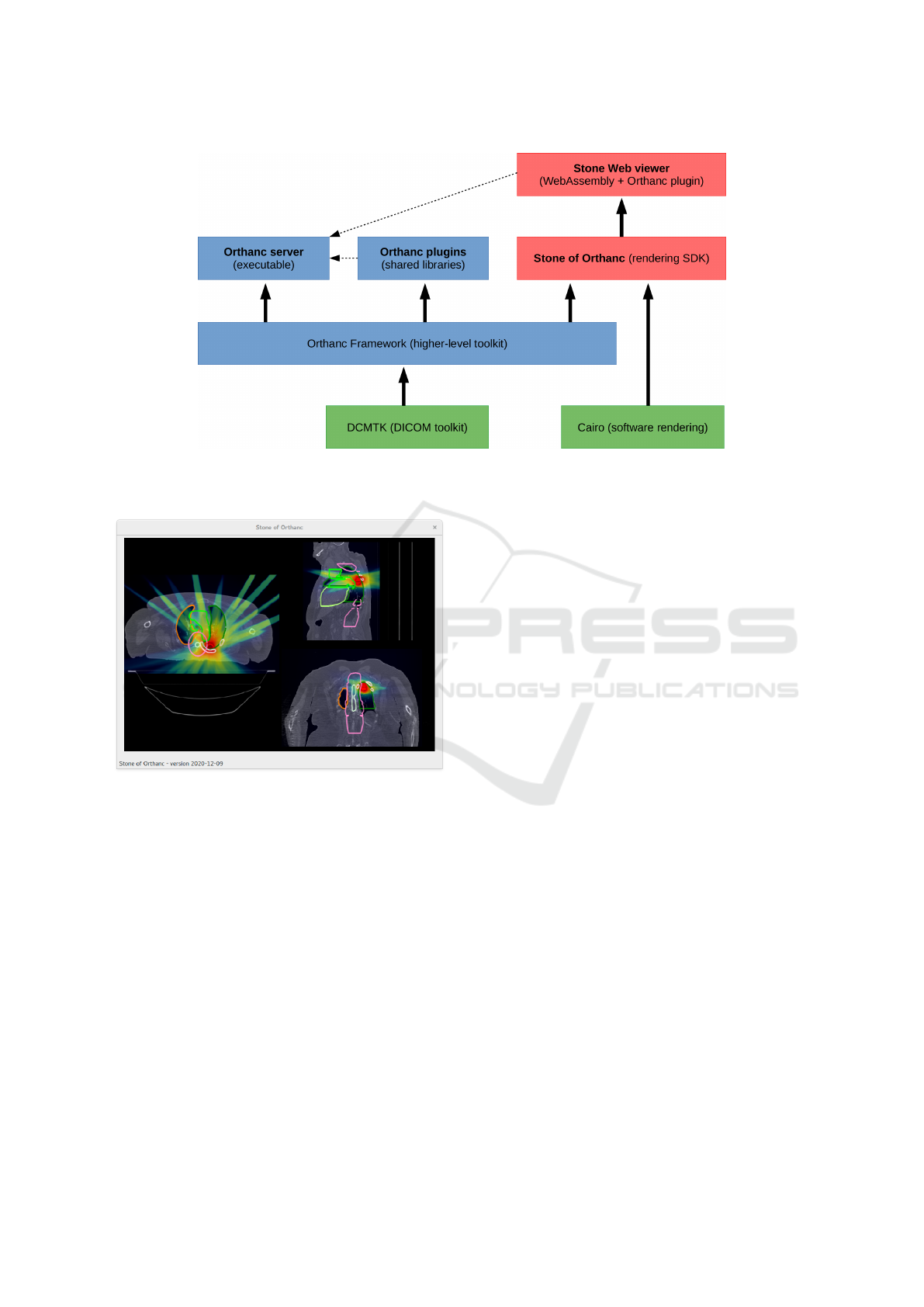
Figure 4: Overview of the high-level architecture of the Stone Web viewer. The green boxes indicate third-party libraries,
the blue boxes represent components provided by the Orthanc project, and the red boxes correspond to the Stone of Orthanc
project.
Figure 5: Desktop application for patient education in the
context of external radiotherapy.
stance, the contours of each volume are specified as
a list of polygons on the individual axial slices. As a
consequence, the rendering of one specific slice in the
axial projection simply consists in drawing the poly-
gons that intersect with this plane of interest. The
sagittal and coronal projections are more complex to
deal with. We first select the polygons that intersect
with the given sagittal or coronal slice. Each of those
intersections define a 2D box in the plane of interest.
The final rendering is done by drawing a set of poly-
gons that corresponds to the union of all those 2D
boxes, for which well-known computational geome-
try algorithms are available (Rivero and Feito, 2000).
On the other hand, the extraction of an axial, sagit-
tal or coronal slice out of the CT-scan volume stored
in RAM whose voxels are indexed by (i, j, k) integers
is straightforward: It consists in setting i, j or k to
the value of interest, then implementing nested loops
over the two other dimensions. The same algorithm
can be applied to the RT-STRUCT volume, as long as
its axes are aligned with those of the CT-scan volume.
Section 3.4 describes a solution if these two volumes
are not aligned.
Note that besides radiotherapy, the Stone of Or-
thanc library can be used to render similar images in
the field of nuclear medicine. For instance, Figure 6
illustrates another desktop application that provides
an advanced rendering of a PET-CT-scan using a lay-
out that is commonly encountered in professional ap-
plications. Evidently, those two applications could be
ported to Web environments thanks to WebAssembly.
3.4 Real-time Volume Reslicing
As explained in Section 2.2, Stone of Orthanc doesn’t
use the GPU if extracting one slice out of a 3D volume
along an arbitrary cutting plane: Volume reslicing is
done entirely in RAM. This raises the question of the
impact of this technical choice on the performance.
To evaluate the performance, a simple WebAssembly
application was developed that is depicted in Figure 7.
The mouse was used in order to change the orientation
of the cutting plane.
From an algorithmic point of view, the cutting
plane is specified by one 3D point indicating the ori-
gin of the plane, and by two orthonormal 3D vectors
indicating the directions of the X and Y axes in the
plane. The intersection of the source volumetric im-
age (which is a rectangular cuboid) with the cutting
plane is first computed. If non-empty, this intersection
Rendering Medical Images using WebAssembly
49
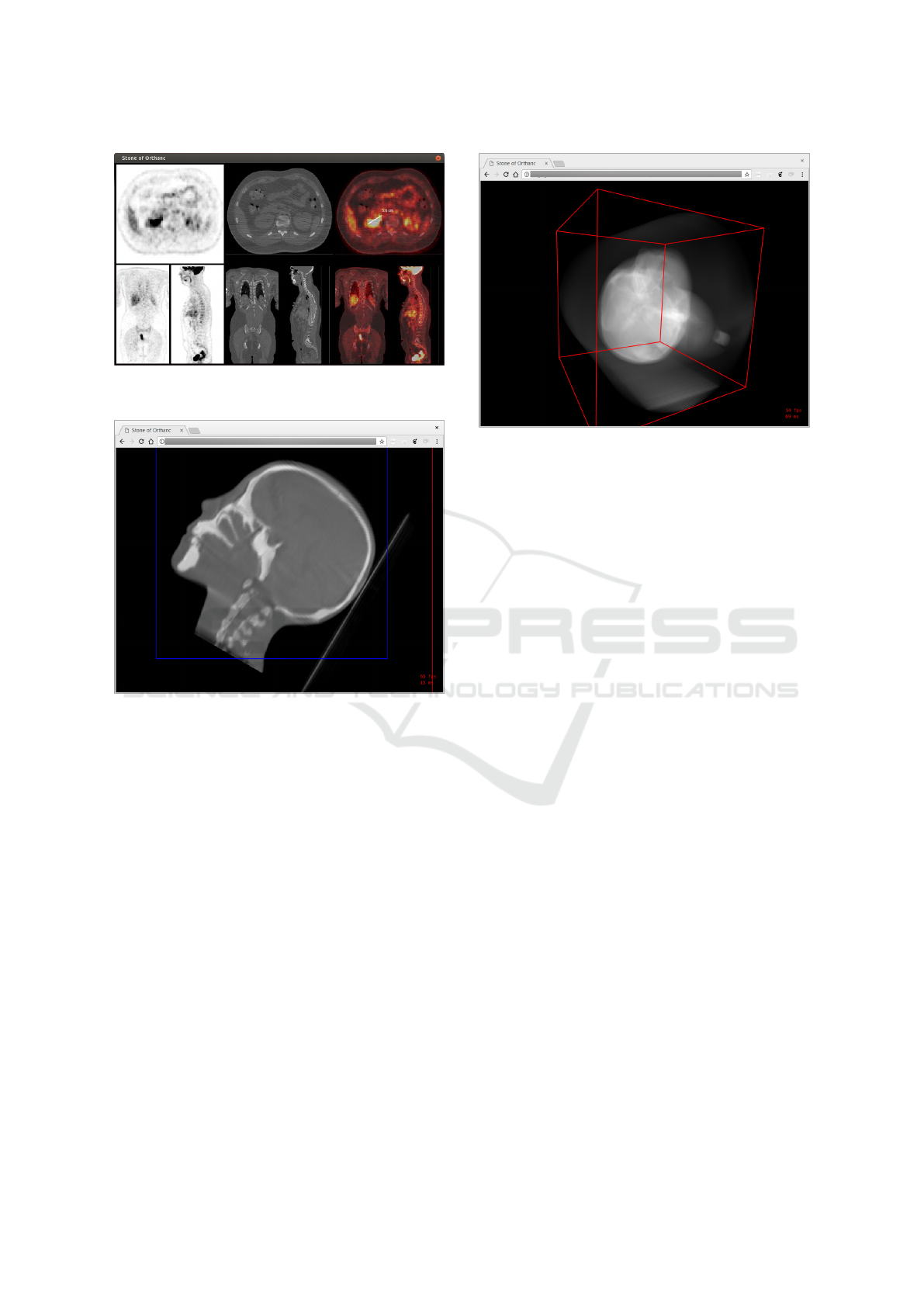
Figure 6: Desktop application for the rendering of PET-CT-
scans using a clinical-like layout.
Figure 7: Real-time reslicing of a 3D volume by a Web
browser.
defines a polygon with 3 to 6 vertices in the cutting
plane. Secondly, the bounding box of this polygon
in the reference system of the cutting plane defines
the physical extent of the 2D image to be generated.
The width and height of the extracted image is derived
from the desired pixel spacing. Finally, the extracted
image is filled by looping over its pixels, looking for
the value of the voxel in the source volume that is the
closest to the coordinates of each 2D pixel on the 3D
cutting plane. Bilinear or trilinear interpolation can
possibly be applied.
On a standard computer (Intel Core i7-3770 at
3.4GHz), the rendering of one slice of size 512 ×
512 × 502 voxels in 16bpp takes about 25 millisec-
onds (40 frames per second), which is fully compat-
ible with real-time requirements. On an old, entry-
level smartphone (Samsung Galaxy A3 2017), the
same rendering takes about 100 milliseconds (10
frames per second), which is still usable in practice.
Figure 8: Web rendering of a digitally reconstructed ra-
diograph (DRR) from a CT-scan. The projection is com-
puted client-side by the Web browser in near-real-time (a
few frames per second).
3.5 Digitally Reconstructed
Radiographs
The Stone of Orthanc library also features some com-
putational geometry primitives to deal with volume
rendering. It notably contains C++ classes in order to
generate a digitally reconstructed radiograph (DRR)
from a CT volume, which can be useful in the context
of radiotherapy. The DRR is the 2D radiograph that
would be imaged using a virtual X-ray camera cen-
tered at a given 3D point with a given focal length.
DRR can be computed by raytracing each voxel
of the CT volume according to the perspective trans-
form, which is a costly computation. Stone of Or-
thanc optimizes this computation by using the “shear-
warp transform” (Lacroute and Levoy, 1994). The in-
tuitive idea behind shear-warp is to postpone as much
as possible the application of the perspective transfor-
mation, and to apply it only once to a single so-called
“intermediate image” (this is the “warp” step). This
intermediate image is created from the summation of
the individual slices of the volume after the applica-
tion of an affine transform in which the X and Y axes
are left uncoupled, which enables a fast computation
(this is the “shear” step that is separately applied to
each slice). The shear-warp transform provides an ef-
ficient algorithm for volume rendering.
Figure 8 shows a WebAssembly application that
computes a DRR in near-real-time. On a standard
computer, the full rendering of a 512 × 512× 502 vol-
ume in 16bpp takes about 1 second. While moving
the virtual camera, the number of slices used for the
rendering can be reduced in order to improve speed,
BIOIMAGING 2022 - 9th International Conference on Bioimaging
50

thus enhancing the user experience. The GPU could
evidently be used to speed up this computation by im-
plementing an appropriate Oracle command. Besides
DRR, the shear-warp algorithm could also be applied
for MIP rendering by putting the virtual camera at in-
finity, and by replacing the summation by a “max”
operation.
4 CONCLUSIONS
This paper introduces Stone of Orthanc as a library
to render medical images, in a way that is compati-
ble with desktop, mobile and Web applications. Stone
of Orthanc is fully written in C++, and leverages the
emerging WebAssembly standard if targeting Web ap-
plications. Sample applications of Stone of Orthanc
are described as well. In particular, the Stone Web
viewer is introduced as the first free and open-source
teleradiology solution that uses WebAssembly. The
source code of the Stone of Orthanc library is publicly
released under the LGPL license, whereas the Stone
Web viewer is licensed under the AGPL
6
.
As shown in this paper, WebAssembly can be used
to maximize code reuse in applications for medical
imaging. The same approach could be used in many
other scientific fields, where C++ code is traditionally
used for numerical computations.
Future work will focus on the exploitation of
Stone of Orthanc for artificial intelligence applica-
tions in radiology. In particular, we plan to work on
the rendering of DICOM SR (structured reports), DI-
COM SEG (image segmentation) and DICOM GSPS
(grayscale softcopy presentation state), as the outputs
of AI algorithms. The integration of MPR render-
ing and whole-slide imaging (Jodogne et al., 2017)
directly within the Stone Web viewer will also be in-
vestigated. Finally, performance of volume rendering
will be improved, for instance by taking advantage of
the GPU and Web workers in the Oracle.
ACKNOWLEDGMENTS
The author wishes to thank Benjamin Golinvaux,
Alain Mazy and Osimis for their contributions to the
development of the Stone of Orthanc project.
6
The source code of the Stone of Orthanc project is
available at: https://hg.orthanc-server.com/orthanc-stone/
REFERENCES
Cheng, P. M., Montagnon, E., Yamashita, R., Pan, I.,
Cadrin-Chênevert, A., Perdigón Romero, F., Char-
trand, G., Kadoury, S., and Tang, A. (2021). Deep
learning: An update for radiologists. RadioGraphics,
41(5):1427–1445. PMID: 34469211.
Costa, C., Ferreira, C., Bastião, L., Ribeiro, L., Silva,
A., and Oliveira, J. L. (2011). Dicoogle – an open
source peer-to-peer PACS. Journal of Digital Imag-
ing, 24(5):848–856.
Gamma, E., Helm, R., Johnson, R., and Vlissides, J. M.
(1994). Design Patterns: Elements of Reusable
Object-Oriented Software. Addison-Wesley Profes-
sional, 1st edition.
Haas, A., Rossberg, A., Schuff, D. L., Titzer, B. L., Holman,
M., Gohman, D., Wagner, L., Zakai, A., and Bastien,
J. F. (2017). Bringing the Web up to speed with We-
bAssembly. In Cohen, A. and Vechev, M. T., editors,
Proceedings of the 38th ACM SIGPLAN Conference
on Programming Language Design and Implementa-
tion, PLDI 2017, Barcelona, Spain, June 18-23, 2017,
pages 185–200. ACM.
Jodogne, S. (2018). The Orthanc ecosystem for medical
imaging. Journal of Digital Imaging, 31(3):341–352.
Jodogne, S., Lenaerts, E., Marquet, L., Erpicum, C., Grei-
mers, R., Gillet, P., Hustinx, R., and Delvenne, P.
(2017). Open implementation of DICOM for whole-
slide microscopic imaging. In VISIGRAPP (6: VIS-
APP), pages 81–87.
Kikinis, R., Pieper, S. D., and Vosburgh, K. G. (2014). 3D
Slicer: A Platform for Subject-Specific Image Anal-
ysis, Visualization, and Clinical Support, pages 277–
289. Springer New York, New York, NY.
Lacroute, P. and Levoy, M. (1994). Fast volume rendering
using a shear-warp factorization of the viewing trans-
formation. In Proceedings of the 21st Annual Con-
ference on Computer Graphics and Interactive Tech-
niques, SIGGRAPH ’94, pages 451–458, New York,
NY, USA. ACM.
National Electrical Manufacturers Association (2021). ISO
12052 / NEMA PS3, Digital Imaging and Communi-
cations in Medicine (DICOM) standard.
http://www.dicomstandard.org.
Rivero, M. and Feito, F. (2000). Boolean operations on
general planar polygons. Computers & Graphics,
24(6):881–896. Calligraphic Interfaces: Towards a
new generation of interactive systems.
Rosset, A., Spadola, L., and Ratib, O. (2004). OsiriX: An
open-source software for navigating in multidimen-
sional DICOM images. Journal of Digital Imaging,
17(3):205–216.
Schroeder, W., Martin, K., and Lorensen, B. (2006). The
Visualization Toolkit: An object-oriented approach to
3D graphics. Kitware, Inc., 4 edition.
Urban, T., Ziegler, E., Lewis, R., Hafey, C., Sadow, C.,
Van den Abbeele, A. D., and Harris, G. J. (2017).
LesionTracker: Extensible open-source zero-footprint
web viewer for cancer imaging research and clinical
trials. Cancer Research, 77(21):e119–e122.
Warnock, M. J., Toland, C., Evans, D., Wallace, B., and
Nagy, P. (2007). Benefits of using the DCM4CHE DI-
COM archive. Journal of Digital Imaging, 20(Supp.
1):125–129.
Rendering Medical Images using WebAssembly
51
