
Adopting CRISPR-mediated Genomic Editing Technique on the
Treatment of Lung Cancer: Using Revolutionary Genomic Editing
Technique to Treat Serious Human Disease
Sixian Chen
a
Beijing Huijia Private School, Beijing China
Keywords: CRISPR, Lung Cancer, EGFR Gene, EML4-ALK Fusion Gene, TSLC1, P107 and P130.
Abstract: CRISPR system was first discovered in E. coli in 1987 by Japanese scientist Yoshizumi Ishino and his team.
Since its emergence, it has been widely applied for a variety of medical use, including lung cancer. Traditional
treatments of lung cancer gradually lost in their effectiveness by inducing drug resitance. Therefore, more
influencing and innovative technologies are needed urgently, and CRISPR-mediated genomic editing
technique is one of them. CRISPR system helps scientist to construct tumor model, to identify certain lung
cancer related genes as well as deleting or repairing those cancer genes which can be done by targeting specific
genes and either inhibit or activate its function. The applications of CRISPR system are now developed in a
flying speed that it already moves to the clinical testing stage. Ideally, within a decade, the adoption of
CRISPR system on treating lung cancer can be popularized widely. This review summarizes the
characteristics of lung cancer and the application of CRISPR system on treating lung cancer. Potential target
genes related with lung cancer will be discussed including EFGR, EML4 fusion gene, TSLC1 etc. The
application of CRISPR system on deleting various types of cancer gene will be introduced, too. In order to
generalize CRISPR technology into human cases, more in-depth investigations of the usage of this system are
necessary for future studies.
1 INTRODUCTION
Lung cancer is a common type of cancer happened in
lungs that leads to roughly 25% of death among all
cancer cases. Its morbidity rate is ranked at the top
level among a lot of developing countries including
China, Europe and so on. There’re mainly two types
of lung cancer – non-small cell lung cancer (NSCLC)
and small cell lung cancer (SCLC). NSCLC takes up
approximately 80% - 85% of all lung cancer cases
(National Comprehensive Cancer Network). There’re
a variety of subtypes within NSCLC including
adenocarcinoma, large cell carcinoma, squamous cell
carcinoma and so on. Although each of them damages
different parts of the lungs, they’re collectively
categorized as non-small cell lung cancer since they
possess similar treatment as well as prognoses. On the
other hand, SCLC, or oat cell cancer occupies 10% -
15% lung cancer cases. It generally grows in a more
rapid rate compared with NSCLC. The metastatic
a
https://orcid.org/0000-0003-4132-7707
speed is also faster in SCLC patients that by the time
the cancer is discovered, it has already diverted to
other organs or tissues. It’s also possible for tumor
cells to be metastasized to the lungs from other parts
of the body, but in this case, the tumor will be named
after the primary cancer site instead of lung cancer
(Niederhuber et al. 2020).
Due to its low surviving rate, the treatment of lung
cancer is always the focus of clinical research. The
pathology of lung cancer is mostly due to the
mutation of certain genes. This mainly includes the
transformation of proto-oncogene into oncogene and
the inactivation of tumor suppressor gene. For the
past decade, people have tried a huge number of
therapeutic pathways to treat lung cancer,
chemotherapy is one of the most common fashion.
However, despite the various types of chemotherapy,
many patients developed drug resistance after
receiving chemotherapy which lower the efficiency
of the treatment significantly. Traditional way of
168
Chen, S.
Adopting CRISPR-mediated Genomic Editing Technique on the Treatment of Lung Cancer: Using Revolutionary Genomic Editing Technique to Treat Serious Human Disease.
DOI: 10.5220/0011228700003444
In Proceedings of the 2nd Conference on Artificial Intelligence and Healthcare (CAIH 2021), pages 168-174
ISBN: 978-989-758-594-4
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
therapy is already behind the times. To invent new
creative way of treatment, it’s crucial to understand
the causes, mechanisms, and pathologies of lung
cancer. Among all possible reasons, genetic mutation
is considered as the most essential cause of lung
cancer. There’re numerous types of gene that might
induce lung cancer after being either activated or
inhibited. Considering the significant role of mutated
genes play in lung cancer, scientists start to
investigate cancer gene therapy, a therapy that
specifically designed for editing human genome.
Cancer gene therapy is a broad concept that
include any treatment related with active cancer
genes. In the past few years, using gene editing
technologies to treat lung cancer has taken a lot of
researchers’ attentions. CRISPR system is one of
them. CRISPR system is an efficient tool used to edit
the genome of cells. The essential components of this
system include single-guide RNA (sgRNA) and
nuclease in which sgRNA guide the nuclease to cause
a DNA cleavage at the targeted site. This enables
researchers to edit the removed site, normally by
introducing a DNA repair. There are several other
types of gene editing technologies including zinc-
finger endonucleases (ZFNs), transcription activator-
like effector nuclease (TALENs) and so on.
Compared with them, CRISPR system is more
accurate and straightforward. It also allows
researcher to target or edit any type of gene.
There’s a huge amount of research aimed to
investigate lung cancer and the application of
CRISPR system on treating lung cancer. This review
will focus on discussing the pathology of lung cancer,
the traditional treatment of lung cancer as well as the
application of gene editing technology, specifically
CRISPR system on treating lung cancer.
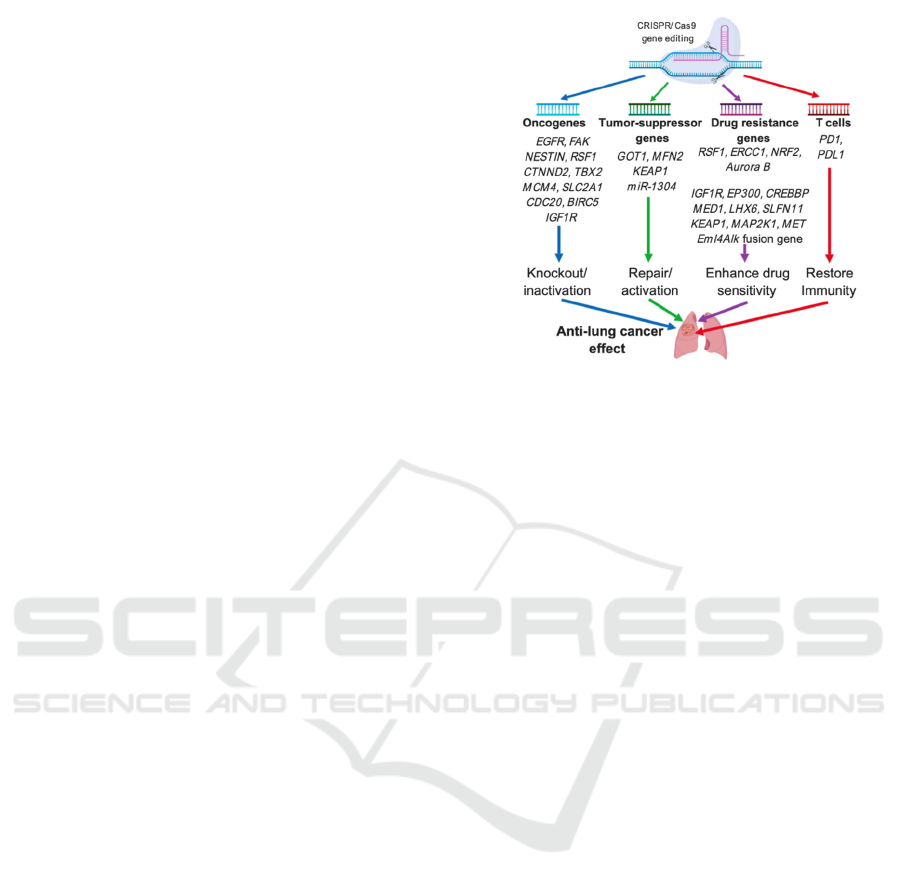
2 PATHOLOGY OF LUNG
CANCER
The pathogenesis of lung cancer is mostly caused by
the mutation of protooncogenes into oncogenes,
inactivation of tumor suppressor genes, insensitivity
of cancerous treatment as well as dysfunction of
immune system (Fig 1) (Jiang, Lin, & Zhao 2019)
Figure 1: Summary of 4 main pathological causes of lung
cancer.
2.1 Mutations of Protooncogene
Cancers are basically caused by mutation of specific
genes. Gene that works in its normal state is called
protooncogene which is in charge of regulating the
progress of cell division. In cancer, protooncogene
will mutate into oncogene which will significantly
disturb the normal machinery of cell division. This
will lead to an uncontrollable proliferation of cancer
cells. Below shows several common types of
oncogenes related with lung cancer.
2.1.1 Epidermal Growth Factor Receptor
(EGFR)
Epidermal growth factor receptor (EGFR) gene
mutation is associated with 13% cases of lung cancer.
It’s commonly detected in non-smoker or Asian
patients with NSCLC (Soda et al. 2007). EFGR gene
is located at chromosome 7. It is belonged to a class
of receptor tyrosine kinases named HER/erbB. The
homodimerization or heterodimerization of this class
of receptors will induce a tyrosine kinase activity.
This process will start a cascade of reactions which
will eventually couple the receptor to the pathway of
downstream signaling. This signaling will cause
various cancerous phenomenon like decreased
apoptosis (Koivunen et al. 2008). The expression of
EGFR gene is the main inducing factor for rapid cell
proliferation, angiogenesis, oncogenesis, and other
cancerous symptoms. EGFR mutation is mostly
happened when there’s a deletion in exon 19 or a
missense in exon 21 (Fig 2). In the missense mutation
circumstance, one of the thymine molecules is
substituted by a guanine molecule (Koo et al. 2017).
Adopting CRISPR-mediated Genomic Editing Technique on the Treatment of Lung Cancer: Using Revolutionary Genomic Editing
Technique to Treat Serious Human Disease
169
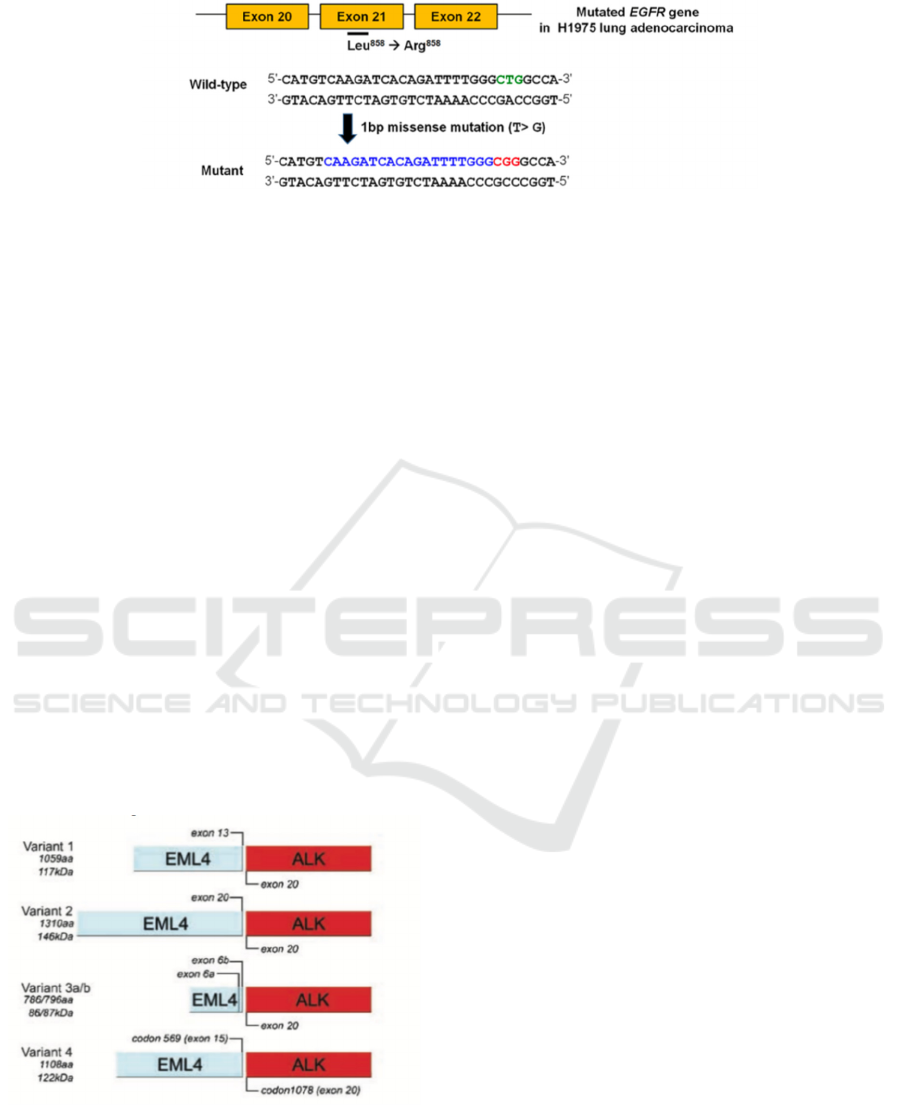
Figure 2. Mutation of EGFR gene due to the missense of exon 21. The thymine molecule is replaced by a guanine molecule
which turn the code for amino acid leucine into arginine.
2.1.2 EML4-ALK Fusion Gene
Anaplastic lymphoma kinase (ALK) is a type of
enzyme discovered from chromosomal translocation
that will induce the formation of fusion protein. In
this fusion, COOH-terminal donated by ALK is
combined with a NH2 terminal from other genes.
Recently, it is discovered that the fusion of ALK with
echinoderm microtubule- associated protein-like 4
(EML4) might lead to the onset of lung cancer.
Similarly, both ALK and EML4 are located at
chromosome 2 and are separated by 12Mb (millions
of base pairs). There’re a variety of types of variants
of EML4-ALK (Fig 3). For instance, when exon 20-
29 of ALK is fused with either exon 1-13 or exon 1-
20 of EMLK. The previous one is referred to variant
1 while the latter one is referred to variant 2. Both
of the variants are responsible for initiation and
maintenance of lung cancer. Expression of EML4-
ALK gene was inhibited by injection TAE684 which
is an ALK kinase inhibitor in mice sample. This
causes a cease in the size of lung tumor which further
suggests that lung cancer is related with the mutation
of EML4-ALK gene.
Figure 3: Example of 4 different types of EML4-ALK
fusion gene variants with their mutated site and specific
locations.
2.2 Tumor Suppressor Gene
Tumor suppressor gene is a type of gene that can
prevent the growth of tumor. Inactivation of tumor
suppressor gene will lead to a rapid growth of cancer
cells.
2.2.1 TSLC1
A majority of tumor suppressor activities are located
at a 100-kb segment 11q23.2. Kuramochi et al.
conducted a genetic research by adopting yeast
artificial chromosome (YACs) at chromosome 11
(
Kuramochi et al. 2001)
. The study aimed to localize the
existence of tumor suppressor genes in a small
section at 11q23.2. By transferring the overlapped
YACs region from human and nude mice, researchers
have successfully localized a tumor suppressor gene
at the central 700-kb segment of a 1.6-Mb YAC
which is known as TSLC1. Scientists have found that
the expression of TSLC1 is either reduced or absent
in NSCLC as well as several other types of lung
cancer cell lines. Loss of 11q23.2 which cause the
deletion of one allele of TSLC1 was found in 40%
cases of SCLC. The expression of TSLC1 is even
more inhibited with the promoter methylation in
those cell lines. As expected, by reactivating TSLC1,
there’s a significant suppression of malignant
phenotype in lung cancer cells.
2.2.2 p107 and p130
Ng, S. R. et al. (2020) aimed to use CRISPR system
to model lung cancers that are caused by the mutation
of tumor suppressor gene by targeting and
reactivating tumor suppressor gene that are once
dormant. To do this, they target 2 members of
retinoblastoma protein, p107 and p130. The mutation
of retinoblastoma protein is found to happen in 6% of
SCLC patients. Researcher made sgRNA to target
p107 and p130 genes. The validity of those
sgRNAs
was tested in vitro by Western blot to ensure if there’s
a decrease in retinoblastoma protein level.
CAIH 2021 - Conference on Artificial Intelligence and Healthcare
170

To test whether the system is worked in mice or
not, researcher infect mouses with Ad5-USEC
vectors which will express the sgRNA that target
p107 or p130 gene. Then, they conducted a in vivo
bioluminescence imaging to monitor the
circumstance of lung tumor in animal’s body. After a
period of time, researchers detected an acute level of
luciferase activity in both p107 infected and p130
infected animals which is accordant with the
accelerated tumor progression. The average survival
rate within those mouses have decreases
significantly, too. This verifies the tumor suppressing
function of p107 and p130 gene.
2.3 Insensitivity toward
Chemotherapeutic Drugs
In order to treat cancer, chemotherapy is an effective
and frequently used therapeutic method.
Unfortunately, the resistance of patient’s body toward
chemotherapeutic drugs has increased a lot from the
past few years. In such condition, some of the
previously prevailed market drugs are not as effective
as they were used to be For instance, cisplatin-
containing regimens is a commonly used
chemotherapeutic method to treat lung cancer, while
the sensitivity of this cytotoxic drug to NSCLC
patients still remain indistinct. Morodomi et al.
conducted a research to investigate the sensitivity of
chemotherapeutic drugs toward lung cancer patient
with different types of gene mutation (Morodomi et
al. 2014). The study found that patients with EML4-
ALK fusion gene is less sensitive to cytotoxic
chemotherapy compared with patient with EFGR
gene mutation. By injecting chemotherapeutic drugs
to patients with different gene mutations, it is found
that the response rate is highest in patients with EGFR
and lowest in patients with EML4-ALK fusion gene.
2.4 Single Agent Chemotherapy and
Combination Chemotherapy
Maio et al. conducted a meta-analysis aimed to
compare the effectiveness of single agent
chemotherapy and combination chemotherapy as a
second-line treatment (Di Maio et al. 2009). The
result shows that as a second-line treatment, the
response rate of patients toward combination
chemotherapy is significantly higher than who
received single agent chemotherapy. However, the
overall surviving rate was not improved. In addition,
most of the patients will end up by developing
resistance to those drugs. Therefore, an innovative
and effective way of treating lung cancer is in urgent
needs.
3 TRADITIONAL TREATMENTS
Due to lung cancer’s high prevalence rate among the
world, a variety of innovative and effective therapies
are in urgent need.
3.1 Chemotherapy
Chemotherapy is the initial treatment that SCLC
patient usually received. It is effective in relieving
various lung cancer complications like bronchial
obstruction, pleural effusion, tumor metastasis and so
on. It is a preferable palliative treatment. Platinum
drugs are the most commonly used chemotherapeutic
drugs on treating lung cancer. It works by bind with
the N7 atom on adenine and guanine to stop DNA
replication and in this way, induce the process of
apoptosis (Rossi & Di 2016). There’re several types
of chemotherapy.
4 ADOPT CRISPR SYSTEM IN
THE TREATMENT OF LUNG
CANCER
CRISPR Cas9 technology is a revolution technique
that allows scientists to target and edit a specific
sequence of genes precisely (
Jiang & Doudna 2017).
Theoretically, it’s possible to delete any cancerous
genes by adopting CRISPR technology. This fresh
idea of treating lung cancer by using “genetic
scissors” has become to a hot topic to conduct
research about recently. In the following section,
different mechanisms of using CRISPR technology to
edit cancer genes will be discussed.
4.1 Delete Cancer Gene
Crispr system takes advantages from those
characteristics of cancer to fight against lung cancer.
By deleting oncogene, activating tumor suppresser
gene, or enhancing the sensitivity of
chemotherapeutic drugs, the syndrome of lung cancer
can be greatly relieved.
In order to use CRISPR Cas9 system to target this
mutated gene, it’s necessary to make an assumption
that there’s a protospacer- adjacent motif (PAM)
sequence which is a nucleotide sequence targeted by
Cas9 nuclease beside the missense gene. By
Adopting CRISPR-mediated Genomic Editing Technique on the Treatment of Lung Cancer: Using Revolutionary Genomic Editing
Technique to Treat Serious Human Disease
171
transferring an oncogenic mutant specific Cas9 using
adenovirus as a media, the mutated gene can be
targeted and deleted with high accuracy, which leads
to a significant reduction of tumor production.
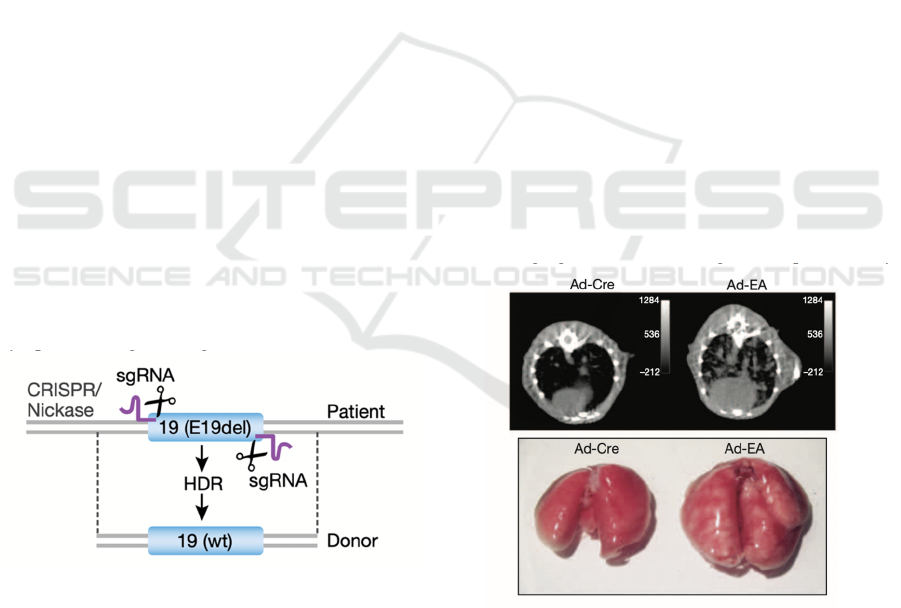
4.1.1 Repairing EGFR Gene
In order to treat patients by repairing EGFR gene, the
biopsy sample of patient’s tumor will be first
obtained. The mutated EGFR gene will be identified
from the biopsy and the correspondent single guide
RNA (sgRNA) which is the RNA that will guide the
endonuclease will be designed. The designed sgRNA
will target a
specific region on the mutated exon, e.g.,
L858R in exon 21, E19del in exon 19 and so on.
Then, the sgRNA will guide Cas9 nickase to the
target region which will make a single strand breaks
in on each opposite side of the mutated exon (Fig 4).
Next, the donated healthy DNA sequence will be
substituted to the removed sequence by homology-
directed repair (HDR) where its left and right arms
are connected with the cut, mutated DNA sequence.
The deletion of mutated exon and replacement with
healthy DNA sequence will deracinate the mutated
sequence and thus, stop the progression of lung
cancer. While it is also possible to conduct a non-
homologous end-joining (NHEJ) in which the
mutated sequence is deleted, and rest of the DNA
sequences are rejoined. By those two means, a stop
codon introduced by HDR or indel made by NHEJ
will destroy the translation of EGFR protein. As the
result, the translated protein will lose its normal
function, and therefore will not cause any oncogenic
symptoms (Tang & Shrager 2016).
Figure 4: CRISPR Cas9 nickase cutting the opposite side of
the mutated sequence.
4.1.2 Rearranging EML4-ALK Fusion Gene
Maddalo et al. (2014) conducted an experiment on
adult mice aimed to investigate the efficiency of
CRISPR Cas9 system on rearranging EML4–ALK
oncogene. The study is unusual since it did the
verification in a regressive way rather than a
progression way. This means that instead of
removing the EML4–ALK oncogene in mouses with
lung cancer, they chose to introduce lung tumor into
mouses body. If researcher can indeed add EML4-
ALK fusion gene into mouses genome, it implies that
it’s also possible to remove the oncogene away from
their genome.
They’ve genetically engineered mouses genome
to simulate the most common type of EML4–ALK
variant in NSCLC cases. They did this by introducing
a double stranded DNA that breaks at specific regions
in this case, intron 14 of EML4 gene and intron 19 of
ALK oncogene. Next, in order to express CAS9 and
sgRNA, researcher genetically engineered the
plasmid started from tandem U6 promoters. Then,
they made a recombinant adenovirus (Ad-EA) by
introducing the CAS9 nuclease and sgRNA into an
adenoviral shuttle vector to target the EML4 and
ALK loci. Numerous mouses were infected by Ad-
EA which leads to a speedy production of EML4-
ALK inversion. This is exactly the pathological
causes of NSCLC. After a month of infection, mouses
lungs started to appear several small lesions. By the
time of 6-8 weeks infection, the lungs tumor was
large enough to be easily seen by necropsy and micro-
computed tomography (Fig 5). The result shows that
there’s an obvious appearance and enlargement of
lungs tumor in mouses lungs. This indicated the
succession of rearranging EML4-ALK
fusion gene. It
also implies the feasibility of treating lung cancer by
rearranging EML4-ALK oncogene in a positive way.
Figure 5: The above shows the micro-computed
tomography of mouses lung tumor after 6-8 weeks of Ad-
EA infection, while the below image shows the necropsy of
mouses lung tumor. Ad-Cre is another type of infection that
has the similar mechanism with Ad-EA infection.
CAIH 2021 - Conference on Artificial Intelligence and Healthcare
172

5 CONCLUSIONS
Overall, CRISPR technology is a booming and
innovative gene editing technology. It has an
excellent applicational potential on treating and
investigating lung cancer. CRISPR system can help
scientists to construct tumor model, to study the
pathology of lung cancer, to discover certain drug
resistance and so on. It also enables researcher to
target specific oncogene or tumor suppressor gene.
By deleting or reactivating those genes, cancer
symptoms can be greatly relieved. Recently, there’re
a huge amount of research of adopting CRISPR
system on treating mouses with lung cancer. In order
to apply this system on curing genuine human cases,
more studies are in urgent needs. CRISPR system
must pass several strict assessments before they’re
applied into clinical trials. In addition, there’re a lot
of ethical issues raised in testing the efficiency of
CRISPR system on human samples. More animal
studies are required before it is used in clinical
circumstances.
However, it is necessary to aware the potential
defects of CRISPR technology. Since human genome
is an extremely large system, it is possible for
CRISPR system to cut other genes that are not
targeted by the nucleases which might cause an
unpredictable effect.
This off-targeted effect can be reduced by
improving guide RNA. Studies show that the length
of guide RNA may induce certain type of mutations.
Appropriate length of guide RNA is necessary for an
optimum genome-editing efficiency. Research also
reveals that specific chemical modification of guide
RNA, like introducing 2ʹ-O-methyl-3ʹ-
phosphonoacetate in the sugar backbone of guide
RNA, can significantly reduce the rate of off target
cleavage.
It is also possible to reduce off-target effect by
improving the delivery system of CRISPR
technology. For instance, adeno viruses have the
potential of integrating into target cell genome in a
more
meager way, which will restrict off-target
influence. In addition, deliver CAS9 protein and
guide RNA together as a ribonucleoprotein complex
will reduce the probability of missing the target gene,
too. Yet, those methods are only be proven by a few
studies. In order to further decrease the rate of off-
target effect, more empirical evidence is needed.
Until now, substantial amount of research has
done on animal sample. Once the technique has
proven to be safe enough, clinical research should be
conducted on human sample in order to adopt
CRISPR technology on treating genuine human lung
cancer. However, researchers should always be aware
of the ethics involved in clinical research and should
strictly follow the ethical guidelines.
REFERENCES
Di Maio, M., Chiodini, P., Georgoulias, V., Hatzidaki, D.,
Takeda, K., Wachters, F. M., ... & Gridelli, C. (2009).
Meta-analysis of single-agent chemotherapy compared
with combination chemotherapy as second-line
treatment of advanced non-small-cell lung cancer. J
Clin Oncol, 27(11), 1836-1843.
Jiang, C., Lin, X., & Zhao, Z. (2019). Applications of
CRISPR/Cas9 technology in the treatment of lung
cancer. Trends in molecular medicine, 25(11), 1039-
1049.
Jiang, F., & Doudna, J. A. (2017). CRISPR–Cas9 structures
and mechanisms. Annual review of biophysics, 46,
505-529.
Koivunen, J. P., Mermel, C., Zejnullahu, K., Murphy, C.,
Lifshits, E., Holmes, A. J., ... & Jänne, P. A. (2008).
EML4-ALK fusion gene and efficacy of an ALK kinase
inhibitor in lung cancer. Clinical cancer research,
14(13), 4275-4283.
Koo, T., Yoon, A. R., Cho, H. Y., Bae, S., Yun, C. O., &
Kim, J. S. (2017). Selective disruption of an oncogenic
mutant allele by CRISPR/Cas9 induces efficient tumor
regression. Nucleic acids research, 45(13), 7897-7908.
Kuramochi, M., Fukuhara, H., Nobukuni, T., Kanbe, T.,
Maruyama, T., Ghosh, H. P., ... & Murakami, Y.
(2001). TSLC1 is a tumor-suppressor gene in human
non-small-cell lung cancer. Nature genetics, 27(4),
427-430.
Maddalo, D., Manchado, E., Concepcion, C. P., Bonetti, C.,
Vidigal, J. A., Han, Y. C., ... & Ventura, A. (2014). In
vivo engineering of oncogenic chromosomal
rearrangements with the CRISPR/Cas9 system. Nature,
516(7531), 423-427.
Morodomi, Y., Takenoyama, M., Inamasu, E., Toyozawa,
R., Kojo, M., Toyokawa, G., ... & Ichinose, Y. (2014).
Non-small cell lung cancer patients with EML4-ALK
fusion gene are insensitive to cytotoxic chemotherapy.
Anticancer research, 34(7), 3825-3830.
Ng, S. R., Rideout, W. M., Akama-Garren, E. H., Bhutkar,
A., Mercer, K. L., Schenkel, J. M., ... & Jacks, T.
(2020). CRISPR-mediated modeling and functional
validation of candidate tumor suppressor genes in small
cell lung cancer. Proceedings of the National Academy
of Sciences, 117(1), 513-521.
Niederhuber JE, et al., eds. Cancer of the lung: Non-small
cell lung cancer and small cell lung cancer. In:
Abeloff's Clinical Oncology. 6th ed. Elsevier; 2020.
https://www.clinicalkey.com. Accessed Jan. 13, 2020.
Non-small cell lung cancer. National Comprehensive
Cancer Network.
https://www.nccn.org/professionals/physician_gls/def
ault.aspx. Accessed Jan. 13, 2020.
Adopting CRISPR-mediated Genomic Editing Technique on the Treatment of Lung Cancer: Using Revolutionary Genomic Editing
Technique to Treat Serious Human Disease
173

Rossi, A., & Di Maio, M. (2016). Platinum-based
chemotherapy in advanced non-small-cell lung cancer:
optimal number of treatment cycles. Expert review of
anticancer therapy, 16(6), 653-660.
Soda, M., Choi, Y. L., Enomoto, M., Takada, S.,
Yamashita, Y., Ishikawa, S., ... & Mano, H. (2007).
Identification of the transforming EML4–ALK fusion
gene in non-small-cell lung cancer. Nature, 448(7153),
561-566.
Tang, H., & Shrager, J. B. (2016). CRISPR/Cas‐mediated
genome editing to treat EGFR‐mutant lung cancer: a
personalized molecular surgical therapy. EMBO
molecular medicine, 8(2), 83-85.
CAIH 2021 - Conference on Artificial Intelligence and Healthcare
174
