
Brain MRI Images Pre-processing of Heterogeneous Data-sets for Deep
Learning Applications
S. Ostellino
a
, A. Benso
b
and G. Politano
c
Politecnico di Torino, Computer Science and Automation Department, Torino, Italy
Keywords:
Multiple Sclerosis, MRI, Imaging, Pre-processing, Deep Learning, Data Preparation, Heterogeneous
Data-sets, Real Clinical Data.
Abstract:
Automatic segmentation of tissues and lesions is a very important step in any Artificial Intelligence pipeline de-
signed to analyze medical images (especially MRI). This is particularly true for brain MRI images of patients
affected by neurological pathologies like Multiple Sclerosis (MS). To perform well, cutting edge Artificial
Intelligence approaches like Deep Learning need a huge amount of training data. Unfortunately, available
data-sets of MRI medical images often lack annotations, standardized acquisition protocols, formats and di-
mensions. This heterogeneity in the data-sets makes it often very difficult to use and integrate different data-
sets in the same pipeline. Available image pre-processing tools have specific requirements and might not be
adequate for extensive usage with heterogeneous data-sets. This paper presents an on-going work on a com-
prehensive and consistent brain MRI images pre-processing pipeline for Deep Learning applications enabling
the creation of a congruous data-set. The pipeline was tested with the public available ISBI2015 data-set.
1 INTRODUCTION
Magnetic resonance imaging (MRI) is a non-invasive
and fundamental diagnostic and monitoring tool for
many of the existing neurological conditions. Among
them, Multiple Sclerosis (MS) is a chronic autoim-
mune disease for which MRI is particularly impor-
tant as the disease needs to be carefully monitored
with at least one MRI per-year. The progression of
MS is variable between patients and a good monitor-
ing is crucial for correct therapeutic choices: MRI
is a support during disease diagnosis and follow-
up, together with others indicators of disease sta-
tus (Inojosa, 2021). MRI allows to visualize differ-
ent brain tissues depending on the chosen acquisi-
tion sequence and acquisition protocol. The modal-
ities that allow MS monitoring are the T1-weighted
(T1w), that allows easy annotation of healthy tissues,
and T2-weighted (T2w) and FLAIR images that are
used for detecting inflammatory lesions, indicators of
disease activity. Images are typically visually exam-
ined by neuro-radiologists that fill a written clinical
report that in many cases is affected by intra-and-
a
https://orcid.org/0000-0002-6275-3214
b
https://orcid.org/0000-0003-3433-7739
c
https://orcid.org/0000-0001-5268-9899
inter reader variability; therefore, a lot of attention
is now being paid to tools and methodologies for the
automatic segmentation of tissues and lesions (Kaur,
2021) (Zeng, 2020). The current problem with Deep
Learning pipelines, is that not only they require very
large training sets, but also they enforce strict require-
ments in the input data-sets’ format, size, and image
quality. This makes it often impossible to use differ-
ent data-sets as inputs for the same pipeline because
the differences in the data-sets can be easily result in
biases in the segmentation results.
The purpose of this paper is to present a compre-
hensive pre-processing pipeline able to prepare raw
MRI brain images data-sets (with the corresponding
lesions masks, if present) so that they can be directly
fed into Deep Learning architectures. The pipeline
has been implemented entirely in Python and partic-
ular attention has been paid in giving the possibility
to directly access each functions’ parameters to allow
further customizations/optimizations (see 3.1).
2 PROBLEMS DEFINITION
Several issues can negatively influence the perfor-
mances of a segmentation strategy: not only non-
brain tissues are a source of errors and need to be
Ostellino, S., Benso, A. and Politano, G.
Brain MRI Images Pre-processing of Heterogeneous Data-sets for Deep Learning Applications.
DOI: 10.5220/0010828500003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 3: BIOINFORMATICS, pages 115-120
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
115

removed, but MS lesions come in different locations
and sizes. Moreover, brain MRI images suffer the
presence of noise artifacts, non-uniformities, and are
affected by the intrinsic differences in the anatomy of
human brains; the lack of annotated data (MRI im-
ages without a corresponding lesion mask) is also se-
rious limitation that can cause misclassification prob-
lems, and result in a reduction of performances in
lesion identification, both in Machine Learning and
Deep Learning approaches. To overcome these prob-
lems, Artificial Intelligence requires very large train-
ing sets to correctly ”learn” to identify the relevant
features (tissues/lesions) on the image. Unfortunately,
on top of the physiological variability of the human
brains and MS lesions, different data-sets also come
in different formats, have been generated by differ-
ent equipment that introduce different artifacts, have
different dimensions, number of slices, file formats.
On the other hand, Deep learning methods require, as
input, images with a certain standard images’ file for-
mat (such as PNG), so it is not possible to feed a deep
learning architecture directly with an image stored
in a typical medical image format (such as NIfTI or
DICOM). Existing software and tools for image pre-
processing have some limitations. Firstly, they are of-
ten maintained by separated groups: when updating to
a new version of one of these tools, versioning prob-
lems and inconsistencies may occur if such update is
not supported by the other tools or plugins, such as
CBS Tools, JIST, TOADS-CRUISE, and BrainSuite.
Secondly, they lack in easy customization, and have
often strict requirements in terms of settings, making
it difficult to simply obtaining homogenous and weel
structured data-sets. All the mentioned problems of-
Figure 1: Steps of the proposed pipeline.
ten force scientists to rely on a single data-set, hope-
lessly affecting the quality of the Machine Learning
or Deep Learning pipeline performance.
3 MATERIALS AND METHODS
Pre-processing is crucial for any data-analysis
pipeline, and this is particularly true for medical im-
ages. The proposed pipeline (summarized in Figure
1) includes the following typical pre-processing steps
for brain MRI images (which are usually performed
by separate and independent tools):
• MRI sequence selection: consists in selecting
the MRI modality that will be processed. Our
pipeline focuses on T2w images, as MS lesions
are mostly visible in such modality;
• Image registration: registering an image means,
in this case, matching it with a reference model.
It is a key step and a prerequisite for all applica-
tions that want to compare data-sets among sub-
jects or across time (Toga, 2019); raw MRI im-
ages are not registered and may have different
spacing and slice resolution (Alam, 2016). The
registration step consists in a set of transformation
of the raw image that optimises a similarity index
with the reference image. The registered image
is obtained linearly interpolating the initial image
domain into the new domain, as image files con-
sist of a variable number of slices, each slice cor-
responding to a different longitudinal brain sec-
tion: when the number of slices in the original im-
age is not consistent with the number of images in
the atlas, the missing slices are interpolated. Our
pipeline registers each MRI image and then saves
it back as NIfTI files; moreover, this step makes
sure the MRI image contains the same number of
slices as the reference image by (if necessary) in-
terpolating missing slices;
• Brain-extraction: also known as skull-stripping,
it is a step which removes tissues that are not
of interest, such as skull and dura mater. In-
cluding non-brain tissues is a known source of
errors (Rehman, 2020); there are several possi-
ble brain-extraction methods, for instance rely-
ing on deep learning techniques or on traditional
morphological operations (Kalavathi, 2016). The
brain-extraction method proposed in this paper is
an adaptation of the method proposed by Gam-
bino et al. (Gambino, 2011), and uses a combina-
tion of morphological operations;
• Bias field correction and noise reduction: it cor-
rects the bias field, that is a low frequency inten-
sity nonuniformity present in the image data as
inhomogeneity and illumination nonuniformity.
• Final data-set creation: images (and correspond-
ing masks) are saved back as NIfTI files and as
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
116

PNG files in the correct format required by the
chosen neural network pipeline.
The pipeline was tested on the raw longitudinal
T2w images of the public avaiable ISBI2015 data-set
that consists in MRI (acquired at 1 to 4 different time
points) images of 5 subjects diagnosed with differ-
ent MS subtypes. Lesions masks of two independent
readers are also given. The data-set includes, besides
the raw images, the images that were processed when
creating the ISBI2015 data-set with the MIPAV soft-
ware, integrated with several plugins such TOADS-
CRUISE plugins. We used the latter images as a com-
parison to evaluate the results of our pipeline. The
MIPAV package, as many others such as CBS Tools,
JIST, and TOADS-CRUISE, are maintained by sepa-
rate groups: when a new version of one is released,
instabilities may occur if such update is not supported
by the other tools or plugins.
The proposed pipeline addresses this limitation by
using only stable Python libraries, chosen based on
the clarity of their documentation and on their perfor-
mances:
• ANTsPy for Registration, Brain extraction, and
Bias Field Correction;
• Dicom2nifti for DICOM to NIfTI conversion;
• SimpleITK for NIfTI files storage.
As previously pointed out, using different data-
sets acquired with different purposes and in differ-
ent centers, is fundamental for developing accurate
and efficient automatic segmentation methods based
on Deep Learning.
3.1 Pipeline Description
ANTsPy library was chosen for implementing the
most important steps of the pipeline because of its
good performances and extensive documentation. To
give the reader an idea, Table 1 shows the NIfTI to
NumPy (NumPy Python library) conversion times of
three common libraries on a MacBook Pro laptop.
This time is important because in each MRI data-set
there are thousands of images to be converted.
Table 1: Libraries performances.
ITK time 4.375 seconds
NIBABEL time 2.902 seconds
ANTS time 1.713 seconds
It is possible to easy access specific parameters as
shown in Table 2 thus allowing the pipeline steps to
be customized as well as optimized. Such parameters
directly affect the core points of the processing func-
tions.
Image Conversion
Image format conversion is fundamental to improve
the ability to generalize and work with more diversi-
fied data. The present pipeline supports, if needed,
the conversion from a series of DICOM images be-
longing to a single scan into a single NIfTI file, as the
DICOM format is another widespread medical image
format.
Table 2: Pipeline parameters.
Registration Transform, atlas
Brain extraction Iterations, kernels
Bias field correction Correction parameters
Image Registration
Images were registered to a reference atlas that was
chosen accordingly to the registration step proposed
by the ISBI data-set guidelines as a matter of con-
sistency: as they performed manual segmentation on
FLAIR images with the help of T1w and T2w im-
ages, and as there is no matching standard atlas for
FLAIR images, we rigidly registered T2w images to
the corresponding reference T2 standard atlas in the
MNI space
1
, so that the correspondence between im-
ages and lesion masks was obtained. For the purposes
of this paper and its future implementations for the
development of segmentation via deep learning, only
T2w images are considered at this point (Abderrahim,
2020). The atlas that was used is the ICBM Average
Brain linearly transformed to Talairach space, adapted
for use with the MNI Linear Registration Package.
The pipeline is designed to easily choose to use dif-
ferent atlases too, as it is sufficient to download and
import the desired file. Registration is done by deter-
mining the transformation needed to match the source
image with the target atlas, optimizing the similarity
index between them. The registered image is then ob-
tained linearly interpolating the initial image domain
into the new domain. The registration step is particu-
larly important when different images are put together
in a comprehensive set. Due to different acquisition
parameters and settings, the number of slices (being
each slice a section of the brain) between different
image files can vary: registration makes it possible
to uniform the number of slices to an atlas, conse-
quently matching the resolution of the image to atlas
resolution (in this case with a resolution of 1mm
3
) via
interpolation, so that the different scans match each
other in terms of anatomical references and in terms
of number of images per-scan. This step is extremely
1
http://nist.mni.mcgill.ca/icbm-152lin/
Brain MRI Images Pre-processing of Heterogeneous Data-sets for Deep Learning Applications
117
important to reduce the input variability, which is fun-
damental in the development of neural networks. As
an example, the original ISBI2015 T2w NIfTI files
contain 70 slices, while the registered image consists
of 181 slices.
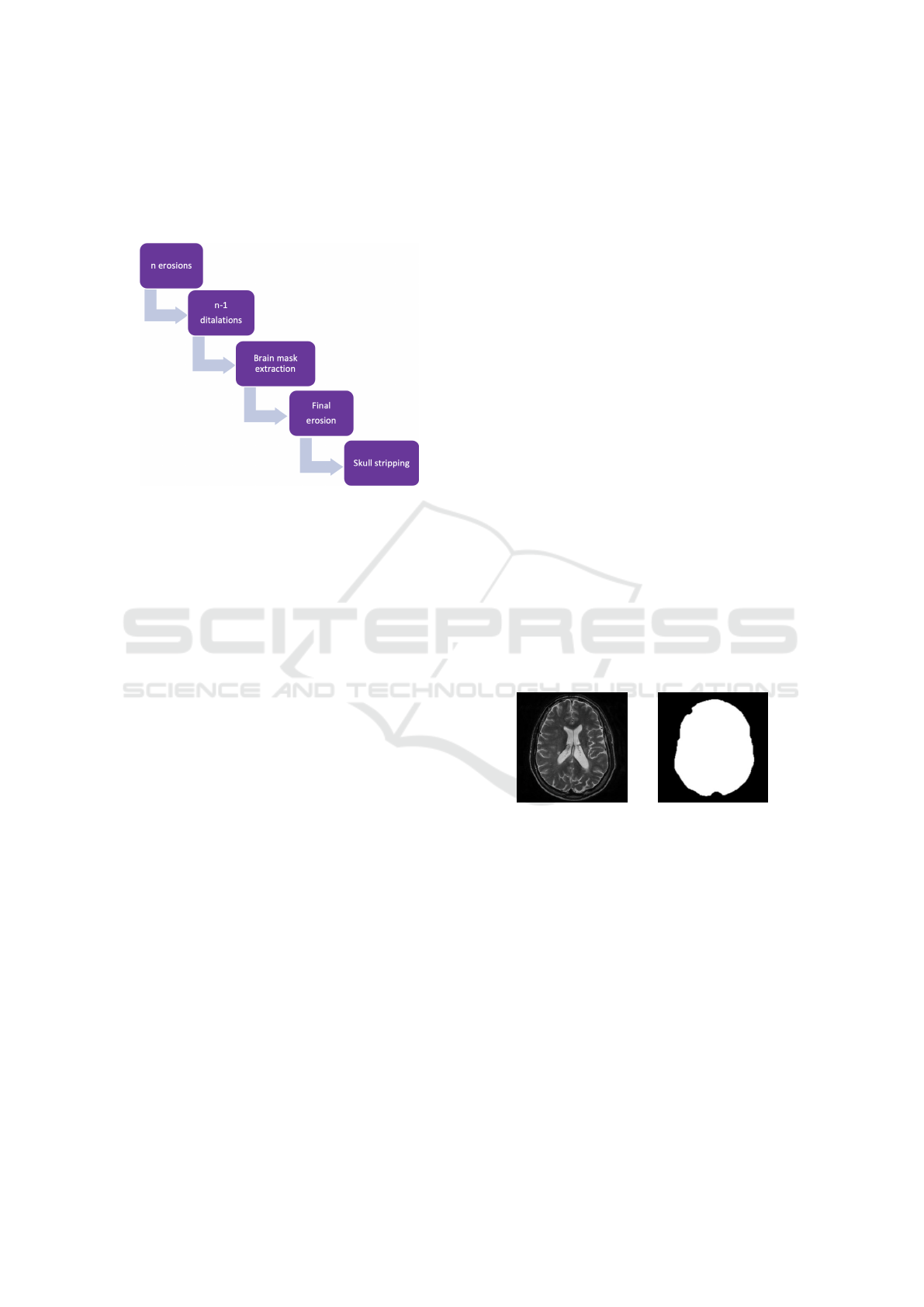
Figure 2: Brain extraction steps.
Brain Extraction
The steps that compose the brain extraction are briefly
summarized in Figure 2. Such steps were imple-
mented adapting the findings of Gambino et al. (Gam-
bino, 2011). Skull-stripping is usually implemented
on T1w images, but in our pipeline it is adapted to
work directly on T2w images, without requiring inter-
mediate steps that would need to further register T2w
images to T1w images, in order to apply the brain
mask obtained with the T1w images to the T2w scan.
Brain extraction develops in five simple steps; again
all parameters such as the number of iterations or the
features of morphological operators can be easily cus-
tomized (see Table 2):
1. n (n=3) erosions with a cross kernel,
2. n-1 dilatations with a cross kernel,
3. brain mask extraction,
4. final erosion,
5. the original image is multiplied by the brain mask,
in order to obtain the brain.
Such parameters can be adapted in order to obtain
better performances as future implementations, and
most of them can be adapted in order to be able to
work independently from the MRI image modalities.
This is not true, for example, for tools such as MIPAV
MP2RAGE that, in order to obtain the skull stripping
of T2w images, require both T1w images and T2w
images as input.
Bias Field Correction
The N4 Bias Field Correction method (Tustison,
2010) was applied for estimating and correcting the
bias, giving as output the corrected image.
Deep Learning Data-set Creation
Once the images are processed, it is possible to con-
vert them into PNG files that can be then organised in
folders, and directly fed to a neural network. As not
all the images might be needed (for example, those
for which the pre-processing has failed, or those that
only partially show the cerebellum), the pipeline also
allows to store only images of interest, selecting their
corresponding identification number. Particular atten-
tion was paid to preserve image orientations during
the creation of the data-set, as multiple conversions
between formats is needed for the pipeline itself to
work.
3.2 Working Example
The steps previously described are exemplified in
Figure 3 and in Figure 4 where an image from the
ISBI2015 data-set was processed and then saved as
.png file for later use.
(a) Original image. (b) Brain mask.
Figure 3: Extraction of the brain.
Figure 3 shows a slice of the image that is fed to
the pipeline for the pre-processing: the brain mask
represents the result of the brain-extraction step. Such
mask is then multiplied to the initial slice, giving the
desired brain-stripped result.
In Figure 4 the lesion mask (coloured in blue) is
superimposed over the skull-stripped brain, and such
mask will serve as ground truth for a segmentation
algorithm. The masks were given together with the
ISBI2015 dataset, and they were obtained by two ex-
pert readers that annotated the MRIs previously pre-
processed with the MIPAV software. The correspon-
dence of such lesion masks with the brain images pro-
cessed with our pipeline indicates that it works as ex-
pected and gives comparable results.
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
118

(a) Processed. (b) With lesions.
Figure 4: Skull-stripping.
3.2.1 Processing Time
We report here the processing times of the execu-
tion of the pipeline to the data-set. The processing
times
2
of the registration step and of the rest of the
process are separately reported, as the registration is
performed on the original raw image, while brain-
extraction and bias field correction are done on the
registered image.
Table 3: Performances with 1 MRI scan.
Registration 4.5 sec.
Processing 143.52 sec.
Values in Table 3 refer to the processing of a single
NIfTI file that corresponds to a T2w MRI scan of a
subject. Each NIfTI file contains, once it is registered
with the atlas, 181 images.
4 DISCUSSION
Deep Learning pipelines require large training sets,
and have strict requirements in the input format, size,
and image quality. Moreover, Deep Learning per-
formances benefit from homogenous and weel struc-
tured data-sets. This makes it often impossible to
use different data-sets directly as inputs because their
differences can be easily result in biases in the seg-
mentation results, affecting performances. This pa-
per presents a comprehensive pre-processing pipeline
able to prepare raw MRI brain images data-sets so
that they can be directly fed into deep learning ar-
chitectures. The pipeline has been implemented en-
tirely in Python and it guarantees the possibility to
directly access each functions’ parameters to allow
further customizations/optimizations. Several other
approaches recommended for MRI images process-
ing have strict constrains in terms of the images that
they can process: some are limited to 3D images,
2
Running on a MacBook Pro - macOS Bis Sur - 2.6GHz
Intel Core i7 6 core
and some require specific MRI modalities to function
properly. The presented pipeline differs from other
existing software as it incorporates, with flexibility
and customizability, all the steps that are needed when
preparing heterogenous data-sets for deep learning
application, from the raw MRI scan (in different im-
age formats) to the image that will be the input of a
deep learning algorithm. Furthermore, it differs from
other known tools such as BrainSuite as it can be di-
rectly and easily incorporated into the development of
any automatic medical images analysis systems based
on Deep Learning architectures, without limiting it-
self to a processing or visualising tool for MRI im-
ages and, above all, relying on stable Python libraries
without depending on many plug-ins that can cause
inconsistencies and versioning problems. This ap-
proach can help with the task of data preparation and
image pre-processing, that cannot be ignored or un-
derestimated when constructing data-sets for Machine
Learning (and in particular Deep Learning) pipelines.
Future implementations will include an optimization
of the performances to include as input more MRI
modalities and formats, increasing its versatility, and
the incorporation of the pipeline as the backbone of
an innovative Deep Learning architecture targeted for
application in real clinical practice.
5 CONCLUSIONS
Handling the variability of MRI medical images
needs an efficient pre-processing pipeline aimed at
solving practical issues that include but are not limited
to raw images format and dimensions, physiological
differences, image artifacts. This paper introduces a
pipeline to fill the gap between heterogeneous data-
sets and their practical integration and usability in the
same Artificial Intelligence pipeline. We successfully
tested the pipeline to show how it helps filling the gap
between heterogeneous data-sets and their practical
use. Future work will include extensive comparative
evaluations of the pipeline, and an optimization of the
performances to include as input more MRI modali-
ties and formats.
Finally, it is important to point out that the pro-
posed approach is not necessarily strictly related to
brain MRI images, but could be easily adapted to
other MRI scans, such as chest imaging.
REFERENCES
Abderrahim, Marwa, e. a. (2020). Comparative study of rel-
evant methods for mri/x brain image registration. The
Brain MRI Images Pre-processing of Heterogeneous Data-sets for Deep Learning Applications
119

Impact of Digital Technologies on Public Health in
Developed and Developing Countries, page 338–47.
Alam, Fakhre, e. a. (2016). Evaluation of medical image
registration techniques based on nature and domain of
the transformation. Journal of Medical Imaging and
Radiation Sciences, 47(2):178–93.
Gambino, Orazio, e. a. (2011). Automatic skull stripping in
mri based on morphological filters and fuzzy c-means
segmentation. 2011 Annual International Conference
of the IEEE Engineering in Medicine and Biology So-
ciety, page 5040–43.
Inojosa, Hernan, e. a. (2021). Should we use clinical tools to
identify disease progression? Frontiers in Neurology,
11:1890.
Kalavathi, P., e. V. B. S. P. (2016). Methods on skull strip-
ping of mri head scan images — a review. Journal of
Digital Imaging, 29(3):365–79.
Kaur, Amrita, e. a. (2021). State-of-the-art segmentation
techniques and future directions for multiple sclerosis
brain lesions. Archives of Computational Methods in
Engineering, 28(3):951–77.
Rehman, Hafiz Zia Ur, e. a. (2020). Conventional and deep
learning methods for skull stripping in brain mri. Ap-
plied Sciences, 10(5):1773.
Toga, A. W., e. P. M. T. (2019). The role of image regis-
tration in brain mapping. Journal of Big Data, 19(1-
2):3–24.
Tustison, Nicholas J., e. a. (2010). N4itk: Improved n3 bias
correction. IEEE transactions on medical imaging,
29(6):1310–20.
Zeng, Chenyi, e. a. (2020). Review of deep learning ap-
proaches for the segmentation of multiple sclerosis
lesions on brain mri. Frontiers in Neuroinformatics,
14:55.
APPENDIX
The code of the presented pipeline can be downloaded
from GitHub at the following address: https://github.
com/aSofworkOrange/BrainMRI-preproc
BIOINFORMATICS 2022 - 13th International Conference on Bioinformatics Models, Methods and Algorithms
120
