
Multi-Objective Bees Algorithm for Feature Selection
Natalia Hartono
1,2 a
1
Department of Mechanical Engineering, University of Birmingham, Edgbaston, Birmingham B15 2TT, U.K.
2
Department of Industrial Engineering, University of Pelita Harapan, MH Thamrin Boulevard 1100, Tangerang, Indonesia
Keywords: Bees Algorithm, Classification, Feature Selection, Machine Learning, Multi-objective.
Abstract: In machine learning, there are enormous features that can affect learning performance. The problem is that
not all the features are relevant or important. Feature selection is a vital first step in finding a smaller number
of relevant features. The feature selection problem is categorised as an NP-hard problem, where the possible
solution exponentially surges when the number of n-dimensional features increases. Previous research in
feature selection has shifted from single-objective to multi-objective because there are two conflicting
objectives: minimising the number of features and minimising classification errors. Bees Algorithm (BA) is
one of the most popular metaheuristics for solving complex problems. However, none of the previous studies
used BA in feature selection using a multi-objective approach. This paper aims to present the first study using
the Multi-objective Bees Algorithm (MOBA) as a wrapper approach in feature selection. The MOBA
developed for this study using basic combinatorial BA with combinatorial of swap, insertion and reversion as
local operators with Non-Dominated Sorting and Crowding Distance to find the Pareto Optimal Solutions.
The performance evaluation using nine Machine Learning classifiers shows that MOBA performs well in
classification. Future work will improve the MOBA and use larger datasets.
1 INTRODUCTION
The main issue in machine learning and data mining
is that there are immense features that are often
redundant and unrelated that lead to poor
performance of classification accuracy (Hammami et
al., 2019; Al-Tashi et al., 2020a; Jha and Saha, 2021).
The curse of dimensionality is a term coined by
Gheyas and Smith (2010) to describe a large number
of features (dimension) that leads to a large search
space size even though not all of the features are
relevant. To handle this problem, the researcher uses
feature selection. Researchers emphasise the benefit
of feature selections: reducing redundant data,
improving accuracy, and reducing the complexity of
the algorithm, thus increasing the algorithm training
speed (Xue et al., 2015; Hancer et al., 2018; Al-Tashi
et al., 2020a). Feature selection is known as an NP-
complete problem (Gheyas and Smith, 2010).
Albrecht (2006) provide the mathematical proof of
NP-complete of this feature selection problem. The
possible feature subsets are 2
n
with n features, which
is unrealistic to find the best subset using an
a
https://orcid.org/0000-0003-2314-1394
exhaustive search (Gheyas and Smith, 2010; Hancer
et al., 2018).
There are four known methods for feature
selection: filter method, wrapper method, hybrid
method, and embedded method (Jha and Saha, 2021).
The most widely used methods are the filter and
wrapper method. The difference between the filter
and wrapper lies in evaluating feature subsets, where
the wrapper uses classifiers in the evaluation process
(Al-Tashi et al., 2020a). Xue et al. (2015) point out
that the wrappers method is slower than filters but
yields better classification performance. Similarly,
other researchers also noted that the wrapper yields
more promising results than the filter approach
(Jimenez et al., 2017; Hancer et al., 2018; Hammami
et al., 2019). In addition, in their multi-objective
feature selection systematic literature review, Al-
Tashi et al. (2020a) reports that 84% of the articles
use the wrapper method, whereas 13% and 2% use the
filter method and hybrid method, respectively.
Adding to that, they also identify that the wrapper
method is exceptionally preferred by the researchers
because of the better performance compared to the
filter method.
358
Hartono, N.
Multi-Objective Bees Algorithm for Feature Selection.
DOI: 10.5220/0010754200003113
In Proceedings of the 1st International Conference on Emerging Issues in Technology, Engineering and Science (ICE-TES 2021), pages 358-369
ISBN: 978-989-758-601-9
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Initial research for feature selection uses a single-
objective (SO) approach; however, the multi-
objective (MO) approach has gained attention in
recent years due to the ability to yield trade-off
solutions between two objectives (Wang et al., 2020).
In the literature review, the researcher has argued that
feature selection has at least two conflicting
objectives, for example, minimising the error rate of
classification and minimising the number of features
(Vignolo et al., 2013; Kozodoi et al., 2019; Al-Tashi
et al., 2020b). The MO approach is different from SO
because the best solution for one objective may not be
the best solution for the other objectives. The MO
approach gives different solutions that give trade-off
that balance between the objectives. Xue et al. (2015)
also compare the single-objective and multi-objective
approaches and concludes that multi-objective is
preferred over single-objective.
As explained earlier, it is impractical to search for
all possible solutions to find the best solution; the use
of metaheuristic in feature selection has attracted the
attention of researchers. Metaheuristics are well-
known for their ability to find a near-optimal solution
in a shorter computational time, and they have been
used in numerous studies. For example, Genetic
Algorithm and Particle Swarm Optimisation and its
MO version Non-dominated Sorting Genetic
Algorithm (NSGA-II) and MO-PSO are popular
metaheuristics used in feature selection. Regarding
the metaheuristic technique, one of the population-
based metaheuristics with a robust solution in the
continuous and combinatorial domain is Bees
Algorithm (BA). BA, inspired by the foraging activity
of honeybees to find nectar sources, was introduced
by Pham et al. (2005) and gained popularity due to its
ability to solve complex problems in faster
computational time and wide application in
engineering, business, bioinformatics (Yuce et al.,
2013; Hussein et al., 2017).
BA was used to solve SO feature selection. From
2007 to 2021, sixteen previous research articles are
used single-objective BA for feature selection. The
first research of single objective BA for feature
selection is in semiconductor manufacturing by Pham
et al. (2007), and the latest research in liver disease
case study by Ramlie et al. (2020). However, to date,
there is no single research using multi-objective BA
for feature selection. Hancer et al. (2018) also point
out in their study that multi-objective research for
feature selection is still in its early stages. Similar to
that, Kozodoi et al. (2019) said that the literature on
MOFS is lacking. Al-Tashi et al. (2020) supported
this view in their systematic literature review of
multi-objective feature selection (MOFS); they
provide 38 articles from 2012 to 2019. Further
literature search to find new articles until April 2021
shows that no previous studies use MOBA; thus,
MOBA’s potential for MOFS has not been
investigated. The previous studies in MOFS and the
research position of this study are depicted in Table
1.
Initially, BA’s development is in the continuous
domain and yields good results because of the balance
between local and global search architecture;
however, in the combinatorial domain, the approach
is different due to different neighbourhood concepts;
thus, it needs a different approach in the local search
operator (Koc, 2010). Koc (2010) developed the
combinatorial BA using simple-swap and insertion as
local search operator with application in a single
machine scheduling problem. Ismail et al. (2020)
improve the local search operator using swap,
reverse, and insertion with the Travelling Salesman
Problem (TSP). The results show that the best local
search operator uses a combination of those three
operators (swap, reverse and insertion) rather than
using single operators. This version developed by
Ismail et al. (2020) is called basic combinatorial BA.
The second application of basic combinatorial BA in
Vehicle Routing Problem by Ismail et al. (2021).
Zeybek et al. (2021) improved the combinatorial BA
called VPBA-II using the Vantage Point (VP)
strategy proposed by Zeybek and Koc (2015). The
VPBA-II put the Vantage Point Tree (VPT) in the
initial solution and global search while keeping the
same local search operator with previous research by
Ismail et al. (2020). The difference between the basic
combinatorial BA and VPBA-II lies in the initial
solution and global search architecture, where the
former uses random permutation while the VPBA-II
uses VPT.
The current work is the first study using the Multi-
objective Bees Algorithm (MOBA) for feature
selection. The aim is to use a wrapper method using
MOBA for better classification with Pareto Optimal
Solutions that reduces the number of features while
minimising the error rate of classification. As
aforementioned, the wrapper method and multi-
objective approach produce better results, and this
study employs this approach in the development of
BA. In addition, the MOBA uses the same local
operators as basic combinatorial BA. As Al-Tashi et
al. (2020a) point out, the MOFS is gaining traction in
machine learning and data mining research due to its
enormous number of features. They suggest that the
area of MOFS still has a wide improvement
possibility in the future regarding improvement
of
accuracy, reduced computational time, the search
Multi-Objective Bees Algorithm for Feature Selection
359
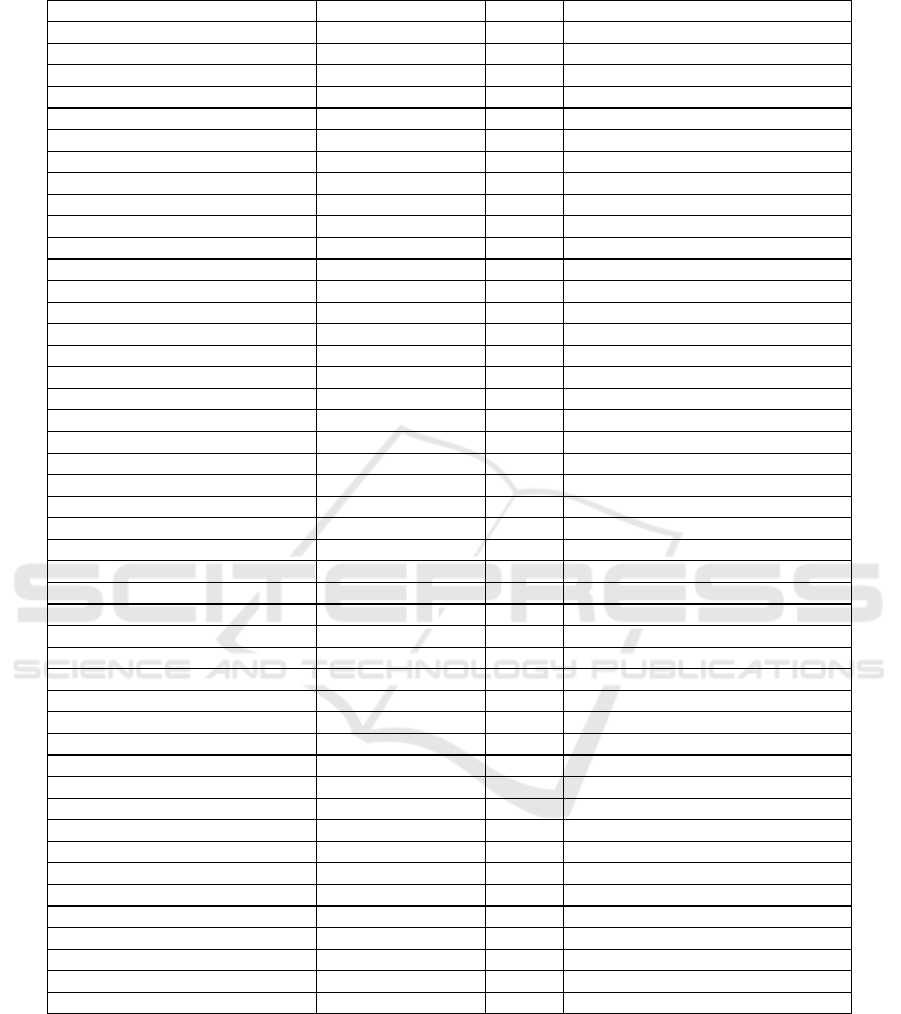
Table 1: Previous Research and Research Position.
mechanism, the number of objectives, and
evaluation measure. Therefore, the contribution of
this study is two-fold. First, the current work is the
first study using MOBA for feature selection.
Secondly, this study used more than three Machine
Learning classifiers to measure the feature subsets'
performance.
2 METHODS (AND MATERIALS)
As explained earlier, the objective of MOBA for
feature selection in this study comprises of two
objectives: minimise the number of features and
minimise the error rate of classification.
Author Search Technique Evaluation Dataset
Xue et al. (2012) NSBPSO & CMDBPSO Filter UCI
Vignolo et al. (2013) MOGA Wrapper Essex Face Database
Mukhopadhyay and Maulik (2013) NSGA-II Wrapper Medical Dataset
Xue et al. (2013a) NSGA-II and SPEA2 Filter UCI
Xue et al. (2013b) MOPSO Wrapper UCI
Xia et al. (2014) MOUFSA Wrapper UCI
de la Hoz et al. (2014) NSGA-II Wrapper NSL-KDD
Tan et al. (2014) MmGA Wrapper UCI
Khan and Baig (2015) NSGA-II Wrapper UCI
Wang et al. (2015) MECY-FS Filter UCI
Han and Ren (2015) MO-MIFS & NSGA-II Wrapper (Own) Real
Kundu and Mitra (2015) NSGA-II Wrapper UCI
Kimovski et al. (2015) MOEA Wrapper BCI (Own)
Yong et al. (2016) MOPSO Wrapper UCI
Sahoo and Chandra (2017) MOGWO Wrapper (Own) Real
Mlakar et al. (2017) MODE Wrapper CK, MMI, JAFFE
Zhu et al. (2017) I-NSGA-III Wrapper NSL-KDD
Peimankar et al. (2017) MOPSO Wrapper DGA
Jiménez et al. (2017) ENORA Wrapper Kaggle
Sohrabi and Tajik (2017) NSGA-II & MOPSO Wrapper (Own) Real
Deniz et al. (2017) MOGA Wrapper UCI
Zhang et al. (2017) MOPSO Wrapper MULAN
Das and Das (2017) MOEA/D Wrapper UCI
Kizilos et al. (2018) MO-TLBO Wrapper UCI
Amoozegar and Minaei-Bidgoli (2018) MO-PSO Filter UCI
Hancer et al. (2018) MO-ABC Wrapper UCI
Dashtban et al. (2018) MO-Bat Wrapper Cancer Dataset
Lai (2018) MOSSO Hybrid Medical Dataset
Cheng et al. (2018) MOFSRank Wrapper LETOR
Kozodoi et al. (2019) NSGA-II Wrapper Credit Scoring (Kaggle)
González et al. (2019) NSGA-II Wrapper BCI (Own)
Sharma and Rani (2019) MOSHO & SSA Wrapper Cancer Dataset
Zhang et al. (2020) MOFSBDE Wrapper UCI
Nayak et al. (2020) FAEMODE Filter UCI
Al-Tashi et al. (2020) Grey Wolf Wrapper UCI
Wang et al. (2020) MO ABC Wrapper UCI
Rostami et al. (2020) MO PSO Wrapper Medical Dataset
Rodrigues et al. (2020) ABO Wrapper UCI
Rathee and Ratnoo (2020) NSGA-II + CHC Wrapper UCI
Khammasi and Krichen (2020) NSGAII and LR Wrapper NSL-KDD, UNSW-NB15, CIC-IDS2017
Kou et al. (2021) NSGA-II Wrapper (Own)
Karagoz et al. (2021) NSGA-II Wrapper MIR-Flickr and WMS
Jha and Saha (2021) MO PSO Filter UCI
Hu and Zhang (2021) PSOMOFS Wrapper UCI & Real (Own)
Hu et al. (2021) Grey Wolf Wrapper IEEE CEC3014
This paper MOBA Wrapper UCI
ICE-TES 2021 - International Conference on Emerging Issues in Technology, Engineering, and Science
360
The equation for multi-objective feature selection
as follows:
f
(
x
) = min(
f
1
(
x
),
f
2
(
x
)) (1)
where
f
1
=
F
s
(2)
and
f
2 = (ωtrain.Ftrain) + [(1 − ωtrain).Fval](3)
Fs denotes the Number of Feature Selected, ωtrain
denotes the weighting factor for training set in cross-
validation. For this study, the ωtrain set at 0.8. The
classification error on the training set is Ftrain, and
Fval is the classification error on the validating set.
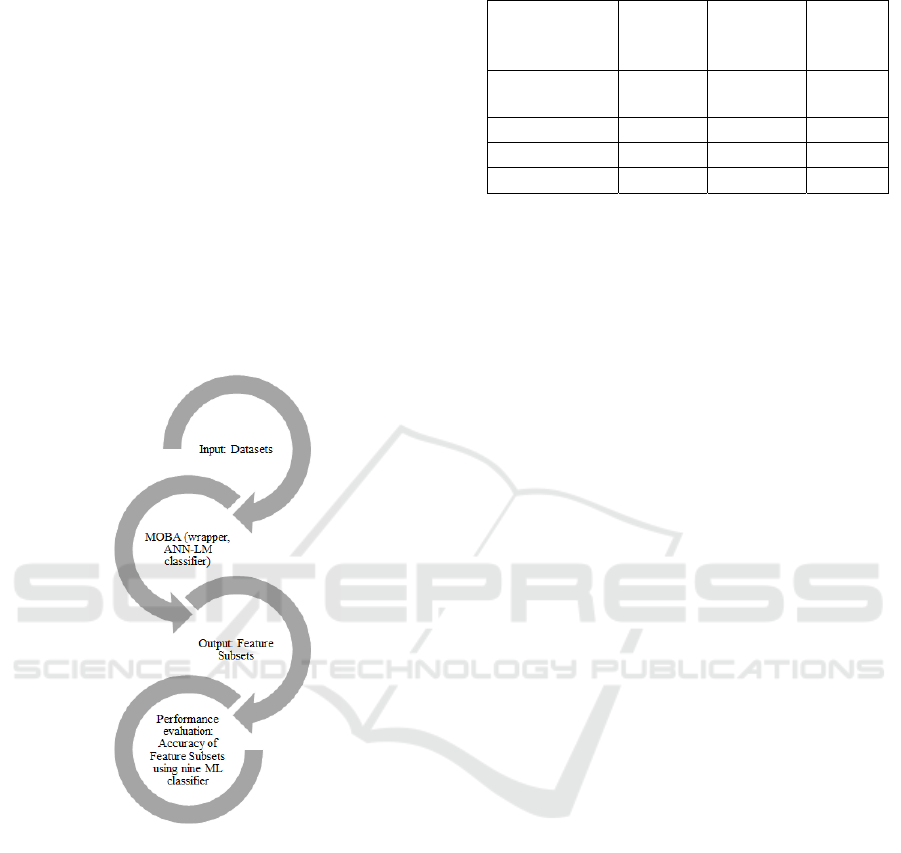
The research steps for this study are depicted in
Figure 1, followed by the description of each step.
Figure 1: Research Steps in current work.
As depicted in Table 1, 89% of previous MOFS
studies use benchmark datasets, and only 11% use
their own datasets. The most widely used benchmark
datasets are the UCI Machine Learning Repository
(University of California). Due to the fact that
benchmark datasets are popular and concerning data
availability, this study uses the UCI Machine
Learning Repository, detailed in Table 2. The dataset
has a balanced distribution of classes.
Table 2: Benchmark Data Description.
Dataset
Number
of
Features
Number
of
Instances
Classes
Pima Indian
Diabetes
8 768 2
Breastcancer 9 699 2
Wine 13 178 3
Sonar 60 208 2
In this study, the MOBA for feature selection was
developed using the best local operator from basic
combinatorial BA. The combination of swap,
insertion and reserve introduced by Ismail et al.
(2020) was chosen for MOBA’s development. As a
wrapper-based method, the MOBA needs a classifier
to calculate the error of the classification. Al-Tashi et
al. (2020b) point out that the Artificial Neural
Network (ANN) is known as a superior classifier due
to its speed in classification. Moreover, Baptista et al.
(2013) suggest that one of the best ANN training
algorithms is Levenberg-Marquardt (LM)
backpropagation. The MOBA utilises ANN to
calculate the classification error, which in this study
uses LM backpropagation with 10 hidden layers and
a 0.8 learning rate. The MOBA parameter for this
study is as follows: 20 number of scout bees (n), 10
number of elite bees (nep), 5 number of best bees
(nsp), 1 number of elite sites (e) and 5 number of best
sites (m), maximum iteration 50. The MOBA
algorithm flowchart for this study is presented in
Figure 2.
The experiment runs 10 times using Matlab 2020a
in the University of Birmingham’s BEAR Cloud
service for each dataset. The results are Pareto
Optimal Solutions in the form of a feature subset that
balanced the two objectives. The performance
measurement for feature subsets generated by
MOBA, nine supervised Machine Learning (ML)
Techniques, is used to compare the accuracy of the
full features and the feature subsets. The ML
techniques are Medium Tree (MT), Linear
Discriminant (LD), Quadratic Discriminant (QD),
Gaussian Naive Bayes (GNB), Kernel Naive Bayes
(KNB), Linear Support Vector Machine (L-SVM),
Quadratic SVM (Q-SVM), Medium KNN (M-KNN),
and Cosine KNN (Co-KNN) with 10-fold cross-
validation.
Multi-Objective Bees Algorithm for Feature Selection
361
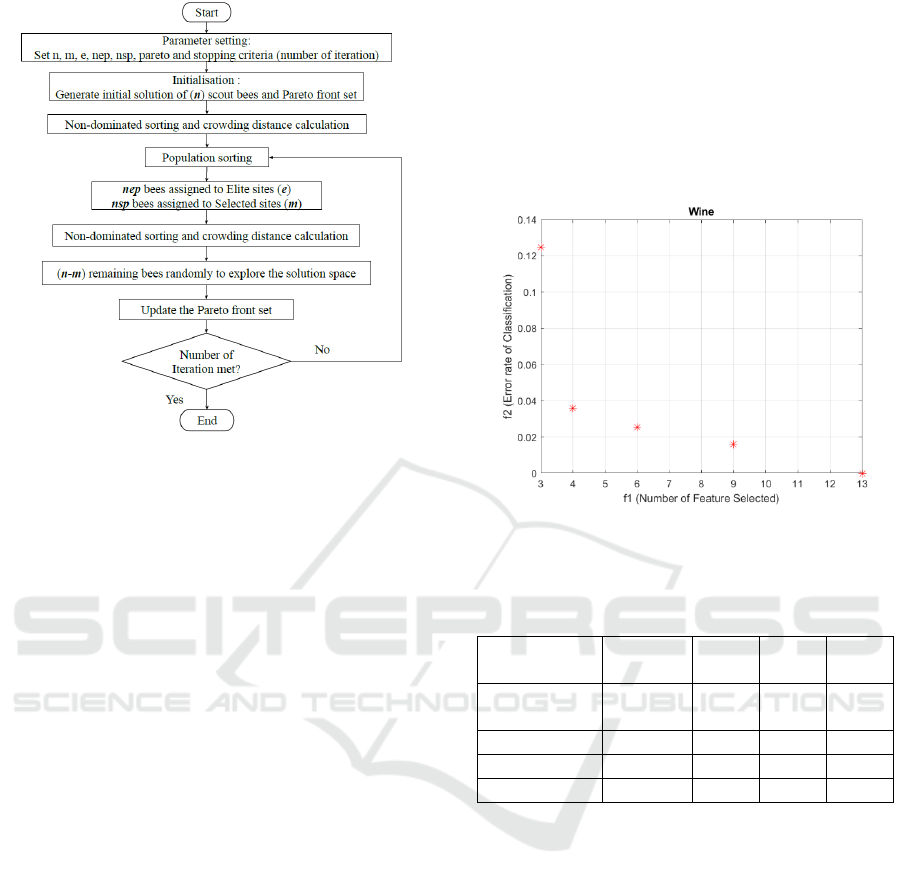
Figure 2: MOBA flowchart for this study.
3 RESULTS AND DISCUSSION
The benefit of using the MO approach is that
decision-makers will have more options to choose
from the Pareto Frontier. For example, figure 3
depicts the Pareto Optimal Solution from one of the
experiments performed on Wine Datasets. As can be
seen, the higher the number of features selected, the
lower the classification error. As a result, the
decision-maker could pick one subset for
classification calculations, saving time on the
experiments.
Table 3 provides the average results from 10 runs
for each dataset. It is apparent from this table that
MOBA is able to reduce the number of features by
more than 50%. The average ratio of selected features
ranges from 0.38 to 0.45. The average error of the
feature subset ranges from 0.05 to 0.17. Interestingly,
the bigger dataset shows lower errors.
As described in the previous section, the selected
features trained using nine ML classifiers and Pima
Indian Diabetes, Breastcancer, Wine and Sonar
presented in Table 4, 5, 6, and 7, respectively. The
MOBA feature subsets and the accuracy of the Pima
Indian Diabetes Dataset can be seen in Table 4. It
shows that most of the feature subsets generated by
MOBA for this dataset yield the same or better
accuracy (in bold) with a smaller number of features
than the accuracy of full features for all nine ML
training. The interesting finding in Table 5 is that
accuracy using Medium Tree and Coarse KNN is
higher than using all features. What stands out in
Table 6 is that in the 9th run, the feature subsets with
eight features (ratio equal to 0.6154) yield 100%
classification accuracy when trained using QD. Table
7 shows that the classification accuracy in each
feature subset is all higher in Medium KNN. Thus,
overall results from four benchmark datasets indicate
that feature subsets generated by MOBA yield good
performance for classification.
Figure 3: Pareto Optimal Solution on Wine Dataset.
Table 3: Average Size of Selected Features (f1), Average
Error of the Selected Features (f2) and Average Ratio of
Selected Features.
Dataset
Total
Features
Mean
f1
Mean
f2
Mean
Ratio
Pima Indian
Diabetes
8 3.05 0.17 0.38
Breastcance
r
9 3.68 0.17 0.41
Wine 13 5.57 0.05 0.43
Sona
r
60 26.95 0.09 0.45
This study confirms previous studies that not all
the features are relevant for classification, and a
reduced dimensionality can achieve similar or higher
classification accuracy. Furthermore, results show
that MOBA performs well in classification accuracy.
ICE-TES 2021 - International Conference on Emerging Issues in Technology, Engineering, and Science
362
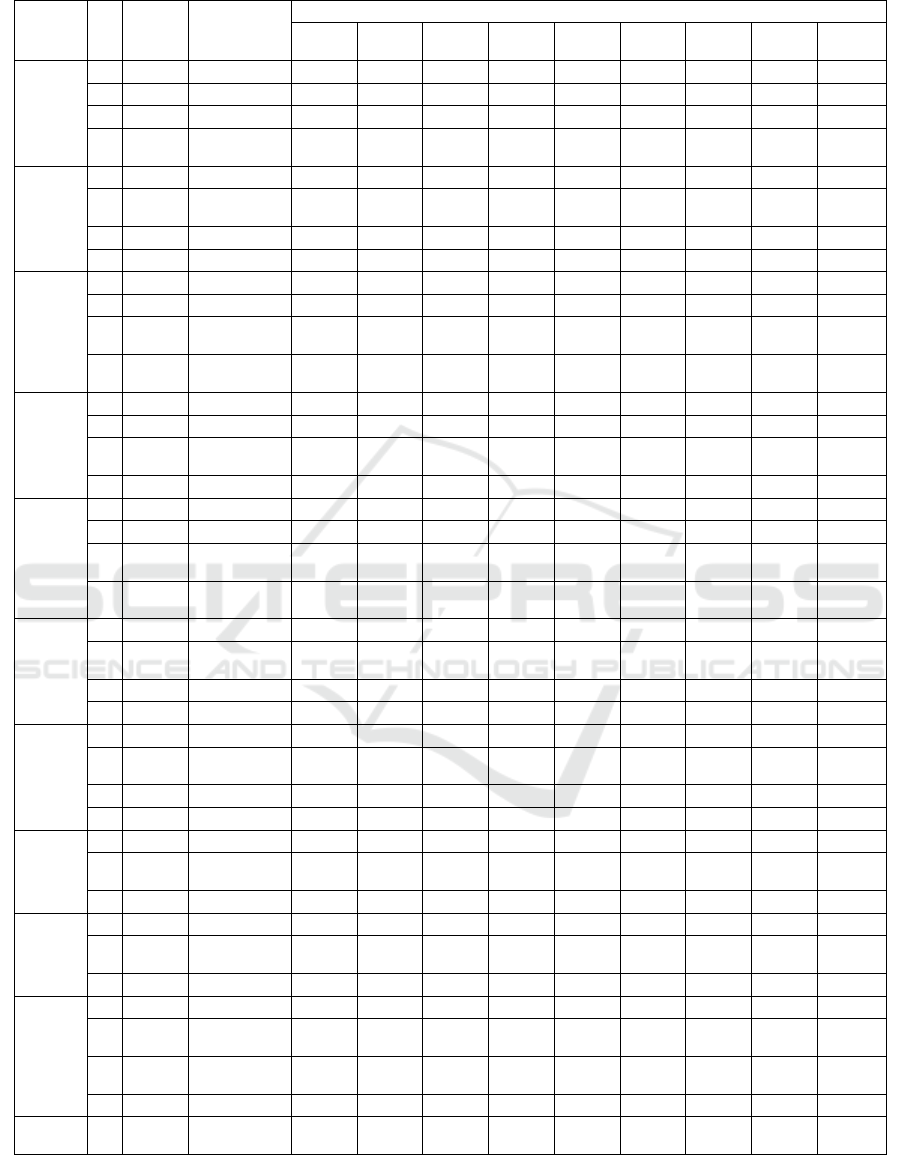
Table 4: MOBA result and classification accuracy on Pima Indian Diabetes Dataset.
Run f1 f2
Feature
subsets
Accuracy
MT LD QD GNB KNB
L
SVM
Q
SVM
M
KNN
Coarse
KNN
1
1 0.2193 F3 62.9% 65.1% 64.7% 64.7% 64.6% 65.1% 60.0% 59.5% 65.0%
3 0.1569 F2, F4, F6 72.8% 76.6% 75.1% 76.0% 75.3% 76.2% 75.1% 71.9% 75.1%
2 0.2081 F1, F4 66.3% 67.1% 67.3% 67.3% 66.4% 65.1% 66.9% 64.2% 68.0%
5 0.1450
F1, F2, F5,
F6, F8
73.6% 76.3% 75.4% 75.1% 74.0% 76.4% 76.7% 75.7% 75.9%
2
1 0.1996 F8 64.8% 65.6% 66.4% 66.4% 65.6% 65.0% 44.1% 62.5% 64.6%
5 0.1531
F2, F3, F4,
F7, F8
71.6% 76.7% 74.7% 77.0% 75.1% 76.3% 76.6% 72.8% 74.5%
3 0.1599 F1, F2, F7 72.3% 76.2% 75.4% 75.7% 76.0% 76.3% 75.5% 75.4% 76.0%
2 0.1929 F4, F8 65.5% 65.1% 66.1% 64.5% 67.3% 65.1% 64.5% 64.2% 67.2%
3
2 0.1934 F1, F8 66.5% 66.3% 65.8% 66.9% 66.9% 65.1% 64.6% 65.9% 66.8%
3 0.1512 F2, F7, F8 70.7% 75.3% 74.9% 75.1% 75.3% 74.3% 75.4% 73.4% 76.4%
6 0.1463
F1, F2, F4,
F6, F7, F8
73.2% 77.3% 74.7% 76.3% 77.5% 77.1% 76.0% 75.7% 75.0%
4 0.1470
F2, F6, F7,
F8
73.3% 77.5% 76.4% 77.9% 77.3% 77.5% 77.7% 75.5% 77.6%
4
1 0.1997 F8 64.8% 65.6% 66.4% 66.4% 65.6% 65.0% 44.1% 62.5% 64.6%
2 0.1701 F2, F4 71.9% 73.7% 74.7% 74.6% 74.6% 74.2% 71.1% 71.0% 74.7%
5 0.1488
F1, F2, F4,
F6, F7, F8
72.9% 76.8% 75.1% 75.8% 77.1% 77.2% 77.0% 75.7% 76.0%
3 0.1531 F2, F6, F8 73.7% 76.8% 76.2% 77.0% 76.8% 76.7% 76.3% 74.7% 76.3%
5
2 0.2045 F1, F4 66.3% 67.1% 67.3% 67.3% 66.4% 65.1% 66.9% 64.2% 68.0%
3 0.1540 F2, F5, F8 73.4% 73.3% 75.3% 73.2% 70.4% 74.1% 76.0% 72.7% 76.7%
5 0.1449
F2, F3, F5,
F6, F8
73.8% 77.3% 75.1% 76.2% 73.2% 77.2% 77.0% 75.5% 75.8%
4 0.1497
F2, F5, F7,
F8
70.3% 75.3% 74.1% 74.7% 72.7% 75.7% 76.7% 74.7% 75.9%
6
1 0.2201 F3 62.9% 65.1% 64.7% 64.7% 64.6% 65.1% 63.2% 59.5% 65.0%
4 0.1512
F2, F3, F6,
F8
73.6% 76.7% 75.9% 77.2% 75.8% 76.6% 77.7% 75.8% 76.0%
3 0.1768 F5, F6, F8 70.3% 68.0% 69.7% 69.0% 70.6% 68.2% 69.7% 71.5% 68.0%
2 0.1950 F1, F8 66.5% 66.3% 65.8% 66.9% 66.9% 65.1% 64.6% 65.9% 66.8%
7
1 0.2162 F7 64.8% 65.6% 66.4% 66.4% 65.6% 65.1% 41.9% 62.5% 64.6%
4 0.1533
F1, F2, F7,
F8
70.2% 75.8% 75.7% 74.7% 74.7% 75.7% 76.2% 76.4% 75.4%
3 0.1889 F5, F7, F8 66.5% 65.9% 67.6% 67.8% 69.3% 65.1% 67.3% 66.5% 68.5%
2 0.1893 F5, F6 66.1% 67.2% 68.8% 67.6% 67.4% 65.1% 55.6% 67.3% 65.1%
8
2 0.1936 F4, F8 65.5% 65.1% 66.1% 64.5% 67.3% 65.1% 64.5% 64.2% 67.2%
4 0.1503
F2, F4, F6,
F8
74.0% 76.7% 75.4% 76.8% 77.3% 77.2% 77.2% 75.5% 76.6%
3 0.1620 F2, F3, F7 69.5% 74.2% 74.3% 75.4% 74.7% 74.1% 74.9% 73.7% 74.3%
9
1 0.1696 F2 71.2% 74.7% 75.0% 75.0% 74.2% 74.6% 47.0% 70.2% 74.0%
5 0.1529
F1, F2, F3,
F6, F8
74.2% 77.0% 75.7% 75.7% 76.0% 76.8% 78.0% 75.5% 75.1%
3 0.1540 F2, F5, F6 72.1% 75.7% 74.9% 75.1% 71.1% 75.9% 75.5% 74.9% 75.0%
10
2 0.1584 F2, F6 72.8% 77.1% 76.0% 76.8% 75.5% 76.0% 74.2% 73.8% 75.0%
6 0.1443
F1, F2, F3,
F4, F6, F8
74.2% 77.0% 74.5% 76.2% 75.3% 76.4% 77.6% 77.1% 74.2%
5 0.1527
F1, F2, F5,
F6, F8
73.6% 76.3% 75.4% 75.1% 74.0% 76.4% 76.7% 75.7% 75.9%
3 0.1555 F2, F3, F8 72.1% 74.2% 75.3% 74.6% 74.6% 75.0% 77.0% 74.2% 74.3%
All
features
8 - F1-F8 74.2% 77.5% 73.4% 75.3% 73.0% 77.1% 76.7% 72.9% 73.8%
Multi-Objective Bees Algorithm for Feature Selection
363
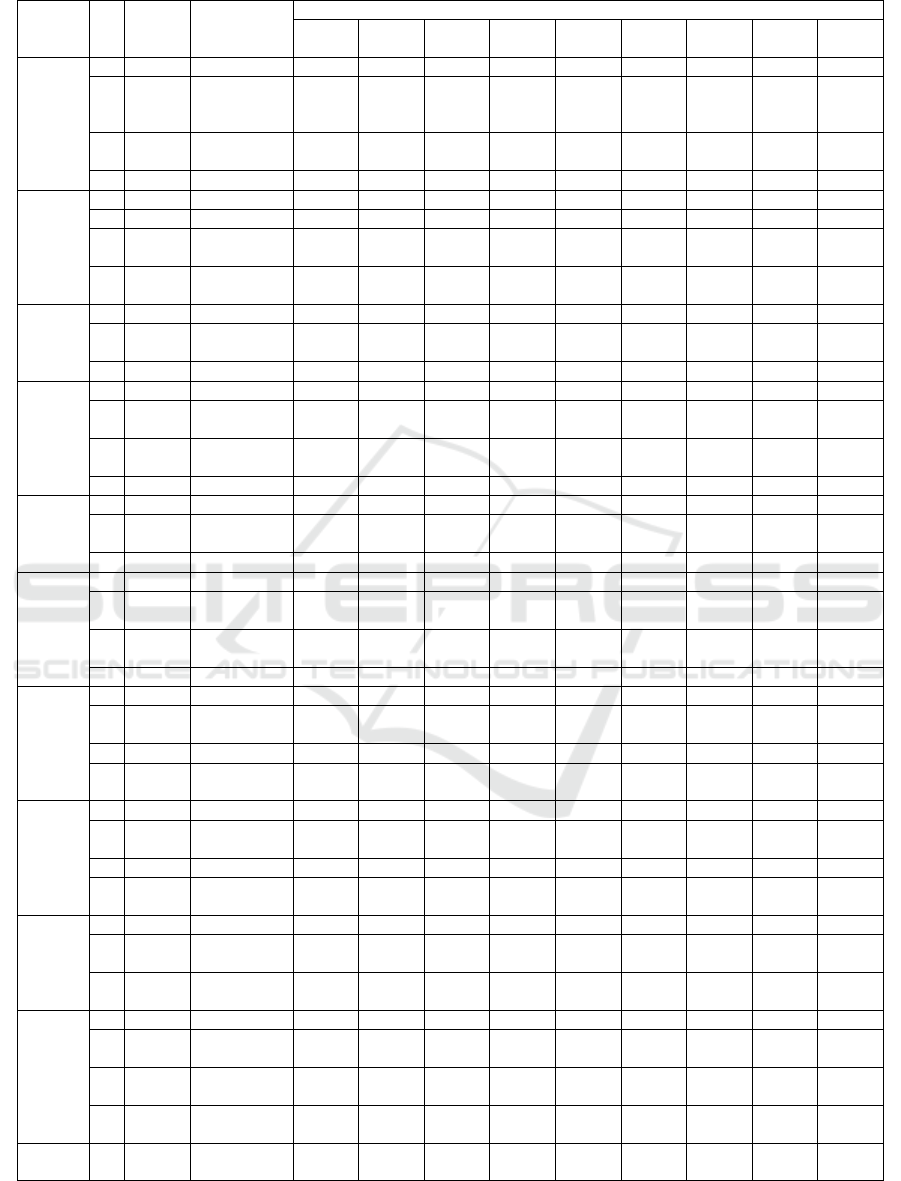
Table 5: MOBA result and classification accuracy on Breastcancer Dataset.
Run f1 f2
Feature
subsets
Accuracy
MT LD QD GNB KNB
L-
SVM
Q-
SVM
M-
KNN
Co-
KNN
1
2 0.1839 F3, F4 68.9% 70.3% 71.3% 71.7% 70.3% 71.3% 67.8% 69.2% 70.3%
7 0.1560
F1, F3, F4,
F5, F6, F7,
F8
67.5% 74.1% 72.0% 74.5% 70.6% 72.0% 75.5% 73.4% 70.3%
4 0.1614
F1, F4, F5,
F6
73.1% 74.8% 72.0% 72.7% 71.7% 72.4% 74.5% 74.5% 70.3%
3 0.1770 F3, F6, F7 71.7% 71.7% 69.9% 70.3% 70.3% 70.3% 69.6% 70.3% 70.3%
2
1 0.2067 F8 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 68.5% 70.3%
2 0.1758 F5, F6 75.2% 75.9% 71.0% 71.3% 70.3% 71.0% 76.2% 70.3% 70.3%
6 0.1496
F1, F5, F6,
F7, F8, F9
71.7% 73.8% 72.4% 71.7% 71.3% 71.0% 74.5% 75.2% 70.3%
5 0.1581
F1, F3, F4,
F5, F6
67.8% 73.1% 71.7% 73.4% 72.7% 72.0% 74.8% 73.4% 70.3%
3
2 0.1909 F3, F9 71.0% 67.5% 68.5% 69.2% 68.5% 70.3% 69.2% 67.5% 70.3%
5 0.1569
F4, F5, F6,
F7, F9
75.5% 75.2% 69.9% 72.7% 72.4% 72.0% 76.6% 72.0% 70.3%
3 0.1698 F3, F4, F6 71.3% 73.4% 72.7% 72.7% 73.1% 71.7% 75.9% 72.7% 70.6%
4
2 0.2006 F2, F8 71.0% 70.3% 68.9% 70.3% 70.3% 70.3% 70.3% 65.0% 70.3%
6 0.1421
F1, F3, F5,
F6, F7, F9
72.0% 73.4% 71.3% 71.7% 71.7% 71.0% 74.5% 73.8% 70.3%
5 0.1677
F4, F5, F6,
F7, F8
73.4% 74.8% 74.1% 73.8% 73.4% 72.0% 74.5% 73.8% 70.3%
3 0.1835 F1, F4, F9 67.1% 72.7% 69.9% 72.0% 72.0% 71.7% 67.5% 69.2% 70.3%
5
2 0.1833 F4, F5 69.6% 72.0% 72.0% 72.0% 71.7% 72.0% 69.9% 71.3% 70.3%
5 0.1608
F2, F4, F5,
F7, F9
72.4% 72.7% 69.2% 70.6% 71.3% 72.0% 73.4% 72.7% 70.3%
3 0.1657 F3, F5, F6 76.2% 75.5% 70.3% 71.7% 71.0% 71.3% 76.2% 74.5% 70.3%
6
2 0.1862 F1, F6 72.7% 72.4% 69.2% 72.0% 71.0% 70.3% 72.0% 72.4% 70.3%
6 0.1534
F1, F2, F3,
F5, F6, F8
71.0% 75.2% 72.4% 71.3% 71.3% 71.3% 76.2% 72.0% 70.3%
4 0.1630
F2, F4, F5,
F6
73.8% 74.5% 73.8% 72.7% 73.4% 72.0% 75.2% 74.8% 70.3%
3 0.1774 F4, F6, F7 73.8% 75.2% 74.5% 73.1% 73.1% 71.7% 75.2% 74.1% 70.3%
7
1 0.2085 F7 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 70.3%
6 0.1540
F2, F4, F5,
F7, F8, F9
73.1% 72.4% 70.6% 71.0% 71.7% 72.0% 71.7% 73.1% 70.3%
2 0.1709 F4, F6 74.8% 74.1% 73.4% 72.7% 73.4% 71.7% 75.5% 74.8% 71.7%
4 0.1649
F4, F6, F7,
F8
73.4% 74.8% 73.8% 73.8% 72.0% 71.7% 74.8% 74.5% 70.3%
8
1 0.2079 F8 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 70.3% 68.5% 70.3%
5 0.1566
F1, F3, F5,
F6, F9
72.0% 74.1% 71.7% 71.7% 72.7% 70.3% 75.5% 71.3% 70.3%
3 0.1773 F2, F5, F6 74.1% 75.9% 71.3% 71.7% 70.3% 71.3% 76.2% 75.2% 70.3%
4 0.1723
F2, F3, F4,
F5
72.0% 71.0% 72.4% 72.0% 70.3% 72.0% 72.7% 68.2% 70.3%
9
3 0.1738 F1, F4, F7 74.8% 72.4% 71.7% 71.7% 70.6% 71.7% 74.5% 74.1% 70.3%
5 0.1488
F3, F5, F6,
F7, F8
72.0% 75.9% 72.4% 70.6% 72.0% 71.3% 75.2% 73.4% 70.3%
4 0.1630
F2, F4, F6,
F9
73.1% 73.8% 71.3% 73.1% 72.0% 71.7% 74.5% 72.7% 70.3%
10
2 0.1824 F3, F4 68.9% 70.3% 71.3% 71.7% 70.3% 71.3% 67.8% 69.2% 70.3%
6 0.1506
F3, F4, F5,
F6, F7, F9
70.3% 73.8% 69.9% 73.1% 72.7% 72.0% 76.6% 73.1% 70.3%
5 0.1659
F3, F4, F5,
F6, F8
71.7% 74.5% 73.1% 73.1% 72.4% 72.0% 75.5% 72.7% 70.3%
4 0.1748
F4, F5, F7,
F9
72.4% 72.4% 69.6% 70.6% 71.7% 72.0% 73.8% 70.3% 70.3%
All
features
9
F1 - F9 65.4% 74.1% 69.2% 72.0% 72.0% 72.0% 72.4% 72.7% 70.3%
ICE-TES 2021 - International Conference on Emerging Issues in Technology, Engineering, and Science
364
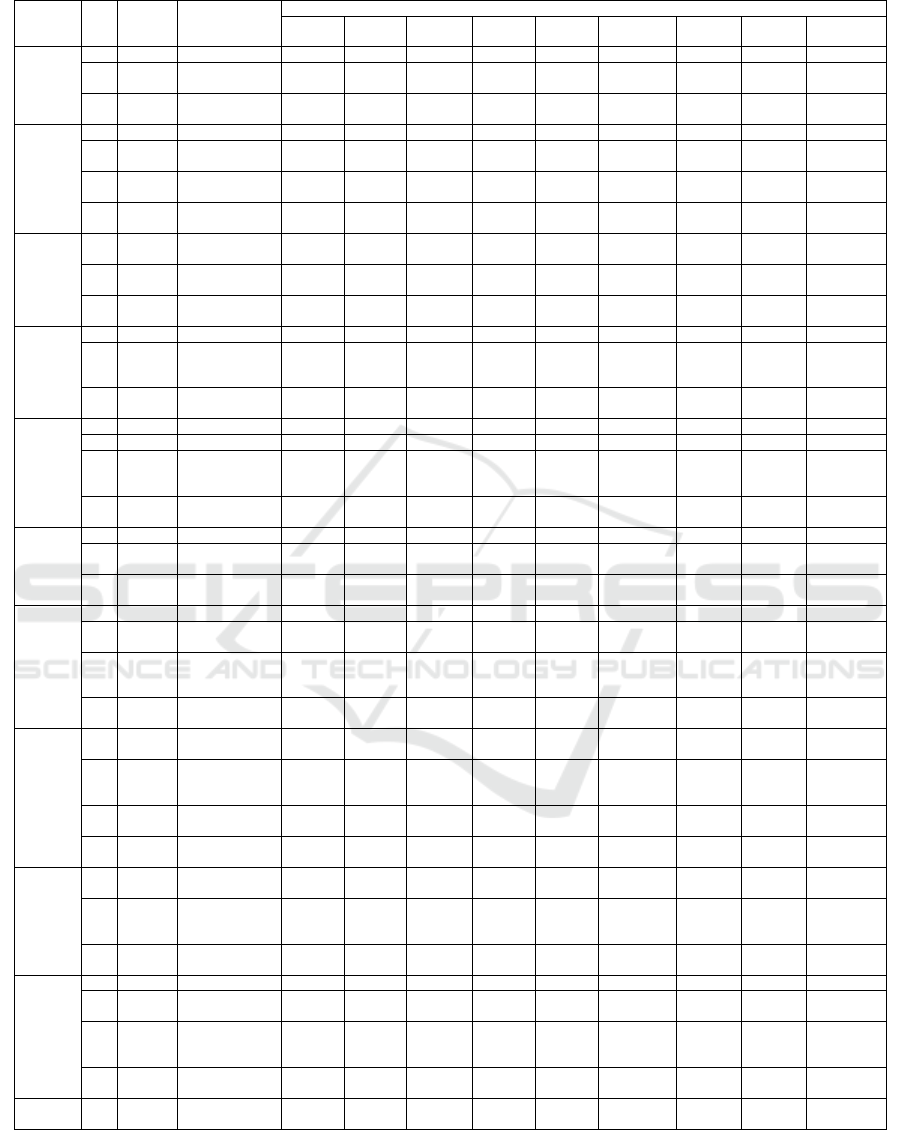
Table 6: MOBA result and classification accuracy on Wine Dataset.
Run f1 f2 Feature subsets
Accurac
y
MT LD QD GNB KNB L-SVM Q-SVM
M-
KNN
Co-KNN
1
2 0.071 F1, F12 88.2% 89.3% 88.2% 88.2% 88.8% 89.3% 87.6% 88.8% 85.4%
7 0.022
F2, F3, F5, F6,
F10, F11, F13
89.9% 94.4% 95.5% 94.9% 94.9% 96.1% 94.4% 93.8% 88.8%
5 0.053
F3, F5, F6, F7,
F11
91.0% 86.0% 93.3% 88.8% 90.4% 87.1% 93.3% 87.6% 77.5%
2
4 0.062 F2, F4, F6, F10 88.2% 88.8% 93.8% 88.2% 93.8% 89.9% 91.6% 90.4% 79.2%
7 0.015
F1, F6, F7, F9,
F11, F12, F13
90.4% 93.8% 98.9% 94.9% 94.9% 95.5% 96.1% 94.9% 69.1%
5 0.025
F2, F6, F7,
F10, F13
92.7% 96.1% 97.2% 94.4% 97.2% 94.9% 97.2% 95.5% 78.7%
6 0.019
F2, F5, F7,
F10, F12, F13
91.6% 94.9% 96.6% 94.9% 95.5% 95.5% 96.1% 95.5% 82.6%
3
4 0.028
F7, F8, F10,
F13
93.3% 93.8% 95.5% 93.8% 94.4% 93.8% 97.2% 93.8% 78.7%
7 0.015
F1, F3, F4, F6,
F7, F12, F13
91.6% 96.1% 97.8% 94.4% 95.5% 97.2% 95.5% 96.1% 77.0%
6 0.027
F1, F3, F6, F7,
F9, F13
89.3% 95.5% 98.3% 93.8% 93.3% 93.3% 94.9% 94.9% 80.9%
4
4 0.111 F2, F6, F9, F13 85.4% 87.1% 87.1% 87.1% 89.3% 87.1% 87.1% 88.8% 71.9%
8 0.015
F1, F2, F3, F6,
F7, F8, F11,
F13
90.4% 95.5% 98.9% 94.9% 94.9% 94.9% 96.6% 94.9% 86.5%
5 0.031
F1, F2, F4,
F12, F13
92.7% 93.3% 93.3% 93.8% 96.1% 94.4% 94.4% 96.1% 84.8%
5
3 0.125 F7, F8, F12 75.3% 82.6% 86.5% 82.6% 83.1% 83.1% 83.7% 83.1% 56.2%
4 0.036 F1, F7, F8, F10 92.7% 92.1% 96.6% 93.3% 94.4% 92.1% 94.9% 93.8% 78.7%
9 0.016
F1, F4, F6, F7,
F8, F10, F11,
F12, F13
88.8% 96.6% 97.8% 97.2% 96.6% 97.2% 97.2% 96.1% 80.3%
6 0.026
F2, F5, F10,
F11, F12, F13
89.3% 93.8% 94.9% 95.5% 96.1% 94.9% 95.5% 95.5% 89.3%
6
2 0.080 F1, F12 88.2% 89.3% 88.2% 88.2% 88.8% 89.3% 87.6% 88.8% 85.4%
7 0.015
F2, F3, F5, F6,
F10, F11, F13
89.9% 94.4% 95.5% 94.9% 94.9% 96.1% 94.4% 93.8% 88.8%
5 0.054
F3, F5, F6, F7,
F11
91.0% 86.0% 93.3% 88.8% 90.4% 87.1% 93.3% 87.6% 77.5%
7
4 0.050 F2, F4, F6, F10 88.2% 88.8% 93.8% 88.2% 93.8% 89.9% 91.6% 90.4% 79.2%
6 0.018
F1, F3, F7, F8,
F10, F12
92.1% 95.5% 97.2% 93.3% 94.9% 95.5% 95.5% 93.8% 89.3%
9 0.002
F1, F3, F4, F5,
F6, F7, F8,
F10, F13
91.6% 98.3% 98.3% 97.8% 97.8% 97.8% 97.2% 97.2% 83.7%
5 0.035
F2, F6, F7,
F10, F13
92.7% 96.1% 97.2% 94.4% 97.2% 94.9% 97.2% 95.5% 78.7%
8
4 0.074
F1, F7, F10,
F13
92.1% 96.6% 96.6% 96.1% 97.2% 94.9% 97.2% 96.1% 83.7%
8 0.013
F1, F3, F5, F8,
F9, F11, F12,
F13
89.3% 96.1% 97.8% 96.6% 96.6% 94.9% 97.2% 94.9% 88.8%
5 0.030
F1, F3, F6,
F12, F13
91.6% 94.9% 93.3% 93.3% 94.9% 93.8% 92.1% 94.9% 82.0%
6 0.022
F4, F6, F8, F9,
F10, F13
93.8% 91.0% 94.9% 93.8% 95.5% 92.7% 91.6% 91.6% 78.1%
9
5 0.026
F2, F6, F10,
F12, F13
93.3% 94.9% 95.5% 93.8% 95.5% 96.1% 94.9% 95.5% 80.3%
8 0.000
F1, F3, F4, F7,
F9, F11, F12,
F13
91.0% 97.2% 100.0% 97.2% 97.8% 98.9% 98.9% 97.8% 82.0%
7 0.018
F1, F4, F5, F9,
F10, F11, F13
91.6% 95.5% 97.2% 98.3% 97.8% 96.1% 95.5% 94.9% 86.5%
10
3 0.111 F3, F11, F12 77.0% 75.3% 80.9% 78.1% 77.5% 72.5% 80.9% 77.5% 77.0%
4 0.031
F1, F3, F11,
F12
91.6% 93.3% 95.5% 92.1% 95.5% 95.5% 95.5% 93.8% 91.0%
9 0.006
F2, F3, F4, F8,
F9, F10, F11,
F12, F13
90.4% 98.3% 97.2% 97.2% 97.2% 98.9% 98.3% 94.9% 91.0%
6 0.016
F2, F7, F9,
F10, F11, F13
92.7% 96.1% 97.2% 95.5% 96.6% 96.1% 96.6% 95.5% 80.3%
All
Features
13 - F1 - F13 88.8% 98.9% 99.4% 97.2% 96.6% 98.3% 96.6% 97.2% 83.7%
Multi-Objective Bees Algorithm for Feature Selection
365
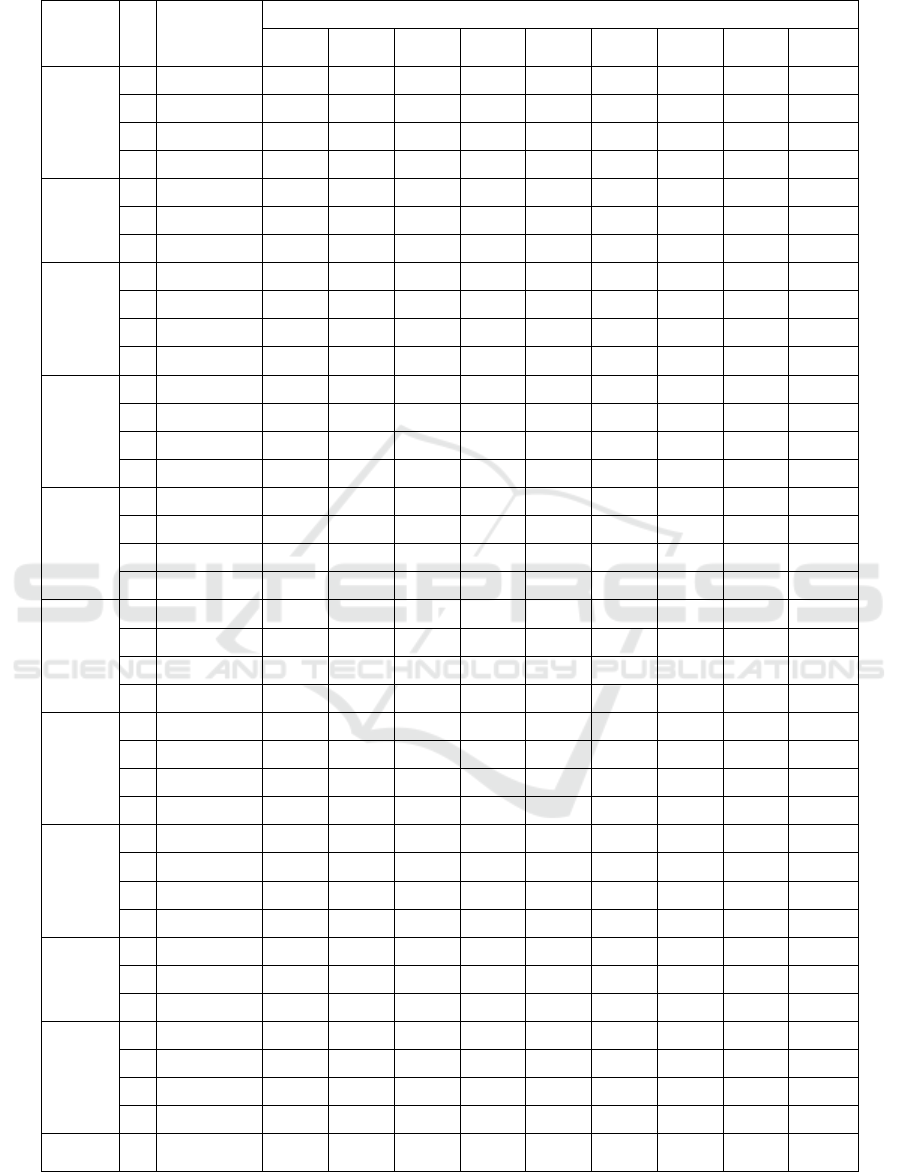
Table 7: MOBA result and classification accuracy on Sonar Dataset.
Run f1 f2
Accuracy
MT LD QD GNB KNB
L-
SVM
Q-
SVM
M-
KNN
Co-
KNN
1
24 0.099570351 74.5% 74.5% 80.8% 67.8% 77.4% 77.4% 82.7% 74.5% 70.2%
28 0.065141175 76.9% 76.4% 83.2% 66.3% 74.5% 75.0% 88.9% 75.5% 67.8%
26 0.082009824 75.0% 73.1% 73.1% 67.8% 76.9% 75.0% 86.1% 73.6% 65.4%
25 0.087133333 69.7% 74.0% 84.6% 66.3% 73.1% 75.5% 80.8% 75.0% 70.2%
2
26 0.119605592 69.2% 69.7% 80.3% 71.2% 78.4% 73.6% 88.9% 78.8% 67.8%
34 0.066256786 66.8% 76.4% 80.8% 63.5% 74.5% 75.0% 85.6% 72.6% 72.1%
27 0.067409799 76.0% 73.6% 79.8% 66.3% 79.8% 75.5% 81.3% 77.4% 70.2%
3
22 0.082342043 67.8% 76.0% 86.5% 68.3% 76.0% 79.8% 83.2% 73.1% 69.2%
30 0.061619488 72.1% 73.6% 77.9% 67.3% 74.5% 71.6% 79.3% 75.5% 69.7%
25 0.073323144 68.8% 72.1% 79.3% 64.9% 75.0% 75.5% 82.2% 78.4% 68.3%
28 0.069419249 77.4% 75.5% 80.8% 65.4% 73.1% 74.5% 81.7% 77.4% 70.2%
4
22 0.156447064 75.0% 74.5% 70.7% 63.9% 70.7% 72.6% 78.8% 74.0% 69.7%
32 0.073525597 74.0% 75.0% 78.4% 65.9% 72.1% 77.9% 85.6% 73.6% 74.0%
26 0.116298846 66.8% 73.1% 81.7% 67.8% 79.3% 76.9% 85.1% 77.4% 69.2%
28 0.081887471 75.5% 75.0% 81.7% 65.4% 74.0% 75.0% 84.1% 75.0% 70.7%
5
22 0.114538244 70.2% 72.6% 73.6% 65.9% 75.0% 75.5% 74.0% 76.4% 71.6%
31 0.062488966 72.6% 75.5% 82.2% 70.7% 76.0% 74.0% 84.1% 76.9% 71.6%
30 0.092796008 70.2% 72.1% 82.2% 67.3% 77.9% 76.0% 82.7% 73.1% 71.6%
26 0.095563806 73.1% 72.6% 81.7% 70.2% 74.0% 76.9% 86.5% 74.5% 66.8%
6
24 0.123323653 68.3% 73.1% 76.4% 72.1% 70.7% 73.6% 81.7% 73.1% 68.8%
33 0.07257117 69.2% 75.0% 82.7% 68.8% 77.4% 77.4% 85.1% 75.0% 68.8%
26 0.080773004 72.1% 73.1% 76.9% 65.4% 72.6% 76.9% 80.8% 73.1% 72.6%
25 0.108421803 70.7% 76.4% 76.4% 70.7% 76.0% 78.8% 84.1% 73.6% 66.3%
7
25 0.106049107 73.1% 76.4% 81.3% 66.8% 79.3% 76.9% 87.0% 77.4% 70.2%
32 0.069255874 72.6% 72.6% 76.0% 70.2% 76.4% 80.3% 86.1% 75.0% 68.3%
29 0.086057013 71.2% 72.6% 78.4% 67.3% 73.1% 74.5% 80.8% 68.3% 66.8%
26 0.098852021 76.9% 75.0% 81.7% 66.3% 75.5% 74.5% 80.8% 71.6% 70.2%
8
22 0.086423399 68.8% 70.7% 81.3% 67.8% 71.6% 72.1% 83.2% 76.0% 66.3%
38 0.06712303 69.7% 73.1% 82.2% 66.8% 78.8% 76.4% 87.5% 73.6% 69.2%
31 0.081604734 67.3% 75.0% 78.4% 67.3% 74.0% 75.5% 80.3% 77.9% 65.9%
32 0.074393338 73.6% 75.0% 80.3% 65.4% 72.6% 72.1% 84.1% 76.4% 72.6%
9
22 0.08962364 73.6% 70.2% 78.4% 66.3% 76.0% 70.2% 79.3% 76.9% 66.3%
30 0.062019176 72.1% 77.4% 79.3% 65.4% 75.5% 78.4% 83.7% 79.8% 69.2%
23 0.089152606 68.3% 70.2% 78.4% 65.4% 73.1% 73.6% 80.8% 74.0% 67.8%
10
25 0.083410711 75.5% 72.1% 76.9% 67.3% 74.0% 75.0% 80.8% 78.8% 73.1%
34 0.071585229 69.7% 74.5% 73.6% 66.8% 73.6% 75.0% 80.8% 75.0% 67.3%
27 0.076225531 62.5% 73.6% 75.5% 70.7% 75.5% 75.0% 79.3% 75.5% 65.9%
26 0.079361732 74.5% 77.4% 80.3% 64.9% 76.9% 80.3% 85.6% 74.0% 70.2%
All
features
60 - 72.6% 76.4% 74.5% 69.2% 76.4% 76.9% 87.0% 72.1% 72.1%
ICE-TES 2021 - International Conference on Emerging Issues in Technology, Engineering, and Science
366

4 CONCLUSIONS
The current work is a wrapper-based Multi-objective
Bees Algorithm (MOBA) for feature selection. The
study aim is to propose the first study of MOBA for
feature selection. The results of four benchmark
datasets confirm earlier research that feature selection
is required to reduce dimensionality and yield
equivalent or superior classification performance.
However, there are limitations and room for
improvement because this is the first MOBA study,
and some issues were not addressed. To begin with,
the optimal parameter for MOBA has not been
considered in this study. Second, the largest feature in
this study is 60 features; thus, this proposed algorithm
has not been tested on larger datasets.
Third, the development of MOBA using basic
combinatorial BA can be improved, for example,
adding the abandonment strategy, which is a strategy
in the standard (continues) BA. Fourth, this study has
not compared with other methods. Future works will
overcome these four limitations.
ACKNOWLEDGEMENTS
The author would like to acknowledge the generous
support from the Indonesia Endowment Fund for
Education (LPDP); Professor DT Pham; and
Industrial Engineering University of Pelita Harapan.
Thank you to the University of Birmingham for
providing flexible resources for intensive
computational work to the University’s research
community through BEAR Cloud Service.
REFERENCES
Albrecht, A. A. (2006). Stochastic local search for the
feature set problem, with applications to microarray
data. Appl. Math. Comput., 183(2), 1148–1164.
Al-Tashi, Q., Abdulkadir, S. J., Rais, H. M., Mirjalili, S., &
Alhussian, H. (2020a). Approaches to multi-objective
feature selection: A systematic literature review. IEEE
Access, 8, 125076–125096.
Al-Tashi, Q., Abdulkadir, S. J., Rais, H. M., Mirjalili, S.,
Alhussian, H., Ragab, M. G., & Alqushaibi, A. (2020b).
Binary multi-objective grey wolf optimiser for feature
selection in classification. IEEE Access, 8, 106247–
106263.
Amoozegar, M., & Minaei-Bidgoli, B. (2018). Optimising
multi-objective PSO based feature selection method
using a feature elitism mechanism. Expert Syst. Appl.,
113, 499–514.
Baptista, F. D., Rodrigues, S., & Morgado-Dias, F. (2013).
Performance comparison of ANN training algorithms
for classification. In 2013 IEEE 8
th
Int. Symp. Intel.
signal processing (pp. 115–120).
Cheng, F., Guo, W., & Zhang, X. (2018). Mofsrank: A
multiobjective evolutionary algorithm for feature
selection in learning to rank. Complexity, 2018.
Das, A., & Das, S. (2017). Feature weighting and selection
with a pareto-optimal trade-off between relevancy and
redundancy. Pattern Recogn. Lett., 88, 12–19.
Dashtban, M., Balafar, M., & Suravajhala, P. (2018). Gene
selection for tumor classification using a novel bio-
inspired multi-objective approach. Genomics, 110(1),
10–17.
De la Hoz, E., De La Hoz, E., Ortiz, A., Ortega, J., &
Martınez-Alvarez, A. (2014). Feature´ selection by
multi-objective optimisation: Application to network
anomaly detection by hierarchical self-organising
maps. Knowledge-Based Systems, 71, 322–338.
Deniz, A., Kiziloz, H. E., Dokeroglu, T., & Cosar, A.
(2017). Robust multiobjective evolutionary feature
subset selection algorithm for binary classification
using machine learning techniques. Neurocomputing,
241, 128–146.
Gheyas, I. A., & Smith, L. S. (2010). Feature subset
selection in large dimensionality domains. Pattern
Recogn., 43(1), 5–13.
Gonzalez, J., Ortega, J., Damas, M., Martın-Smith, P., &
Gan, J. Q. (2019). A new multiobjective wrapper
method for feature selection–accuracy and stability
analysis for bci. Neurocomputing, 333, 407–418.
Hammami, M., Bechikh, S., Hung, C.-C., & Said, L. B.
(2019). A multi-objective hybrid filter-wrapper
evolutionary approach for feature selection. Memet.
Comput., 11(2), 193–208.
Han, M., & Ren, W. (2015). Global mutual information-
based feature selection approach using single-objective
and multi-objective optimisation. Neurocomputing,
168, 47–54.
Hancer, E., Xue, B., Zhang, M., Karaboga, D., & Akay, B.
(2018). Pareto front feature selection based on artificial
bee colony optimisation. Inform. Sciences, 422, 462–
479.
Hu, Y., Zhang, Y., & Gong, D. (2020). Multiobjective
Particle Swarm Optimisation for feature selection with
fuzzy cost. IEEE T.Cybernetics.
Hussein, W. A., Sahran, S., & Abdullah, S. N. H. S. (2017).
The variants of the Bees Algorithm (BA): A Survey.
Artif. Intell. Rev., 47(1), 67–121.
Ismail, A. H., Hartono, N., Zeybek, S., Caterino, M., &
Jiang, K. (2021). Combinatorial Bees Algorithm for
Vehicle Routing Problem. In Macromol. Sy., 396,
2000284.
Ismail, A. H., Hartono, N., Zeybek, S., & Pham, D. T.
(2020). Using the Bees Algorithm to solve
Combinatorial Optimisation Problems for TSPLIB. In
IOP Conf. Ser-Mat SCI., 847, 012027.
Jha, K., & Saha, S. (2021). Incorporation of multimodal
multiobjective optimisation in designing a filter based
Multi-Objective Bees Algorithm for Feature Selection
367

feature selection technique. Appl. Soft Comput., 98,
106823.
Jimenez, F., S´ anchez, G., Garcia, J. M., Sciavicco, G., &
Miralles, L. (2017). Multi-objective evolutionary
feature selection for online sales forecasting.
Neurocomputing, 234, 75– 92.
Karagoz, G. N., Yazici, A., Dokeroglu, T., & Cosar, A.
(2021). A new framework of multiobjective
evolutionary algorithms for feature selection and multi-
label classification of video data. Int. J. Mach. Learn
Cyb., 12(1), 53–71.
Khammassi, C., & Krichen, S. (2020). A NSGA2-lr
wrapper approach for feature selection in network
intrusion detection. Comput. Netw., 172, 107183.
Khan, A., & Baig, A. R. (2015). Multi-objective feature
subset selection using nondominated sorting genetic
algorithm. J. Appl. Res Technol., 13(1), 145–159.
Kimovski, D., Ortega, J., Ortiz, A., & Banos, R. (2015).
Parallel alternatives for evolutionary multi-objective
optimisation in unsupervised feature selection. Expert
Syst. Appl., 42(9), 4239–4252.
Kiziloz, H. E., Deniz, A., Dokeroglu, T., & Cosar, A.
(2018). Novel multiobjective TLBO algorithms for the
feature subset selection problem. Neurocomputing,
306, 94–107.
Koc, E. (2010). Bees Algorithm: theory, improvements and
applications. Cardiff University.
Kou, G., Xu, Y., Peng, Y., Shen, F., Chen, Y., Chang, K.,
& Kou, S. (2021). Bankruptcy prediction for SMEs
using transactional data and two-stage multiobjective
feature selection. Decis. Support Syst., 140, 113429.
Kozodoi, N., Lessmann, S., Papakonstantinou, K.,
Gatsoulis, Y., & Baesens, B. (2019). A multi-objective
approach for profit-driven feature selection in credit
scoring. Decis. Support Syst., 120, 106–117.
Kundu, P. P., & Mitra, S. (2015). Multi-objective
optimisation of shared nearest neighbor similarity for
feature selection. Appl. Soft Comput., 37, 751–762.
Lai, C.-M. (2018). Multi-objective simplified swarm
optimisation with weighting scheme for gene selection.
Appl. Soft Comput., 65, 58–68.
Mlakar, U., Fister, I., Brest, J., & Potocnik, B. (2017).
Multi-objective differential evolution for feature
selection in facial expression recognition systems.
Expert Syst. Appl., 89, 129–137.
Mukhopadhyay, A., & Maulik, U. (2013). An svm-wrapped
multiobjective evolutionary feature selection approach
for identifying cancer-microrna markers. IEEE T.
Nanobiosci., 12(4), 275–281.
Nayak, S. K., Rout, P. K., Jagadev, A. K., & Swarnkar, T.
(2020). Elitism based multiobjective differential
evolution for feature selection: A filter approach with
an efficient redundancy measure. Journal of King Saud
University-Computer and Information Sciences, 32(2),
174–187.
Peimankar, A., Weddell, S. J., Jalal, T., & Lapthorn, A. C.
(2017). Evolutionary multiobjective fault diagnosis of
power transformers. Swarm Evol. Comput., 36, 62–75.
Pham, D., Ghanbarzadeh, A., Koc, E., Otri, S., Rahim, S.,
& Zaidi, M. (2005). The Bees Algorithm. Technical
Note, Cardiff University, UK.
Pham, D., Mahmuddin, M., Otri, S., & Al-Jabbouli, H.
(2007). Application of the Bees Algorithm to the
selection features for manufacturing data. In
International virtual conference on intelligent
production machines and systems (IPROMS 2007).
Ramlie, F., Muhamad, W. Z. A. W., Jamaludin, K. R.,
Cudney, E., & Dollah, R. (2020). A significant feature
selection in the mahalanobis taguchi system using
modified-bees algorithm. Int. J. Eng. Research and
Technology, 13(1), 117-136.
Rathee, S., & Ratnoo, S. (2020). Feature selection using
multi-objective CHC Genetic Algorithm. Procedia
Computer Science, 167, 1656–1664.
Rodrigues, D., de Albuquerque, V. H. C., & Papa, J. P.
(2020). A multi-objective Artificial Butterfly
Optimisation approach for feature selection. Appl. Soft
Comput., 94, 106442.
Rostami, M., Forouzandeh, S., Berahmand, K., & Soltani,
M. (2020). Integration of multiobjective PSO based
feature selection and node centrality for medical
datasets. Genomics, 112(6), 4370–4384.
Sahoo, A., & Chandra, S. (2017). Multi-objective Grey
Wolf optimiser for improved cervix lesion
classification. Appl. Soft Comput., 52, 64–80.
Sharma, A., & Rani, R. (2019). C-HMOSHSSA: Gene
selection for cancer classification using multi-objective
meta-heuristic and machine learning methods. Comput.
Meth. Prog. Bio., 178, 219–235.
Sohrabi, M. K., & Tajik, A. (2017). Multi-objective feature
selection for warfarin dose prediction. Comput. Biol.
Chem., 69, 126–133.
Tan, C. J., Lim, C. P., & Cheah, Y.-N. (2014). A multi-
objective evolutionary algorithmbased ensemble
optimiser for feature selection and classification with
neural network models. Neurocomputing, 125, 217–
228.
Vignolo, L. D., Milone, D. H., & Scharcanski, J. (2013).
Feature selection for face recognition based on multi-
objective evolutionary wrappers. Expert Syst. Appl.,
40(13), 5077–5084.
Wang, X.-h., Zhang, Y., Sun, X.-y., Wang, Y.-l., & Du, C.-
h. (2020). Multi-objective feature selection based on
Artificial Bee Colony: An acceleration approach with
variable sample size. Appl. Soft Comput., 88, 106041.
Wang, Z., Li, M., & Li, J. (2015). A multi-objective
evolutionary algorithm for feature selection based on
mutual information with a new redundancy measure.
Inform. Sciences, 307, 73–88.
Xia, H., Zhuang, J., & Yu, D. (2014). Multi-objective
unsupervised feature selection algorithm utilising
redundancy measure and negative epsilon-dominance
for fault diagnosis. Neurocomputing, 146, 113–124.
Xue, B., Cervante, L., Shang, L., Browne, W. N., & Zhang,
M. (2012). A multi-objective Particle Swarm
Optimisation for filter-based feature selection in
classification problems. Connect. Sci., 24(2-3), 91–116.
ICE-TES 2021 - International Conference on Emerging Issues in Technology, Engineering, and Science
368

Xue, B., Cervante, L., Shang, L., Browne, W. N., & Zhang,
M. (2013a). Multi-objective evolutionary algorithms
for filter based feature selection in classification. Int. J.
Artif. Intell. T., 22(04), 1350024.
Xue, B., Zhang, M., & Browne, W. N. (2013b). Particle
Swarm Optimisation for feature selection in
classification: A multi-objective approach. IEEE T.
Cybernetics, 43(6), 1656–1671.
Xue, B., Zhang, M., & Browne, W. N. (2015). A
comprehensive comparison on evolutionary feature
selection approaches to classification. Int. J. Comp. Int.
Appl., 14(02), 1550008.
Yong, Z., Dun-wei, G., & Wan-qiu, Z. (2016). Feature
selection of unreliable data using an improved multi-
objective PSO algorithm. Neurocomputing, 171, 1281–
1290.
Yuce, B., Packianather, M. S., Mastrocinque, E., Pham, D.
T., & Lambiase, A. (2013). Honey bees inspired
optimisation method: The Bees Algorithm. Insects,
4(4), 646–662.
Zeybek, S., Ismail, A. H., Hartono, N., Caterino, M., &
Jiang, K. (2021). An Improved Vantage Point Bees
Algorithm to solve Combinatorial Optimisation
Problems from TSPLIB. In Macromol. Sy., 396,
2000299.
Zeybek, S., & Koc¸, E. (2015). The Vantage Point Bees
Algorithm. In 2015 7th International joint conference
on computational intelligence (IJCCI) 1, 340–345.
Zhang, Y., Gong, D.-w., Gao, X.-z., Tian, T., & Sun, X.-y.
(2020). Binary differential evolution with self-learning
for multi-objective feature selection. Inform. Sciences,
507, 67– 85.
Zhang, Y., Gong, D.-w., Sun, X.-y., & Guo, Y.-n. (2017).
A PSO-based multi-objective multilabel feature
selection method in classification. Sci. Rep-UK, 7(1),
1–12.
Zhu, Y., Liang, J., Chen, J., & Ming, Z. (2017). An
improved NSGA-III algorithm for feature selection
used in intrusion detection. Knowledge-Based Systems,
116, 74–85.
Multi-Objective Bees Algorithm for Feature Selection
369
