
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
Youngjoo Kim
1,† a
, Mateus Espadoto
2,† b
, Scott C. Trager
3 c
, Jos B. T. M. Roerdink
1 d
and
Alexandru C. Telea
4 e
1
Bernoulli Institute for Mathematics, Computer Science and Artificial Intelligence,
University of Groningen, The Netherlands
2
Institute of Mathematics and Statistics, University of S
˜
ao Paulo, Brazil
3
Kapteyn Astronomical Institute, University of Groningen, The Netherlands
4
Department of Information and Computing Sciences, Utrecht University, The Netherlands
Keywords:
High-dimensional Visualization, Dimensionality Reduction, Mean Shift, Neural Networks.
Abstract:
Dimensionality reduction (DR) methods aim to map high-dimensional datasets to 2D scatterplots for visual
exploration. Such scatterplots are used to reason about the cluster structure of the data, so creating well-
separated visual clusters from existing data clusters is an important requirement of DR methods. Many DR
methods excel in speed, implementation simplicity, ease of use, stability, and out-of-sample capabilities, but
produce suboptimal cluster separation. Recently, Sharpened DR (SDR) was proposed to generically help such
methods by sharpening the data-distribution prior to the DR step. However, SDR has prohibitive computa-
tional costs for large datasets. We present SDR-NNP, a method that uses deep learning to keep the attractive
sharpening property of SDR while making it scalable, easy to use, and having the out-of-sample ability. We
demonstrate SDR-NNP on seven datasets, applied on three DR methods, using an extensive exploration of its
parameter space. Our results show that SDR-NNP consistently produces projections with clear cluster sepa-
ration, assessed both visually and by four quality metrics, at a fraction of the computational cost of SDR. We
show the added value of SDR-NNP in a concrete use-case involving the labeling of astronomical data.
1 INTRODUCTION
The visual analysis of high-dimensional data is chal-
lenging, due to its many observations (also known
as points or samples) and values recorded per obser-
vation (also known as dimensions, features, or vari-
ables) (Liu et al., 2015; Nonato and Aupetit, 2018;
Espadoto et al., 2019). Dimensionality reduction
(DR), also called projection, is particularly suited
for such data. Compared to glyphs (Yates et al.,
2014), parallel coordinate plots (Inselberg and Dims-
dale, 1990), table lenses (Rao and Card, 1994), and
scatterplot matrices (Becker et al., 1996), DR meth-
ods scale visually up to thousands of dimensions
and hundreds of thousands of samples. DR tech-
niques such as t-SNE (Maaten and Hinton, 2008) and
a
https://orcid.org/0000-0002-9677-163X
b
https://orcid.org/0000-0002-1922-4309
c
https://orcid.org/0000-0001-6994-3566
d
https://orcid.org/0000-0003-1092-9633
e
https://orcid.org/0000-0003-0750-0502
†
These authors contributed equally.
UMAP (McInnes and Healy, 2018), to mention the
most popular ones, can segregate data clusters into
well-separated visual clusters, which enables one to
reason about the former by seeing the latter, a prop-
erty generically known as preservation of data struc-
ture (Behrisch et al., 2018).
A recent survey (Espadoto et al., 2019) noted that
many DR techniques score below t-SNE or UMAP
in cluster segregation but have other assets – simple
usage and implementation, computational scalability,
and out-of-sample behavior. Following this, (Kim
et al., 2021) recently proposed Sharpened DR (SDR)
to generically improve the cluster segregation ability
by sharpening the data prior to DR by a variant of the
Mean Shift algorithm (Comaniciu and Meer, 2002).
However, SDR is impractical to use as Mean Shift
is prohibitively expensive in high dimensions. Sep-
arately, Neural Network Projection (NNP) was pro-
posed (Espadoto et al., 2020) to generically imitate
any DR technique with good quality, speed, ease-of-
use, and out-of-sample ability.
In this paper, we combine the cluster segregation
Kim, Y., Espadoto, M., Trager, S., Roerdink, J. and Telea, A.
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks.
DOI: 10.5220/0010820900003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 3: IVAPP, pages 63-76
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
63

abilities of SDR with the speed, ease-of-use, and out-
of-sample abilities of NNP. Our approach jointly ad-
dresses the Mean Shift sharpening step of SDR and
the generic projection step following afterwards. Our
SDR-NNP method has the following features – to our
knowledge, not yet jointly achieved by existing DR
methods:
Quality (C1): We provide better cluster separation
than existing DR methods, as measured by well-
known metrics in the DR literature;
Scalability (C2): Our method is linear in sample and
dimension counts, allowing the projection of datasets
of up to a million samples and hundreds of dimen-
sions in a few seconds on commodity GPU hardware;
Ease of use (C3): Our method produces good results
with minimal or no parameter tuning;
Genericity (C4): We can handle any real-valued (un-
labeled) high-dimensional data;
Stability and Out-of-Sample Support (C5): We can
project new samples for a learned projection without
recomputing it, in contrast to standard t-SNE and any
other non-parametric methods.
We structure this paper as follows: Section 2 dis-
cusses related work on dimensionality reduction. Sec-
tion 3 details our method. Section 4 presents the re-
sults that support our above contributions. Section 5
discusses our method. Finally, Section 6 concludes
the paper.
2 BACKGROUND
Let x = (x
1
,. .. ,x
n
), x
i
∈ R,1 ≤ i ≤ n be an n-
dimensional (nD) real-valued sample, and let D =
{x
j
}, 1 ≤ j ≤ N be a dataset of N samples. A DR
technique is a function
P : R
n
→ R
q
, (1)
where q n, and typically q = 2. The projection P(x)
of a sample x ∈ D is a point p ∈ R
q
. Projecting the
whole set D yields a qD scatterplot denoted next as
P(D).
DR methods aim to satisfy multiple requirements.
Table 1 outlines prominent ones present in several
DR surveys (Hoffman and Grinstein, 2002; Maaten
and Postma, 2009; Engel et al., 2012; Sorzano et al.,
2014; Liu et al., 2015; Cunningham and Ghahramani,
2015; Xie et al., 2017; Nonato and Aupetit, 2018; Es-
padoto et al., 2019). Besides these, DR techniques
also require locality, steerability, and multilevel com-
putation (Nonato and Aupetit, 2018). We do not fo-
cus on such additional requirements as these are less
mainstream.
The quality (Q) and cluster separation (CS) re-
quirements need additional explanations. Projection
quality is assessed by local metrics that measure how
a small neighborhood of points in D maps to a neigh-
borhood in P(D) and/or conversely. Local quality
metrics include the following (see Table 2 for the for-
mal definitions):
Trustworthiness T (Venna and Kaski, 2006): Mea-
sures the fraction of close points in D that are also
close in P(D). T tells how much one can trust that lo-
cal patterns in a projection represent actual data pat-
terns. In the definition (Table 2), U
(K)
i
is the set of
points that are among the K nearest neighbors of point
i in the 2D space but not among the K nearest neigh-
bors of point i in R
n
; and r(i, j) is the rank of the 2D
point j in the ordered-set of nearest neighbors of i in
P(D);
Continuity C (Venna and Kaski, 2006): Measures
the fraction of close points in P(D) that are also close
in D. In the definition (Table 2), V
(K)
i
are the points in
the K nearest neighbors of point i in D but not among
the K nearest neighbors in 2D; and ˆr(i, j) is the rank of
the R
n
point j in the ordered set of nearest neighbors
of i in D;
Neighborhood Hit NH (Paulovich et al., 2008):
Measures how well a projection P(D) separates la-
beled data, in a rotation-invariant fashion. NH is
the number y
l
k
of the k nearest neighbors of a point
y ∈ P(D), denoted by y
k
, that have the same label l as
y, averaged over P(D);
Shepard Diagram Correlation R (Joia et al., 2011):
The Shepard diagram is a scatterplot of the pairwise
distances between all points in P(D) vs the corre-
sponding distances in D. Points below, respectively
above, the main diagonal show distance ranges for
which false neighbors, respectively missing neigh-
bors, occur. The closer the plot is to the main diag-
onal, the better the overall distance preservation is.
The scatterplot’s Spearman rank correlation R mea-
sures this – a value R = 1 indicates a perfect (positive)
distance correlation.
All above metrics are local, i.e., capture preserva-
tion of data structure in D at the scale given by the
neighborhood size K. In practice, what a ‘good’ K
value is for a given dataset D is unknown. K can also
vary locally within D as function of the point density.
At a higher level, projections are used to reason about
the overall data structure in D by creating, ideally, vi-
sual clusters that are as well separated in P(D) as data
clusters are in D, a property called cluster separa-
tion (CS). High-CS projections show, e.g., how many
point clusters exist and how these correlate (or not)
with labels or specific attributes (Nonato and Aupetit,
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
64

2018), or predict how easy it is to train a classifier for
D based on the CS in P(D) (Rauber et al., 2017). In
general, it is hard to design objective metrics for CS
like one does for local quality, because a ‘well sepa-
rated data cluster’ in D is not evident. Hence, CS is
typically assessed on (labeled) datasets D for which
the ground-truth data-separation is well known, e.g.,
MNIST (LeCun and Cortes, 2010).
We next discuss existing DR methods in the light
of the requirements in Tab. 1. We group these into
unsupervised and supervised methods, as follows.
Unsupervised Methods: Principal Component Anal-
ysis (Jolliffe, 1986) (PCA) is simple, fast, out-of-
sample (OOS), and easy-to-interpret, also used as
pre-processing for other DR techniques that require a
moderate data dimensionality n (Nonato and Aupetit,
2018). Being a linear and global method, PCA is de-
ficient on quality and CS, especially for data of high
intrinsic dimensionality.
Techniques such as MDS (Torgerson, 1958),
Landmark MDS (De Silva and Tenenbaum, 2004),
Isomap (Tenenbaum et al., 2000), and LLE (Roweis
and Saul, 2000) with its variations (Donoho and
Grimes, 2003; Zhang and Zha, 2004; Zhang and
Wang, 2007) detect and project the (neighborhood
of the) high-dimensional manifold on which data is
embedded, and can capture nonlinear data structure.
Such methods yield higher quality than PCA, but can
be hard to tune, do not all support OOS, and do not
work well for high-intrinsic-dimensional data.
Force-directed methods such as LAMP (Joia et al.,
2011) and LSP (Paulovich et al., 2008) can yield
good quality, good scalability, and are simple to
use. However, not all force-directed methods have
OOS capability. Clustering-based methods, such as
PBC (Paulovich and Minghim, 2006), share many
features with force-directed methods, such as good
quality, but also lack OOS.
Stochastic Neighborhood Embedding (SNE)
methods, like the well-known t-SNE (Maaten and
Hinton, 2008), have high overall quality and CS
capabilities. Yet, t-SNE has a (high) complexity of
O(N
2
) in sample count, is very sensitive to small data
changes, can be hard to tune (Wattenberg, 2016), and
has no OOS. Tree-accelerated t-SNE (Maaten, 2014),
hierarchical SNE (Pezzotti et al., 2016), approxi-
mated t-SNE (Pezzotti et al., 2017), and various GPU
variants of t-SNE (Pezzotti et al., 2020; Chan et al.,
2018) improve scalability, but are algorithmically
quite complex, and still have sensitivity, tuning, and
OOS issues. Uniform Manifold Approximation and
Projection (UMAP) (McInnes and Healy, 2018) has
comparable quality to t-SNE, is much faster and has
OOS. Still, UMAP is also sensitive to parameter
tuning.
Autoencoders (AE) (Hinton and Salakhutdinov,
2006; Kingma and Welling, 2013) aim to generate
a compressed, low-dimensional representation of the
data in their bottleneck layers by training to repro-
duce the data input at the output. They have similar
quality to PCA and are easy to set up, train, and use,
are fast, and have OOS capabilities. Self-organizing
maps (SOM) (Kohonen, 1997) share with AE the ease
of use, training, and computational scalability. Yet,
both AE and SOM lag behind t-SNE and UMAP in
CS, which is, as explained, essential for interpreting
projections.
Supervised Methods: ReNDA (Becker et al., 2020)
uses two neural networks to implement (1) a nonlinear
generalization of Fisher’s Linear Discriminant Anal-
ysis (Fisher, 1936) and (2) an autoencoder, used for
regularization. ReNDA scores well on predictability
and has OOS, but needs pre-training of each individ-
ual network and has low scalability. Recently, (Es-
padoto et al., 2020) proposed Neural Network Projec-
tions (NNP), where a selected subset D
s
⊂ D is pro-
jected by any DR method to yield a training projec-
tion P(D
s
) ⊂ R
2
. D
s
is fed into a regression neural
network which is trained to output a 2D scatterplot
that aims to replicate P(D
s
). NNP is very fast, simple
to use, generic, and has OOS. NNP’s major limitation
is a lower CS than its training projection.
Semi-supervised Methods: The SSNP
method (Espadoto et al., 2021) takes a mid path
between supervised methods (e.g., NNP) and unsu-
pervised ones (e.g., AE). Like NNP, SSNP has an
encoder-decoder architecture with a reconstruction
target but adds a classification target using either
ground-truth labels from the dataset D or pseudola-
bels computed from D by a clustering algorithm.
SSNP produces 2D projections which look quite
similar to those created by our method described next
in Sec. 3. However, important differences exist:
• Our method consists of two distinct operations:
high-dimensional data sharpening, followed by
projection. SSNP only performs the projection
step;
• SSNP is a semi-supervised method that relies
upon labels or clustering to create the information
to learn from. Our method, similar to NNP, uses a
t-SNE projection of the data to learn from. This is
a fundamental difference between SSNP and our
method;
• The architectures of the neural networks for SSNP
and our method are fundamentally different. In
particular, SSNP uses two different architectures
for training, respectively inference. Our method
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
65
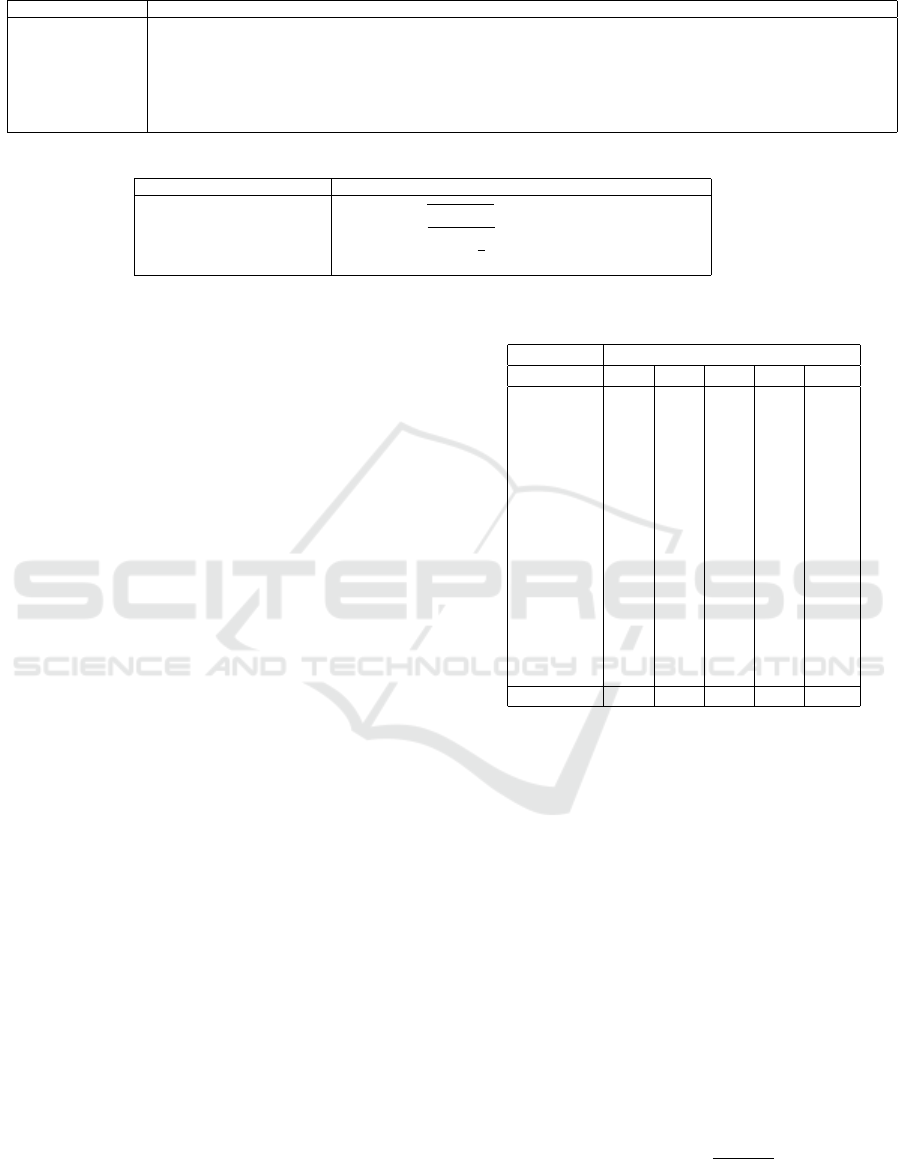
Table 1: Summary of desirable requirements (characteristics) of DR methods.
Requirement name Description (the requirement implies that the method has the following properties):
Quality (Q) Captures local data structures well, as measured by the projection local-quality metrics in Table 2.
Cluster separation (CS) Captures data structures present at larger scales than local structures, e.g. clusters, as visual clusters in the 2D scatterplot.
Scalability (S) Can project datasets of hundreds of dimensions and millions of samples in a few seconds on commodity hardware.
Ease of use (EoU) Has few (ideally: no) free parameters, which are intuitive and easy to tune to get the desired results.
Genericity (G) Can project any (real-valued) dataset, with or without labels.
Out-of-sample (OOS) Can fit new data in an existing projection. OOS projections are also stable – small input-data changes cause only small projection
changes.
Table 2: Local quality metrics for projections. All metrics range in [0,1] with 0 being lowest, and 1 being highest, quality.
Metric Definition
Trustworthiness (T ) 1 −
2
NK(2n−3K−1)
∑
N
i=1
∑
j∈U
(K)
i
(r(i, j) −K)
Continuity (C) 1 −
2
NK(2n−3K−1)
∑
N
i=1
∑
j∈V
(K)
i
(ˆr(i, j) −K)
Neighborhood hit (NH)
1
N
∑
y∈P(D)
y
l
k
/y
k
Shepard diagram correlation (R) Spearman’s rank of (kx
i
− x
j
k,kP(x
i
) − P(x
j
)k),1 ≤ i ≤ N,i 6= j
uses a single architecture for both training and in-
ference.
• A key underlying use-case for SDR-NNP is to en-
hance the separation between unlabeled data clus-
ters so that these can next be labeled by users (see
next Sec. 4.3). This is out of scope of SSNP.
Sharpening Data: Finding clusters of related data
points is a key task in data science, addressed by
tens of clustering methods (Xu and Wunsch, 2005;
Berkhin, 2006). Mean Shift (MS) (Fukunaga and
Hostetler, 1975; Cheng, 1995; Comaniciu and Meer,
2002) is particularly relevant to our work. MS com-
putes the kernel density estimation of a dataset D and
next shifts points in D upstream along the density gra-
dient. This effectively clusters D, with applications in
image segmentation (Comaniciu and Meer, 2002) and
graph drawing (Hurter et al., 2012). Recently, Sharp-
ened DR (SDR) (Kim et al., 2021) used MS for the
first time to assist DR: A dataset D is sharpened by a
few MS iterations, not to be confused with the clus-
tering goal of the original MS. The sharpened dataset
is next projected by a fast, easy-to-use, stable, but po-
tentially low-CS DR method. Sharpening ‘precondi-
tions’ the used DR method to overcome its lack of CS.
Yet, as MS is very slow for high-dimensional data,
this makes SDR impractical for such data.
Table 3 summarizes the DR techniques discussed
above and indicates how they fare with respect to the
requirements discussed earlier in this section. No
reviewed method satisfies all the requirements opti-
mally. We next describe our proposed method SDR-
NNP (Table 3 last row) and show that it scores high
on these requirements.
3 SDR-NNP METHOD
As outlined in Sec. 2, SDR and NNP have comple-
mentary desirable features: SDR produces good clus-
Table 3: Summary of DR techniques in Sec. 2 and their
characteristics from Table 1.
Desirable characteristics of the method
Method Q S EoU G OOS
PCA low high high high yes
MDS mid low low low no
L-MDS mid mid low low no
Isomap mid low low low no
LLE mid low low low no
LAMP mid mid mid high yes
LSP mid mid mid high no
PBC mid mid mid high no
UMAP high high low high yes
t-SNE high low low high no
Autoencoder low high high low yes
SOM low high high low no
ReNDA mid low low mid yes
NNP high high high high yes
SDR high low mid high no
SDR-NNP high high high high yes
ter separation (CS), while NNP is fast, easy to use,
and has OOS ability. Our combined SDR-NNP tech-
nique joins these advantages and works as follows
(see also Fig. 1). We use SDR to sharpen a small data
subset to create an initial 2D projection (Sec. 3.1).
Next, we train NNP on the sharpened data and its 2D
projection (Sec. 3.2) and use it to project the whole
dataset.
3.1 Sharpened Dimensionality
Reduction
SDR has two main components, as follows (for full
details we refer to (Kim et al., 2021)):
Sharpening the Data: Given a dataset D ∈ R
n
, SDR
computes its density using the multivariate kernel
density estimator ρ(x) : R
n
→ R
+
defined as
ρ(x) =
∑
y∈N(x)
L
kx − yk
h
, (2)
where N(x) is the set of k
s
-nearest neighbors of x in
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
66

D; L is a parabolic kernel (Epanechnikov, 1969); and
h is the distance of x to its k
th
s
(farthest) neighbor in
N(x).
Next, SDR shifts points x ∈ D using the update
rule
x
next
= x + α
∇ρ(x)
max(k∇ρ(x)k,ε)
, (3)
where α ∈ [0,1] is a ‘learning rate’ parameter that
controls the shift speed (higher values yield higher
speed) and ε = 10
−5
is a regularization parameter.
After every update (Eqn. 3), the density ρ is com-
puted again using Eqn. 2. This sharpening approach
is called Local Gradient Clustering (LGC) by anal-
ogy with Gradient Clustering (GC) (Fukunaga and
Hostetler, 1975).
SDR has three parameters: T (number of itera-
tions); k
s
(number of nearest neighbors); and α (learn-
ing rate). We use k
s
≥ 50 following (Kim et al., 2021);
setting α and T is discussed in Sec. 4.
Projection: SDR produces a dataset D
s
which is a
sharpened version of the input dataset D. SDR next
projects D
s
by a DR method of choice (typically fast
but not necessarily OOS), called the baseline DR
method below, to obtain a 2D projection P(D
s
). The
data in D
s
are better separated than in D, which allows
a projection P to produce better cluster separation in
P(D
s
) than in P(D).
3.2 Training NNP
The key idea of SDR-NNP is to use SDR (Sec. 3.1) on
a small data subset to obtain P(D
s
). To project the full
dataset D, we next train the NNP regressor (Espadoto
et al., 2020) using D
s
as the high-dimensional in-
put and P(D
s
) as the 2D output. The network has
three fully-connected hidden layers with ReLU acti-
vation (Agarap, 2018), initial weights set to He Uni-
form (He et al., 2015), and an initial bias value set to
0.0001. The output layer has 2 units, one per 2D co-
ordinate, and uses sigmoid activation to constrain out-
put values to [0,1]. We used three different network
sizes, namely, x-small (75, 30, 75 units per layer),
small (150, 60, 150 units per layer) and medium (300,
120, 300 units per layer). We trained the network us-
ing the ADAM optimizer (Kingma and Ba, 2014), as
described in the NNP paper.
Figure 1: Architecture of the SDR-NNP pipeline.
After training, we have a regressor able to mimic
the behavior of SDR for unseen data, thus adding sta-
bility, OOS capability, and computational scalability
to SDR.
4 RESULTS
We measured the performance of SDR-NNP by the
four metrics in Tab. 2 computed for K = 7, in line
with (Maaten and Postma, 2009; Martins et al., 2015;
Espadoto et al., 2019). Note that K, the number
of nearest neighbors used to compute the metrics in
Tab. 2, is smaller than k
s
, the number of nearest met-
rics used to evaluate L (Eqn. 2). Indeed, k
s
needs to be
relatively large to smooth out local noise in the com-
putation of the gradient ∇ρ; in contrast, K is typically
set small to capture more local quality aspects of a
projection.
Evaluation used six publicly available real-
world datasets (Table 4), all being reasonably high-
dimensional and large (tens of dimensions, thousands
of samples), and with a non-trivial data structure. All
dimensions were rescaled to the [0,1] range, to match
NNP’s sigmoid activation function used in its output
layer (Sec. 3). All computed metrics can be found in
the Appendix.
Section 4.1 details the quality of SDR-NNP
trained to mimic SDR in combination with Landmark
MDS (LMDS), PCA, and t-SNE. Section 4.2 studies
the computational scalability of SDR-NNP. Finally,
Section 4.3 presents an application of SDR-NNP to
the analysis of an astronomical dataset.
4.1 Quality on Real-world Datasets
We studied SDR-NNP’s quality with respect to its
parameters (number of iterations T , learning rate α,
training epochs E, and NNP network size) using
LMDS, t-SNE, and PCA as baseline DR methods. A
discussion on the selection of DR methods for SDR
can be found in Sec. 6 from (Kim et al., 2021). For
space reasons we omitted results for all network sizes
other than medium.
Figure 2 shows how the number of iterations T
affects the sharpening of clusters for LMDS and t-
SNE (PCA results omitted for space reasons). For all
datasets, 4 to 8 iterations suffice to have the clusters
sharply defined in the projection. Table 7 in the Ap-
pendix shows quality metrics as functions of T for all
three baseline projections. Increasing T can increase
quality (Air Quality, Reuters with LMDS and PCA)
but generally slightly decreases quality for LMDS
and PCA. For t-SNE, this decrease is visible for all
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
67

Table 4: Datasets used in SDR-NNP’s evaluation.
Dataset name Samples Dimensions Data description
and provenance N n
Air Quality (Vito et al., 2008) 9358 13 Measurements from air sensors used to study and predict air quality
Concrete (Yeh, 1998) 1030 8 Measurements of chemico-physical properties of concrete used to study concrete strength
Reuters (Thoma, 2017) 5000 100 Attributes extracted from news report documents using TF-IDF (Salton and McGill, 1986),
a standard method in text processing. This is a subset of the full dataset which contains data
for the six most frequent classes only. Used to study how features can predict news’ types
(classes)
Spambase (Hopkins et al.,
1999)
4001 57 Text dataset used to train email spam classifiers
Wisconsin (Street et al., 1993) 569 32 Features extracted from images of breast masses used to detect malignant cells
Wine (Cortez et al., 2009) 6497 11 Samples of white and red Portuguese vinho verde used to describe perceived wine quality
No SDR 4 iters 8 iters 12 iters 16 iters 20 iters
Wine Wisconsin Spambase Reuters Concrete Air Quality
(a) LMDS.
No SDR 4 iters 8 iters 12 iters 16 iters 20 iters
Wine Wisconsin Spambase Reuters Concrete Air Quality
(b) t-SNE.
Figure 2: Iteration parameter effect: SDR-NNP learned from LMDS (a) and t-SNE (b) for varying iterations T (columns) and
datasets (rows). Fixed SDR-NNP parameters are α = 0.1, E = 1000 epochs, medium network size.
Table 5: Time measurements for SDR and SDR-NNP (sec-
onds), GALAH dataset. See also Fig. 6.
Samples SDR SDR-NNP inference
1000 0.220 0.108
2000 0.957 0.059
5000 14.799 0.092
10000 157.420 0.149
20000 1302.268 0.414
30000 4736.995 0.355
40000 11058.267 0.727
datasets, which is explainable by the fact that t-SNE
already has a very high quality which is hard to be
learned by NNP (see (Espadoto et al., 2020)). How-
ever, as already argued in (Kim et al., 2021), local
quality metrics will likely decrease when using SDR
to favor visual cluster separation.
Figure 3 shows results for SDR-NNP when vary-
ing the learning rate α for LMDS and t-SNE (PCA
results omitted again for space reasons). Too small
or too large α values tend to affect the scatterplot ad-
versely. Values in the range α ∈ [0.05, 0.1] show the
best results, i.e., a good separation of the projection
into distinct clusters. Table 8 in the Appendix shows
quality metrics as function of α for all three base-
line projections. The effect of α on quality is similar
with that of T with some combinations (Reuters with
LMDS and PCA) showing a slight increase but most
showing a slight decrease for small α values.
Figure 4 shows how the number of training epochs
E affects projection quality. The early stopping strat-
egy proposed in the NNP paper (Espadoto et al.,
2020), which stops training on convergence, defined
as the epoch where the validation loss stops decreas-
ing (roughly E = 60 epochs in practice), does not pro-
duce good results for SDR-NNP – the resulting pro-
jection (Fig. 4a,b leftmost columns) show a fuzzy ver-
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
68
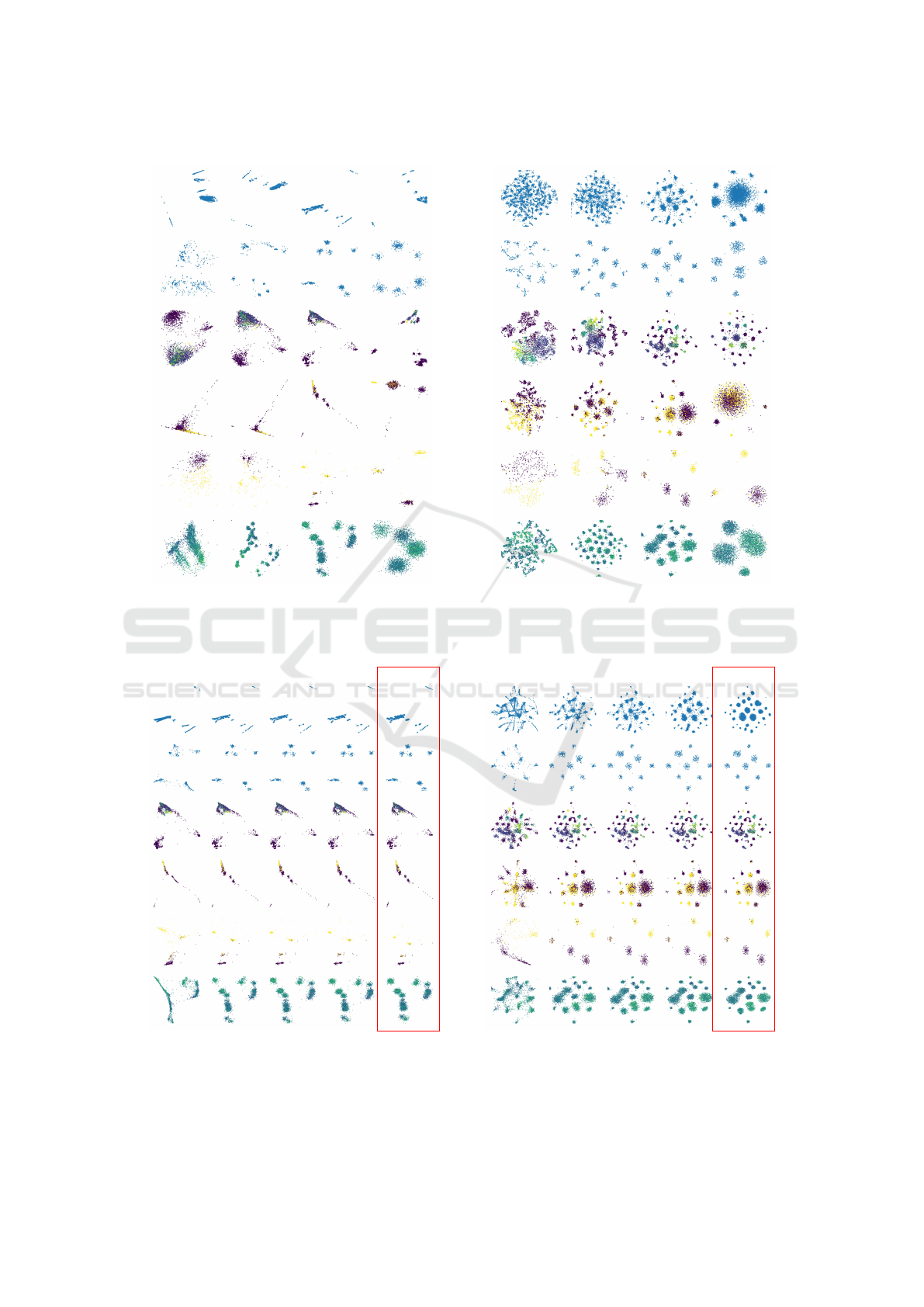
0.01 0.05 0.1 0.2
Wine Wisconsin Spambase Reuters Concrete Air Quality
(a) LMDS.
0.01 0.05 0.1 0.2
Wine Wisconsin Spambase Reuters Concrete Air Quality
(b) t-SNE.
Figure 3: Learning rate effect: SDR-NNP learned from LMDS (a) and t-SNE (b) for varying learning rates α (columns) and
datasets (rows). Fixed SDR-NNP parameters are T = 10 iterations, E = 1000 epochs, medium network size.
Early Stopping 300 ep 1000 ep 3000 ep Training
Wine Wisconsin Spambase Reuters Concrete Air Quality
(a) LMDS.
Early Stopping 300 ep 1000 ep 3000 ep Training
Wine Wisconsin Spambase Reuters Concrete Air Quality
(b) t-SNE.
Figure 4: SDR-NNP learned from LMDS (a) and t-SNE (b) for varying training epochs E (columns) and datasets (rows).
Fixed SDR-NNP parameters are T = 10, α = 0.1, medium network size. Red column shows the training projections.
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
69
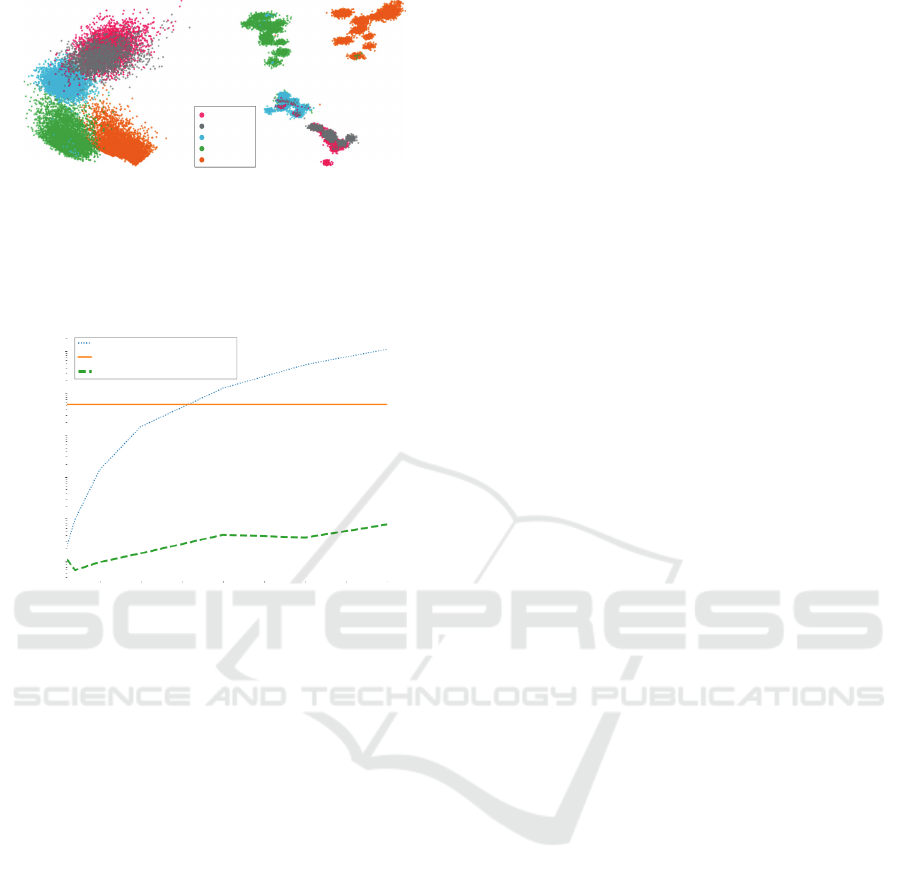
(a) (b)
Running
Dancing
Walking
Standing
Sitting
Figure 5: (a) LMDS and (b) SDR with LMDS baseline
(α = 0.2, T = 10, k
s
= 100) applied to Human Activity Data
(N=24075, n=60), which was originally tested by Kim et
al. (Kim et al., 2021). Points are colored by their labels on
different human activities (sit, stand, walk, run, and dance).
These results demonstrate the advantage of using sharpen-
ing for data that are known to have cluster structures.
number of samples (x1000)
5
10
15
20 25 30 35
40
10
-1
10
0
10
1
10
2
10
3
10
4
time (seconds, log scale)
SDR
SDR-NNP (train 10K + inference)
SDR-NNP (inference only)
Figure 6: Performance of SDR vs SDR-NNP on the
GALAH dataset (time in log scale), 1K to 40K samples.
SDR-NNP trained with 10K samples for 1000 epochs. See
also Tab. 6.
sion of the training projection (Fig. 4a,b rightmost
columns). This is explained by the fact that SDR-
NNP needs to learn both the data sharpening and the
projection P, which needs more effort than learning
just the projection, as NNP did. If we use more train-
ing epochs, Fig. 4 shows that SDR-NNP can repro-
duce the training projection very faithfully. SDR-
NNP produces good results with as little as E = 300
epochs, except for the Air Quality dataset, where
E = 3000 epochs was needed for best results. On av-
erage, E = 1000 epochs produced visually good re-
sults for all datasets and other parameter settings, so
we choose this as a preset value for E.
The projections in Figs. 2–4 deserve some com-
ments. As visible there, varying the T and α pa-
rameters can create artificial oversegmentation – the
appearance of many small clusters in the projection,
which is an artificial cluster separation (CS), see e.g.
Fig. 3b, Reuters, α ≥ 0.1. This effect is strongest,
and undesirable, for baseline projections which al-
ready do have a good CS, such as t-SNE. In contrast,
for projections with a low CS, such as LMDS, artifi-
cial oversegmentation is far less present. Like SDR,
SDR-NNP is best used when combined with baseline
DR methods with a low CS capability.
Separately, as outlined in (Kim et al., 2021), SDR
performs best, and is meant to be used for, datasets
that are known to have cluster structures since sharp-
ening, by construction, will enhance these struc-
tures (Comaniciu and Meer, 2002). SDR-NNP inher-
its these aspects from SDR and, as Fig. 4 shows, can
reproduce SDR highly accurately, and is much faster
(see next Sec. 4.2).
To clarify the above, Fig. 5 shows an example
where SDR shows far better CS compared with a
baseline DR method (LMDS) when applied to real-
world human activity data, which is known to have
distinct and well separated features among different
human motions (sit, stand, walk, run, and dance) (El
Helou, 2020). Even though the points are known to be
well-separated, LMDS produces a relatively low CS
(clusters overlap in Fig. 5a). In contrast, SDR pro-
duces a much higher CS (Fig. 5b), in line with the
ground truth. A second example illustrating the same
point is discussed in detail in Sec. 4.3.
4.2 Computational Scalability
We measured scalability by comparing the execution
time of the original SDR method with SDR-NNP us-
ing samples from the GALAH dataset (described next
in Sec. 4.3) with increasing sizes, namely, 1K, 2K,
5K, 10K, 20K, 30K, and 40K samples. Using more
samples was not practical since SDR already took
over three hours at 40K samples. Figure 6 and Ta-
ble 5 show these figures. For |D
s
| = 10K training
samples and E = 1000 epochs, SDR-NNP takes con-
siderable time to train, i.e., 372.818 seconds (Fig. 6,
orange line). Still, this is already faster than SDR
for 15K samples. In inference mode (after training),
SDR-NNP is orders of magnitude faster than SDR,
taking less than one second to project 40K samples
(Fig. 6, green curve). SDR takes over three hours for
the same data size (Fig. 6, blue curve).
4.3 Case Study: Astronomical Datasets
We applied SDR-NNP to a practical use-case using
real-world astronomical data. We use here the same
subset of 10K samples from the GALactic Archaeol-
ogy with HERMES survey (GALAH DR2) (Buder et
al., 2018) as in Kim et al. to show that our method can
create similar projections to their SDR method. The
original GALAH DR2 dataset consists of various stel-
lar abundance attributes of 342682 stars. Data clean-
ing followed (Kim et al., 2021): first, cross-match
the star ID of GALAH DR2 with Gaia data release
2 (Gaia DR2) to gain additional information on the
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
70
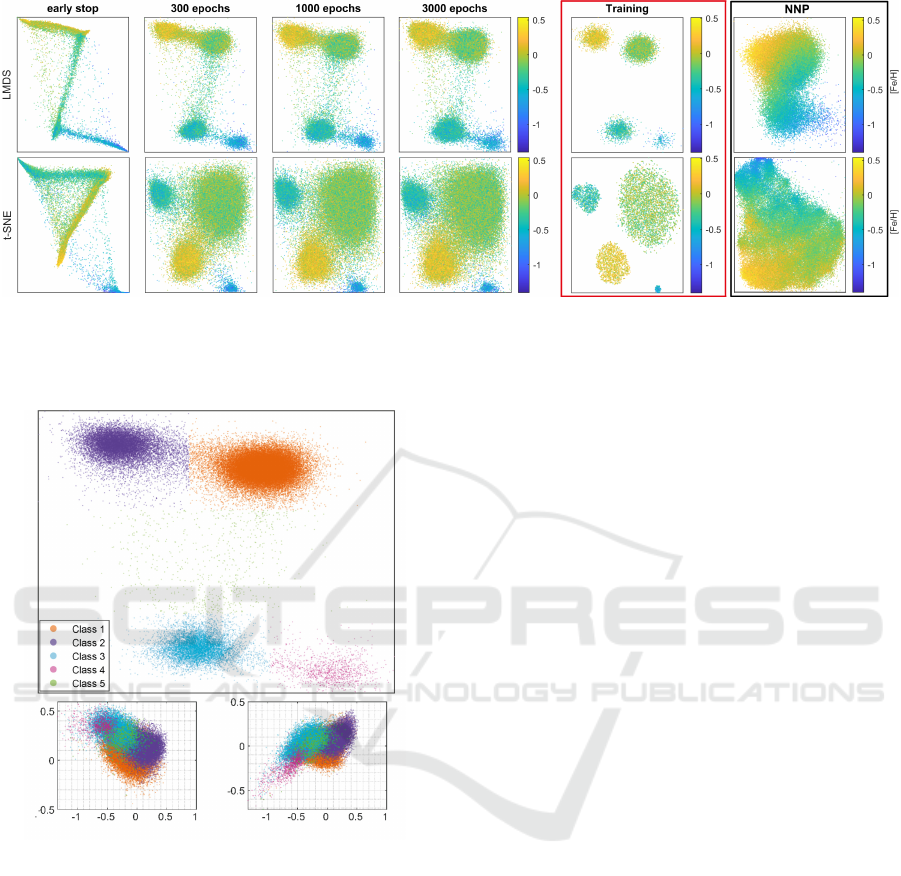
Figure 7: SDR-NNP of 66K samples learned from LMDS (top) and t-SNE (bottom) for different numbers of training epochs
E (four leftmost columns). SDR-NNP parameters are T = 10 iterations, α = 0.18, and medium network size. Red column:
training projection (10K samples). Rightmost column: NNP trained with LMDS and t-SNE instead of SDR applied to the
same test data.
[Mg/Fe]
[Fe/H]
[Cu/Fe]
[Fe/H]
a)
b) c)
Figure 8: Analysis of GALAH DR2 with SDR-NNP
learned from LMDS. (a) Labeling of clusters (classes 1–4)
and outliers (class 5). (b) Tinsley diagram and (c) copper
abundance of stars vs their iron abundance. Astronomers
can infer from (b,c) that class 1 is mostly thin disk stars,
class 2 is mostly metal-rich thick disk stars, classes 5 and
3 are normal thick disk stars, and class 4 is the Gaia Ence-
ladus (GES) in the Milky Way.
stellar kinematics (i.e., 6D phase-space coordinates–
x, y, z, u, v, and w) (Buder et al., 2018; Gaia Collab-
oration, 2016; Gaia Collaboration, 2018); next, ex-
clude stars with implausible values (exceeding 25K
parsec in x, y, and z attributes), having unreliable stel-
lar abundances, or have missing values in any dimen-
sion. From the remaining 76270 samples after pre-
processing, we took the same subset D of 10K stars as
in (Kim et al., 2021), where stars were randomly se-
lected, and compute SDR and the training projection
with the same α = 0.18 (see Sec. 3.1). We trained
SDR-NNP on these 10K stars and used the trained
network to project the remaining 66270 samples.
Figure 7 shows SDR-NNP applied to the 66K test
data with LMDS and t-SNE as baseline DR methods.
Points are colored based on the value of the attribute
[Fe/H], which is of interest to domain experts to ex-
plain possible data clusters. The first four columns
show the SDR-NNP results for varying training epoch
counts E. The red column shows the training projec-
tion P(D
s
) of 10K samples. We see that the structure
of the training projection (four clusters) is well re-
flected by the SDR-NNP results from E = 300 epochs
onwards. The test projections are more fuzzy, but
this is expected, as these contain 66K unseen sam-
ples which, albeit drawn from the same dataset, can-
not perfectly match the four clusters determined by
the 10K training samples. The rightmost column in
Fig. 7 shows the result of the ‘raw’ NNP method,
i.e., trained to imitate LMDS, and t-SNE, without the
sharpening step of SDR, respectively. These results
show clearly far less cluster separation (CS) than ei-
ther the SDR-NNP training projection (red column)
or the inferred SDR-NNP projections (leftmost four
columns). This demonstrates the added value of the
sharpening step: without it, NNP, albeit fast and
OOS-capable, cannot produce useful projections. Ta-
ble 6 in the Appendix shows quality metrics corre-
sponding to the images in Fig. 7 which support the
above observations.
The fact that SDR-NNP shows a good cluster sep-
aration allows astronomers to easily label clusters and
perform further analysis to infer the physical meaning
of stars. To demonstrate this, we manually labeled the
clusters from the SDR-NNP plot learned from LMDS
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
71

to reproduce the same analyses made by Kim et al.
(Fig. 10 in (Kim et al., 2021)) to understand the ori-
gin and location of the stars in each cluster. Figure 8a
shows the manually labeled clusters by one of the au-
thors (astronomy expert). Stars from class 5 are sepa-
rately labeled as outliers. Figures 8b,c are the Tinsley
diagram (Tinsley, 1980) and the copper abundance of
the stars – a tracer of supernovae type 1a – as a func-
tion of their iron abundance, respectively. From these
plots, astronomers are able to identify class-1 stars as
thin-disk stars, class-2 stars as metal-rich thick disk
stars, class-3 and class-5 (outlier) stars as the normal
thick disk stars, and class-4 stars as Gaia Enceladus
(GES) – a group of stars that originated from a galaxy
that merged with the Milky Way several billions years
ago. Importantly, the original SDR method was not
able to perform this analysis and identify class-4 stars
since it could not be applied to the entire dataset due
to its prohibitively low speed.
4.4 Implementation Details
All experiments were run on a dual 16-core Intel
Xeon Silver 4216 at 2.1 GHz with 256 GB RAM
and an NVidia GeForce GTX 1080 Ti GPU with 11
GB VRAM. SDR was implemented in C++ using
Eigen (Guennebaud et al., 2010) for matrix computa-
tions, Nanoflann (Blanco and Rai, 2014) for nearest-
neighbor search, and the implementations of t-SNE
and Landmark MDS from Tapkee (Lisitsyn et al.,
2013). NNP is implemented using the Keras frame-
work (Chollet and others, 2015). The SDR-NNP
code, datasets, and all results discussed above are
publicly available at (Kim et al., 2021a).
5 DISCUSSION
We discuss how SDR-NNP performs with respect to
the criteria laid out in Sec. 1.
Quality (C1): SDR-NNP is able to create projections
which are very similar visually, but also in terms of
quality metrics, to those created by SDR. Importantly,
the strong separation of similar-valued samples, the
key property that SDR promoted, is retained by SDR-
NNP. Combined with properties C2–C4 (which SDR
does not have), this makes SDR-NNP superior to
SDR. Compared to NNP used on the unsharpened
data (Fig. 7), SDR-NNP shows significantly better
cluster separation, which makes it superior to NNP.
Scalability (C2): SDR-NNP is faster than SDR alone
from roughly 15K samples onwards, even when con-
sidering training time. In inference mode (after train-
ing), SDR-NNP is several orders of magnitude faster
than SDR, being able to project tens of thousands of
observations in under a second on a high-end PC. Im-
portantly, SDR-NNP’s speed is linear in the number
of dimensions and samples (a property inherited from
the NNP architecture), and can handle samples in a
streaming fashion, one at a time, i.e, does not need
to hold the entire high-dimensional dataset in mem-
ory. This makes SDR-NNP scalable to large datasets
of millions of samples.
Ease of Use (C3): Once trained, SDR-NNP is
parameter-free. The influence of its hyperparame-
ters T (sharpening iterations), E (number of training
epochs), and α (learning rate) is detailed in Sec. 4.1.
The preset T = 10,E = 1000,α ' 0.2 was shown
to give good results for the entire range of tested
datasets.
Genericity (C4): SDR-NNP is agnostic to the nature
and dimensionality of the input data, being able to
project any dataset having quantitative variables. Ta-
bles 6, 7, and 8 in the Appendix show that SDR-NNP
achieves high quality on datasets of different nature
and coming from a wide range of application domains
(air sensors, civil engineering, text mining, imaging,
and chemistry).
Stability and Out-of-Sample Support (C5): SDR-
NNP inherits the stability and OOS support of NNP,
making it possible to train on a small subset of a
given dataset and then stably project the remaining
data drawn from the same distribution.
Limitations: While inheriting the abovementioned
desirable properties from NNP, SDR-NNP also inher-
its some of its limitations. Its OOS support cannot ex-
tend to datasets of a completely different nature than
those it was trained on – arguably, a limitation that
most machine learning methods have. Also, SDR-
NNP is only as good as the baseline projection P that
was used in the SDR phase. Using a poor quality pro-
jection leads to SDR-NNP learning, and reproducing,
that behavior. Separately, SDR-NNP learns to imi-
tate the sharpening behavior of SDR. While this is of
added value in identifying data structure in terms of
visual structure, as shown by the results in Sec. 4, ap-
plying SDR-NNP on a dataset with little or no clus-
ter structure can create artificial visual oversegmen-
tation in the projection (see Sec. 4.1). The recom-
mended parameter values to prevent oversegmenta-
tion using SDR are discussed further in Sec. 6.5 in
(Kim et al., 2021). Specifying the right amount of
sharpening is dataset- and problem-dependent, thus
considered as future work.
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
72

6 CONCLUSION
We have presented SDR-NNP, a new method for com-
puting projections of high-dimensional datasets for
visual exploration purposes. Our method joins sev-
eral desirable, and complementary, characteristics of
two earlier projection methods, namely NNP (speed,
ease of use, out-of-sample support, ability to imitate
a wide range of existing projection techniques with
a high quality) and SDR (ability to segregate projec-
tions of complex datasets into visually separated clus-
ters of similar observations). In particular, SDR-NNP
removes the main obstacle for practical usage of SDR,
namely, its prohibitive computational time. We have
demonstrated SDR-NNP on a range of datasets com-
ing from different application domains. In particu-
lar, we showed how SDR-NNP can bring added value
in the exploration of a large and recent astronomical
dataset leading to findings which were not achievable
by SDR or NNP alone.
Future work can target several directions. As
SDR-NNP showed that it is possible to learn sharp-
ening methods for high-dimensional data, it is inter-
esting to apply it to other domains beyond projection
where such methods are used, e.g., image segmenta-
tion, graph bundling, or data clustering and simpli-
fication. For the projection use-case, refining SDR-
NNP’s network architecture to accelerate its training
is of high practical interest. Finally, deploying SDR-
NNP as a daily tool for astronomers to analyze their
million-sample datasets is a goal we want to pursue in
the short term.
ACKNOWLEDGMENTS
This work is supported by the DSSC Doctoral Train-
ing Programme co-funded by the Marie Sklodowska-
Curie COFUND project (DSSC 754315), and
FAPESP grant 2020/13275-1, Brazil. The GALAH
survey is based on observations made at the
Australian Astronomical Observatory, under pro-
grammes A/2013B/13, A/2014A/25, A/2015A/19,
A/2017A/18. We acknowledge the traditional
owners of the land on which the AAT stands, the
Gamilaraay people, and pay our respects to elders
past and present. This work has made use of data
from the European Space Agency (ESA) mission
Gaia (https://www.cosmos.esa.int/gaia), processed
by the Gaia Data Processing and Analysis Con-
sortium (DPAC, https://www.cosmos.esa.int/web/
gaia/dpac/consortium). Funding for the DPAC has
been provided by national institutions, in particu-
lar the institutions participating in the Gaia Multilat-
eral Agreement.
REFERENCES
Agarap, A. F. (2018). Deep learning using rectified linear
units (ReLU). arXiv:1803.08375 [cs.NE].
Becker, M., Lippel, J., Stuhlsatz, A., and Zielke, T. (2020).
Robust dimensionality reduction for data visualiza-
tion with deep neural networks. Graphical Models,
108:101060.
Becker, R., Cleveland, W., and Shyu, M. (1996). The visual
design and control of trellis display. J Comp Graph
Stat, 5(2):123–155.
Behrisch, M., Blumenschein, M., Kim, N. W., Shao, L., El-
Assady, M., Fuchs, J., Seebacher, D., Diehl, A., Bran-
des, U., Pfister, H., Schreck, T., Weiskopf, D., and
Keim, D. A. (2018). Quality metrics for information
visualization. Comp Graph Forum, 37(3):625–662.
Berkhin, P. (2006). A survey of clustering data mining tech-
niques. In Grouping Multidimensional Data, pages
25–71. Springer.
Blanco, J. L. and Rai, P. K. (2014). nanoflann: a C++
header-only fork of FLANN, a library for nearest
neighbor (NN) with kd-trees. https://github.com/
jlblancoc/nanoflann.
Buder et al., S. (2018). The GALAH Survey: Second data
release. Mon R R Astron Soc, 478.
Chan, D., Rao, R., Huang, F., and Canny, J. (2018). T-SNE-
CUDA: GPU-accelerated t-SNE and its applications
to modern data. In Proc. SBAC-PAD, pages 330–338.
Cheng, Y. (1995). Mean shift, mode seeking, and clustering.
IEEE TPAMI, 17(8):790–799.
Chollet, F. and others (2015). Keras. https://keras.io.
Comaniciu, D. and Meer, P. (2002). Mean shift: A robust
approach toward feature space analysis. IEEE TPAMI,
24(5):603–619.
Cortez, P., Cerdeira, A., Almeida, F., Matos, T., and Reis,
J. (2009). Modeling wine preferences by data mining
from physicochemical properties. Decis Support Sys,
47(4):547–553.
Cunningham, J. and Ghahramani, Z. (2015). Linear dimen-
sionality reduction: Survey, insights, and generaliza-
tions. JMLR, 16:2859–2900.
De Silva, V. and Tenenbaum, J. B. (2004). Sparse multidi-
mensional scaling using landmark points. Technical
report, Stanford University.
Donoho, D. L. and Grimes, C. (2003). Hessian eigen-
maps: Locally linear embedding techniques for high-
dimensional data. PNAS, 100(10):5591–5596.
El Helou, A. (2020). Sensor HAR recognition app.
www.mathworks.com/matlabcentral/fileexchange/
54138-sensor-har-recognition-app.
Engel, D., Hattenberger, L., and Hamann, B. (2012). A
survey of dimension reduction methods for high-
dimensional data analysis and visualization. In Proc.
IRTG Workshop, volume 27, pages 135–149. Schloss
Dagstuhl.
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
73

Epanechnikov, V. (1969). Non-parametric estimation of a
multivariate probability density. Theor Probab Appl+,
14.
Espadoto, M., Hirata, N., and Telea, A. (2020). Deep
learning multidimensional projections. Inform Visual,
9(3):247–269.
Espadoto, M., Hirata, N., and Telea, A. (2021). Self-
supervised dimensionality reduction with neural net-
works and pseudo-labeling. In Proc. IVAPP.
Espadoto, M., Martins, R., Kerren, A., Hirata, N., and
Telea, A. (2019). Toward a quantitative survey
of dimension reduction techniques. IEEE TVCG,
27(3):2153–2173.
Fisher, R. A. (1936). The use of multiple measurements in
taxonomic problems. Annals of eugenics, 7(2):179–
188.
Fukunaga, K. and Hostetler, L. (1975). The estimation of
the gradient of a density function, with applications in
pattern recognition. IEEE Trans Inf Theor, 21(1):32–
40.
Gaia Collaboration (2016). The Gaia mission. Astronomy
& Astrophysics, 595, A1.
Gaia Collaboration (2018). Gaia Data Release 2-Summary
of the contents and survey properties. Astronomy &
Astrophysics, 616, A1.
Guennebaud, G., Jacob, B., et al. (2010). Eigen v3.
http://eigen.tuxfamily.org.
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Delving
deep into rectifiers: Surpassing human-level perfor-
mance on ImageNet classification. In Proc. ICCV,
pages 1026–1034.
Hinton, G. E. and Salakhutdinov, R. R. (2006). Reducing
the dimensionality of data with neural networks. Sci-
ence, 313(5786):504–507. Publisher: AAAS.
Hoffman, P. and Grinstein, G. (2002). A survey of visual-
izations for high-dimensional data mining. In Infor-
mation Visualization in Data Mining and Knowledge
Discovery, pages 47–82.
Hopkins, M., Reeber, E., Forman, G., and Suermondt, J.
(1999). Spambase dataset. Hewlett-Packard Labs.
Hurter, C., Ersoy, O., and Telea, A. (2012). Graph bundling
by kernel density estimation. Comp Graph Forum,
31(3):865–874.
Inselberg, A. and Dimsdale, B. (1990). Parallel coordinates:
A tool for visualizing multi-dimensional geometry. In
Proc. IEEE Visualization, pages 361–378.
Joia, P., Coimbra, D., Cuminato, J. A., Paulovich, F. V., and
Nonato, L. G. (2011). Local affine multidimensional
projection. IEEE TVCG, 17(12):2563–2571.
Jolliffe, I. T. (1986). Principal component analysis and fac-
tor analysis. In Principal Component Analysis, pages
115–128. Springer.
Kim, Y., Telea, A., Trager, S., and Roerdink, J. B.
T. M. (2021). Visual cluster separation using
high-dimensional sharpened dimensionality reduc-
tion. arXiv:2110.00317 [cs.CV].
Kim, Y., Espadoto, M., Trager, S. C., Roerdink, J. B. T. M.,
and Telea, A. C. (2021a). SDR-NNP implementation
and results. https://github.com/youngjookim/sdr-nnp.
Kingma, D. P. and Ba, J. (2014). Adam: A method for
stochastic optimization. arXiv:1412.6980.
Kingma, D. P. and Welling, M. (2013). Auto-encoding
variational bayes. CoRR, abs/1312.6114. eprint:
1312.6114.
Kohonen, T. (1997). Self-organizing Maps. Springer.
LeCun, Y. and Cortes, C. (2010). MNIST handwritten digits
dataset. http://yann.lecun.com/exdb/mnist.
Lisitsyn, S., Widmer, C., and Garcia, F. J. I. (2013). Tap-
kee: An efficient dimension reduction library. JMLR,
14:2355–2359.
Liu, S., Maljovec, D., Wang, B., Bremer, P.-T., and
Pascucci, V. (2015). Visualizing high-dimensional
data: Advances in the past decade. IEEE TVCG,
23(3):1249–1268.
Maaten, L. v. d. (2014). Accelerating t-SNE using tree-
based algorithms. JMLR, 15:3221–3245.
Maaten, L. v. d. and Hinton, G. (2008). Visualizing data
using t-SNE. JMLR, 9:2579–2605.
Maaten, L. v. d. and Postma, E. (2009). Dimensionality
reduction: A comparative review. Technical report,
Tilburg Univ.
Martins, R. M., Minghim, R., Telea, A. C., and others
(2015). Explaining neighborhood preservation for
multidimensional projections. In Proc. CGVC, pages
7–14.
McInnes, L. and Healy, J. (2018). UMAP: Uniform man-
ifold approximation and projection for dimension re-
duction. arXiv:1802.03426v1 [stat.ML].
Nonato, L. and Aupetit, M. (2018). Multidimensional
projection for visual analytics: Linking techniques
with distortions, tasks, and layout enrichment. IEEE
TVCG.
Paulovich, F. V. and Minghim, R. (2006). Text map ex-
plorer: a tool to create and explore document maps.
In Proc. Information Visualisation, pages 245–251.
IEEE.
Paulovich, F. V., Nonato, L. G., Minghim, R., and Lev-
kowitz, H. (2008). Least square projection: A fast
high-precision multidimensional projection technique
and its application to document mapping. IEEE
TVCG, 14(3):564–575.
Pezzotti, N., H
¨
ollt, T., Lelieveldt, B., Eisemann, E., and
Vilanova, A. (2016). Hierarchical stochastic neighbor
embedding. Comp Graph Forum, 35(3):21–30.
Pezzotti, N., Lelieveldt, B., Maaten, L. v. d., H
¨
ollt, T., Eise-
mann, E., and Vilanova, A. (2017). Approximated and
user steerable t-SNE for progressive visual analytics.
IEEE TVCG, 23:1739–1752.
Pezzotti, N., Thijssen, J., Mordvintsev, A., Hollt, T., Lew,
B. v., Lelieveldt, B., Eisemann, E., and Vilanova, A.
(2020). GPGPU linear complexity t-SNE optimiza-
tion. IEEE TVCG, 26(1):1172–1181.
Rao, R. and Card, S. K. (1994). The table lens: Merging
graphical and symbolic representations in an interac-
tive focus+context visualization for tabular informa-
tion. In Proc. ACM SIGCHI, pages 318–322.
Rauber, P. E., Falc
˜
ao, A. X., and Telea, A. C. (2017). Pro-
jections as visual aids for classification system design.
Inform Visual, 17(4):282–305.
Roweis, S. T. and Saul, L. L. K. (2000). Nonlinear dimen-
sionality reduction by locally linear embedding. Sci-
ence, 290(5500):2323–2326.
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
74

Salton, G. and McGill, M. J. (1986). Introduction to modern
information retrieval. McGraw-Hill.
Sorzano, C., Vargas, J., and Pascual-Montano, A. (2014).
A survey of dimensionality reduction techniques.
arXiv:1403.2877 [stat.ML].
Street, N., Wolberg, W., and Mangasarian, O. (1993). Nu-
clear feature extraction for breast tumor diagnosis. In
Biomedical image processing and biomedical visual-
ization, volume 1905, pages 861–870.
Tenenbaum, J. B., Silva, V. D., and Langford, J. C. (2000).
A global geometric framework for nonlinear dimen-
sionality reduction. Science, 290(5500):2319–2323.
Thoma, M. (2017). The Reuters dataset. https://
martin-thoma.com/nlp-reuters.
Tinsley, B. (1980). Evolution of the stars and gas in galax-
ies. Fundamentals of Cosmic Physics, 5:287–388.
Torgerson, W. (1958). Theory and Methods of Scaling. Wi-
ley.
Venna, J. and Kaski, S. (2006). Visualizing gene interaction
graphs with local multidimensional scaling. In Proc.
ESANN, pages 557–562.
Vito, S. D., Massera, E., Piga, M., Martinotto, L., and Fran-
cia, G. D. (2008). On field calibration of an elec-
tronic nose for benzene estimation in an urban pol-
lution monitoring scenario. Sensors and Actuators
B: Chemical, 129(2):750–757. https://archive.ics.uci.
edu/ml/datasets/Air+Quality.
Wattenberg, M. (2016). How to use t-SNE effectively. https:
//distill.pub/2016/misread-tsne.
Xie, H., Li, J., and Xue, H. (2017). A survey of dimen-
sionality reduction techniques based on random pro-
jection. arXiv:1706.04371 [cs.LG].
Xu, R. and Wunsch, D. (2005). Survey of clustering al-
gorithms. IEEE Transactions on Neural Networks,
16(3):645–678.
Yates, A., Webb, A., Sharpnack, M., Chamberlin, H.,
Huang, K., and Machiraju, R. (2014). Visualizing
multidimensional data with glyph SPLOMs. Comp
Graph Forum, 33(3):301–310.
Yeh, I.-C. (1998). Modeling of strength of high-
performance concrete using artificial neural networks.
Cement and Concrete Research, 28(12):1797–1808.
Zhang, Z. and Wang, J. (2007). MLLE: Modified locally
linear embedding using multiple weights. In Proc.
NIPS, pages 1593–1600.
Zhang, Z. and Zha, H. (2004). Principal manifolds and
nonlinear dimensionality reduction via tangent space
alignment. SIAM J Sci Comput, 26(1):313–338.
SDR-NNP: Sharpened Dimensionality Reduction with Neural Networks
75
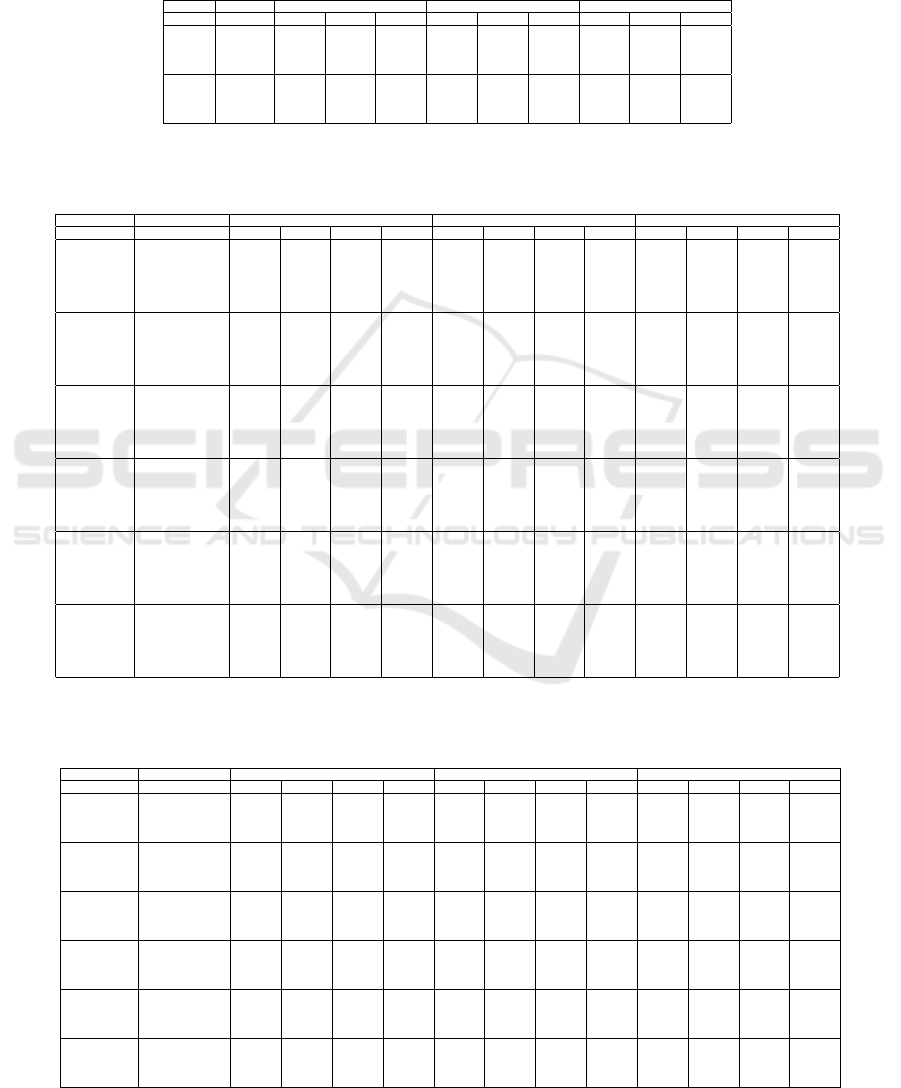
APPENDIX
We next present tables containing full measurements
for the experiments described in Section 4.
Table 6: Metrics for SDR-NNP learned from LMDS, PCA, and t-SNE on the GALAH dataset for train and test samples for
varying number of training epochs E (‘early’ indicates the early-stopping heuristic).
LMDS PCA t-SNE
Mode Epochs T C R T C R T C R
Train
early 0.802 0.877 0.676 0.787 0.853 0.668 0.789 0.870 0.638
300 0.703 0.790 0.629 0.697 0.778 0.617 0.694 0.770 0.496
1000 0.695 0.772 0.615 0.693 0.762 0.607 0.692 0.748 0.481
3000 0.692 0.756 0.600 0.693 0.756 0.601 0.691 0.734 0.447
Test
early 0.775 0.862 0.601 0.754 0.828 0.592 0.756 0.853 0.574
300 0.667 0.768 0.558 0.660 0.759 0.544 0.652 0.755 0.470
1000 0.661 0.749 0.547 0.658 0.746 0.539 0.642 0.734 0.449
3000 0.657 0.737 0.534 0.655 0.739 0.539 0.641 0.720 0.416
Table 7: Metrics for SDR-NNP learned from LMDS, PCA, and t-SNE for different numbers of iterations T and different
datasets. SDR-NNP parameters used: α = 0.1, E = 1000, medium network size. NH values not available for the Air Quality
and Concrete datasets since these are not labeled.
LMDS PCA t-SNE
Dataset SDR iterations T C R NH T C R NH T C R NH
Air Quality
0 0.941 0.992 0.970 0.940 0.992 0.966 0.996 0.996 0.654
4 0.962 0.979 0.963 0.956 0.979 0.963 0.951 0.939 0.396
8 0.954 0.970 0.952 0.942 0.970 0.948 0.945 0.938 0.313
12 0.949 0.970 0.952 0.945 0.970 0.936 0.942 0.942 0.365
16 0.950 0.968 0.943 0.948 0.967 0.930 0.940 0.933 0.317
20 0.954 0.968 0.939 0.949 0.967 0.917 0.943 0.939 0.334
Concrete
0 0.940 0.979 0.744 0.934 0.977 0.736 0.996 0.992 0.479
4 0.938 0.958 0.631 0.934 0.957 0.627 0.952 0.929 0.145
8 0.912 0.944 0.560 0.906 0.943 0.564 0.927 0.918 0.140
12 0.895 0.941 0.556 0.865 0.932 0.558 0.912 0.913 0.167
16 0.884 0.938 0.554 0.872 0.932 0.555 0.904 0.914 0.118
20 0.876 0.935 0.560 0.874 0.934 0.569 0.890 0.910 0.109
Reuters
0 0.817 0.895 0.755 0.724 0.817 0.888 0.754 0.727 0.956 0.960 0.609 0.856
4 0.835 0.913 0.752 0.747 0.833 0.901 0.745 0.743 0.957 0.951 0.405 0.855
8 0.858 0.915 0.713 0.775 0.854 0.906 0.706 0.765 0.950 0.910 0.258 0.845
12 0.883 0.909 0.693 0.803 0.883 0.904 0.687 0.802 0.915 0.855 0.078 0.820
16 0.884 0.910 0.691 0.810 0.882 0.907 0.689 0.804 0.893 0.849 0.102 0.813
20 0.882 0.907 0.691 0.800 0.883 0.907 0.691 0.810 0.893 0.852 0.113 0.820
Spambase
0 0.740 0.909 0.529 0.852 0.747 0.912 0.513 0.849 0.954 0.958 0.408 0.914
4 0.737 0.881 0.463 0.843 0.743 0.877 0.471 0.841 0.873 0.899 0.294 0.882
8 0.723 0.855 0.403 0.838 0.712 0.848 0.383 0.834 0.793 0.845 0.312 0.866
12 0.711 0.845 0.379 0.829 0.704 0.838 0.348 0.830 0.754 0.841 0.324 0.850
16 0.701 0.837 0.370 0.828 0.701 0.837 0.322 0.830 0.744 0.834 0.332 0.845
20 0.710 0.840 0.351 0.833 0.709 0.838 0.311 0.836 0.739 0.828 0.283 0.848
Wisconsin
0 0.895 0.959 0.926 0.941 0.896 0.959 0.928 0.943 0.950 0.939 0.679 0.976
4 0.892 0.915 0.901 0.953 0.888 0.915 0.903 0.958 0.897 0.878 0.557 0.957
8 0.804 0.857 0.785 0.925 0.805 0.856 0.785 0.925 0.814 0.816 0.256 0.925
12 0.790 0.849 0.735 0.930 0.787 0.848 0.736 0.930 0.794 0.805 0.393 0.932
16 0.780 0.847 0.721 0.916 0.780 0.844 0.718 0.922 0.779 0.820 0.432 0.913
20 0.775 0.842 0.707 0.922 0.776 0.841 0.705 0.920 0.778 0.829 0.457 0.921
Wine
0 0.864 0.973 0.839 0.667 0.869 0.972 0.806 0.678 0.986 0.976 0.656 0.702
4 0.867 0.932 0.709 0.669 0.864 0.930 0.686 0.665 0.911 0.894 0.341 0.673
8 0.843 0.916 0.676 0.661 0.843 0.917 0.683 0.661 0.869 0.876 0.283 0.665
12 0.840 0.904 0.646 0.665 0.841 0.905 0.653 0.668 0.846 0.864 0.289 0.668
16 0.845 0.901 0.625 0.664 0.843 0.903 0.635 0.663 0.845 0.865 0.321 0.664
20 0.842 0.899 0.579 0.659 0.846 0.898 0.593 0.664 0.845 0.868 0.292 0.666
Table 8: Metrics for SDR-NNP learned from LMDS, PCA, and t-SNE for different learning rates α. SDR-NNP parameters:
T = 10 iterations, E = 1000 epochs, medium network. NH values not available for the Air Quality and Concrete datasets
since these are not labeled.
LMDS PCA t-SNE
Dataset Learning Rate T C R NH T C R NH T C R NH
Air Quality
0.01 0.971 0.990 0.969 0.968 0.990 0.964 0.958 0.926 0.052
0.05 0.976 0.983 0.963 0.973 0.984 0.964 0.938 0.911 0.175
0.1 0.951 0.969 0.948 0.940 0.969 0.941 0.943 0.939 0.378
0.2 0.866 0.941 0.911 0.862 0.928 0.905 0.824 0.876 0.635
Concrete
0.01 0.959 0.983 0.731 0.950 0.979 0.721 0.994 0.988 0.486
0.05 0.932 0.957 0.601 0.929 0.953 0.601 0.947 0.929 0.219
0.1 0.870 0.933 0.578 0.889 0.935 0.583 0.913 0.918 0.057
0.2 0.858 0.920 0.540 0.859 0.921 0.542 0.857 0.900 0.223
Reuters
0.01 0.822 0.900 0.758 0.729 0.821 0.892 0.755 0.730 0.956 0.960 0.608 0.853
0.05 0.839 0.913 0.737 0.745 0.838 0.903 0.735 0.745 0.955 0.949 0.386 0.849
0.1 0.870 0.910 0.698 0.783 0.871 0.905 0.693 0.790 0.936 0.885 0.159 0.829
0.2 0.866 0.902 0.700 0.784 0.867 0.903 0.693 0.785 0.890 0.848 0.096 0.815
Spambase
0.01 0.755 0.911 0.527 0.860 0.756 0.915 0.523 0.852 0.958 0.942 0.383 0.905
0.05 0.775 0.893 0.415 0.840 0.787 0.894 0.426 0.858 0.851 0.874 0.261 0.874
0.1 0.712 0.843 0.380 0.832 0.704 0.839 0.367 0.828 0.769 0.848 0.341 0.863
0.2 0.604 0.667 0.265 0.760 0.606 0.671 0.263 0.764 0.635 0.676 0.317 0.802
Wisconsin
0.01 0.900 0.960 0.932 0.947 0.898 0.960 0.932 0.949 0.955 0.941 0.635 0.966
0.05 0.868 0.885 0.869 0.946 0.874 0.890 0.870 0.941 0.876 0.861 0.597 0.948
0.1 0.803 0.856 0.757 0.928 0.800 0.851 0.753 0.929 0.802 0.841 0.494 0.927
0.2 0.717 0.764 0.693 0.905 0.718 0.758 0.684 0.918 0.725 0.749 0.596 0.909
Wine
0.01 0.895 0.972 0.811 0.674 0.898 0.971 0.783 0.681 0.983 0.951 0.448 0.696
0.05 0.914 0.944 0.734 0.671 0.920 0.945 0.714 0.670 0.927 0.862 0.135 0.670
0.1 0.837 0.913 0.672 0.658 0.848 0.913 0.672 0.664 0.864 0.882 0.250 0.661
0.2 0.739 0.821 0.479 0.653 0.742 0.825 0.484 0.644 0.744 0.808 0.404 0.646
IVAPP 2022 - 13th International Conference on Information Visualization Theory and Applications
76
