
Machine Learning for Colorectal Cancer Risk Prediction: Systematic
Review
Noura Qarmiche
1 a
, Mehdi Chrifi Alaoui
2 b
,Nada Otmani
3 c
,Samira El Fakir
3d
, Nabil
Tachfouti
3e
, Hind Bourkhime
3f
, Mohammed Omari
3g
, Karima El Rhazi
3h
and Nour El Houda Chaoui
1i
1
Laboratory of Artificial Intelligence, Data Science and Emerging Systems, National School of Applied Sciences Fez, Sidi
Mohamed Ben Abdellah University, Fez, Morocco
2
Laboratory of modelling and mathematical structures, Faculty of Science and Technology fez, Morocco
3
Department of Epidemiology, Clinical Research and Community Health, Faculty of Medicine and Pharmacy of Fez, Sidi
Mohamed Ben Abdellah University, Fez, Morocco
samira.elfakir@usmba.ac.ma, nabil.tachfouti@usmba.ac.ma, hbourkhime@gmail.com, mohammed.omari@usmba.ac.ma,
karima.elrhazi@usmba.ac.ma, houda.chaoui@usmba.ac.ma
Keywords: Machine Learning, Risk, Risk Factor, Risk Assessment, Susceptibility, Prediction, Score, Model, Colorectal
Cancer, Systematic Review
Abstract: Colorectal cancer is one of the world's top five diseases and causes death from cancer. Survival is closely
related to the stage at diagnosis and population-based screening reduces colorectal cancer incidence, and
mortality. Machine learning algorithms have been used to develop risk prediction models in colorectal cancer.
This study reported a systematic review of studies reporting the development of a machine learning model to
predict the risk of colorectal cancer. We performed research on Scopus, Science direct, and web of science
Library. We included original articles reporting or validating machine learning models predicting colorectal
cancer risk, published between 2015 and 2021. We identified nine articles related to eleven distinct models;
three models considered genetic factors only; two models required clinical assessment; the remaining models
are based on nutrition, demographic and lifestyle features. Models were validated by computing accuracy,
sensitivity and air under the roc curve. The most used algorithms are neural networks and logistic regression.
Machine learning models have shown promising performance for colorectal cancer risk prediction. However,
they need to be improved for easy and safe clinical practice use.
1 INTRODUCTION
Colorectal cancer (CRC) is the third most common
cancer among men and the second among women
worldwide, with an estimated 1.8 million new cases
and 881,000 CRC-related deaths per year (Bray et al.,
2018). Survival is strongly related to the stage at
a
https://orcid.org/0000-0002-1786-5049
b
https://orcid.org/0000-0002-6822-847X
c
https://orcid.org/0000-0001-5093-9049
d
https://orcid.org/0000-0003-1623-6176
e
https://orcid.org/0000-0001-7726-9700
f
https://orcid.org/0000-0002-5772-5534
g
https://orcid.org/0000-0003-1289-6206
h
https://orcid.org/0000-0002-8135-9044
i
https://orcid.org/0000-0002-4228-035X
diagnosis (Usher-Smith et al., 2016), and population-
based screening has been shown to significantly
reduce colorectal cancer incidence and mortality.
In addition to the demonstrated benefit of
screening on CRC- related mortality, multiple health
economic models suggest that screening is cost-
effective (Ma & Ladabaum, 2014). Technological
Qarmiche, N., Chrifi Alaoui, M., Otmani, N., El Fakir, S., Tachfouti, N., Bourkhime, H., Omari, M., El Rhazi, K. and Chaoui, N.
Machine Learning for Colorectal Cancer Risk Prediction: Systematic Review.
DOI: 10.5220/0010738100003101
In Proceedings of the 2nd International Conference on Big Data, Modelling and Machine Learning (BML 2021), pages 507-511
ISBN: 978-989-758-559-3
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
507

developments, specifically in statistics and computer
science, have helped to predict the colorectal cancer
risk. A number of models have been developed based
on demographic, life-style, clinical and genetic risk
factors. The application of machine learning
techniques has greatly contributed to improve cancer
prediction. Indeed, many studies have shown that
machine learning models have better accuracy than
statistical models. Our objective is to review existing
machine learning models for colorectal cancer risk
prediction.
2 METHODS
A systematic review, following the recommendations
of the PRISMA 2020 statement(The PRISMA 2020
statement: An updated guideline for reporting
systematic reviews | The EQUATOR Network, s. d.),
was conducted to identify studies reporting the
development of a machine learning model to predict
colorectal cancer risk.
2.1 Search Strategy and Information
Sources
We conducted an exhaustive electronic literature
search for English literature studies of Scopus,
science direct, and web of science from January 2015
to April 2021.
The search strategy was the following: “Machine
learning” AND “Colorectal cancer” AND (Risk OR
“Risk factors” OR “Risk assessment” OR
susceptibility) AND (Model OR Score OR
Prediction). Reference lists of articles included in this
systematic review were consulted to identify more
studies.
2.2 Inclusion and Exclusion Criteria
Articles were included based on passing all the
selection criteria:
Original paper published in a peer- reviewed
journal;
Described the development and validation of
machine learning risk factors for colorectal
cancer
The full article can be obtained in English
One reviewer conducted the search and initially
screened by title and then by abstract to reject
inappropriate articles. Two reviewers independently
examined a random sample of 5% of the articles. Full
text was read for articles whose titles and abstracts
were not sufficient to exclude or include them. The
full text was read for articles whose titles and
abstracts were not sufficient to exclude or include
them. When the decision was still difficult to make,
the articles were discussed by a committee.
Image-based models and statistical models such
as Cox model were excluded.
2.3 Data Extraction
Data extraction was based on the characteristics of
each model: general study characteristics, country,
and year of publication, applied algorithms, model
prediction parameters, model performance,
sensitivity and specificity.
2.4 Data Synthesis and Analysis
In view of the heterogeneity of the existing models
and their small number, we limited to a qualitative
and narrative synthesis method.
3 RESULTS
3.1 Search Results
The literature search returned 426 studies. We
excluded duplicate studies from Scopus, Web of
Science, and Science Direct. We eliminated studies
that did not satisfy our inclusion criteria. We reserved
10 studies for analysis by reading the full text. One
study was rejected for being a statistical model.
Finally, nine articles were included in this review.
The PRISMA diagram for the systematic review
process is illustrated in Figure 1
3.2 Model Development and Validation
Table 1 shows the details of the development of each
model.
The machine learning algorithms used to develop
the 11 risk assessment models were logistic
regression in four (Birks et al., 2017); (Jeon et al.,
2018), Ensemble of decision trees in one(Schneider et
al., 2020), decision trees in one (Hornbrook et al.,
2017) CNN in one(Wang et al., 2019), Gradient
Boosting + random forest in one(Kinar et al., 2016),
ANN in one(Nartowt et al., 2019), Transformer in one
(Amadeus et al., 2021) and Penalized regression+
XGboost in one (Thomas et al., 2020)
BML 2021 - INTERNATIONAL CONFERENCE ON BIG DATA, MODELLING AND MACHINE LEARNING (BML’21)
508
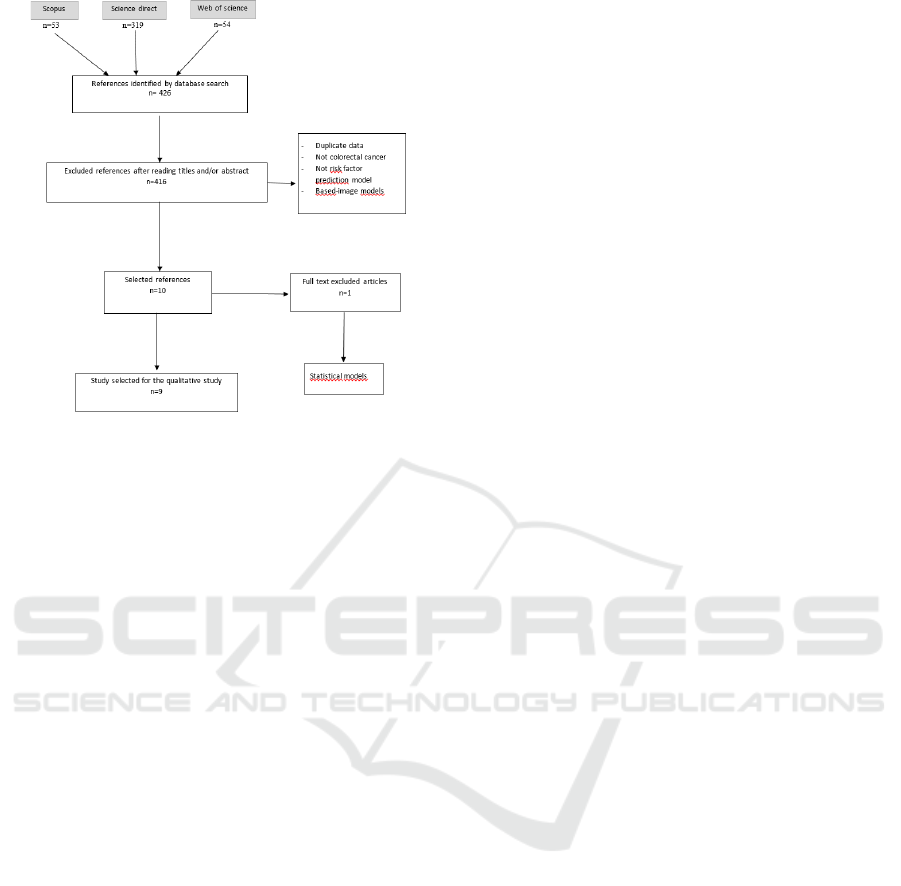
Figure 1: Flow diagram of process of systematic literature
search following PRISMA guidelines
3.3 Study Populations
All included studies used case-control studies to train
the models. Five studies were performed in the United
States (Hornbrook et al., 2017; Jeon et al., 2018;
Nartowt et al., 2019; Schneider et al., 2020; Thomas
et al., 2020), one in Taiwan (Wang et al., 2019), one
in Israël (Kinar et al., 2016), one in the UK(Birks et
al., 2017) and one in Indonesia(Amadeus et al., 2021)
(Table 1).
3.4 Features Selection
Three models used only genetic marker (Amadeus et
al., 2021; Jeon et al., 2018; Thomas et al., 2020), four
models combined laboratory and demographic data,
one model combined lifestyle and family history, one
model used comorbidities and medications history,
one model combined demographics, lifestyle and
medical history and one model combined genetic
marker, lifestyle and family history.
3.5 Models Discrimination
Discrimination, as measured by the Area Under the
Curve (AUC), was reported for 9 of the risk models;
these values were between 0,59 (Jeon et al., 2018) and
0.922(Wang et al., 2019)
3.6 Models Sensitivity and Specificity
Sensitivity is reported for only four models
(Schneider et al., 2020; Wang et al., 2019; Nartowt et
al., 2019; and Birks et al., 2017). These values were
(0.354; 0.837; 0.60 and 0.955) respectively.
Specificity is reported for five models, the values
were between 0,088 (Birks et al., 2017) and 0.99
(Hornbrook et al., 2017)
4 DISCUSSION
To the best of our knowledge, this is the first
systematic review of the machine learning models in
colorectal cancer risk prediction.
It shows that 11 risk models exist for predicting
the risk of developing CRC in asymptomatic
populations. The best discrimination and specificity
model (0.922; 0.837 respectively) were reported for
Wang, 2019.
Given the heterogeneity of the models, we opted
for a qualitative analysis of the results.
This systematic review revealed the need to
standardize and validate the various existing
colorectal cancer prediction models. In order to
implement these models in clinical practice,
interactive and user-friendly frameworks need to be
developed by experts. We have found a lack of
African-developed models; given the continent's
unique biology, nutrition and lifestyle. The
development and validation of an African model is
strongly recommended
5 CONCLUSION
This systematic review revealed the existence of 11
machine learning models for the prediction of
colorectal cancer. The performance of these models
is good, however, further research is necessary before
they could be applied to routine clinical practice.
Machine Learning for Colorectal Cancer Risk Prediction: Systematic Review
509
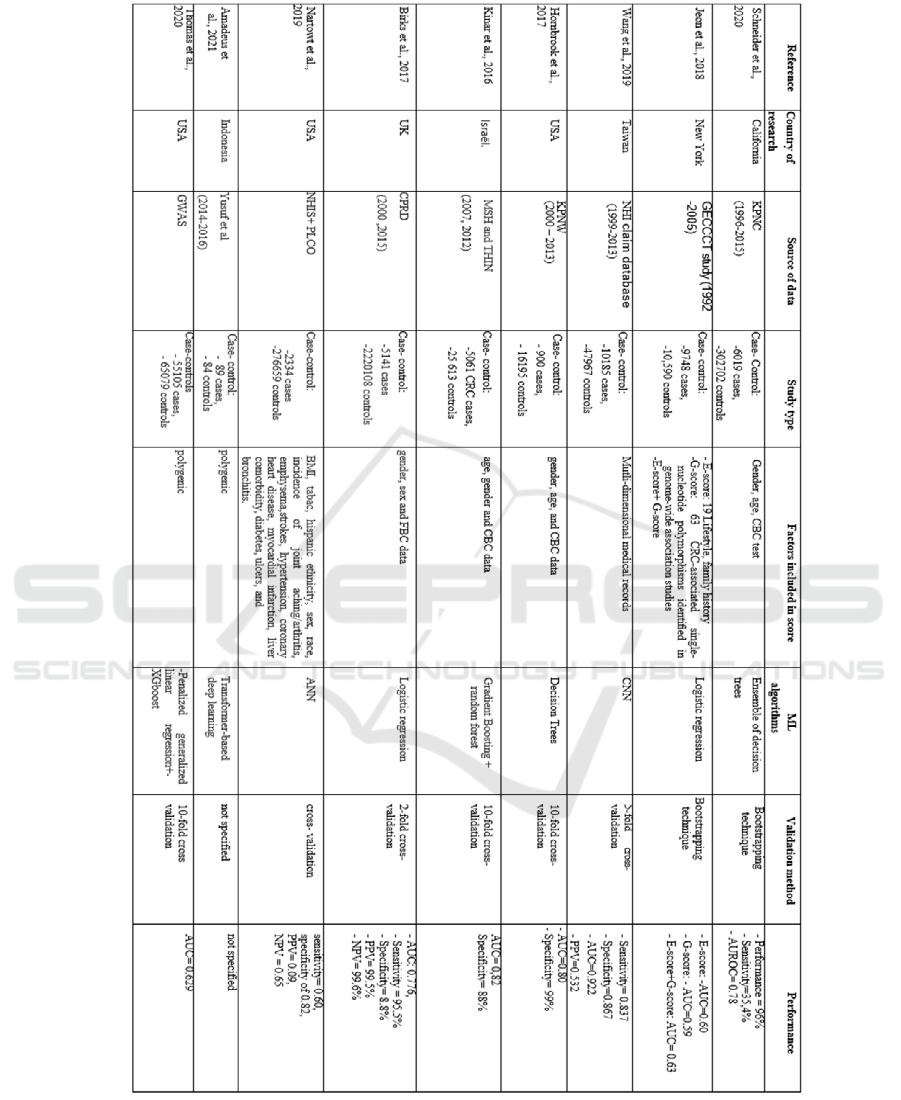
Table 1: Synthesis of the nine studies includes
BML 2021 - INTERNATIONAL CONFERENCE ON BIG DATA, MODELLING AND MACHINE LEARNING (BML’21)
510

REFERENCES
Amadeus, S., Cenggoro, T. W., Budiarto, A., &
Pardamean, B. (2021). A Design of Polygenic Risk
Model with Deep Learning for Colorectal Cancer in
Multiethnic Indonesians. Procedia Computer Science,
179, 632‑639.
https://doi.org/10.1016/j.procs.2021.01.049.
Birks, J., Bankhead, C., Holt, T. A., Fuller, A., & Patnick,
J. (2017). Evaluation of a prediction model for
colorectal cancer : Retrospective analysis of 2.5
million patient records. Cancer Medicine, 6(10),
2453‑2460. https://doi.org/10.1002/cam4.1183
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L.,
Torre, L. A., & Jemal, A. (2018). Global cancer
statistics 2018 : GLOBOCAN estimates of incidence
and mortality worldwide for 36 cancers in 185
countries. CA: A Cancer Journal for Clinicians, 68(6),
394‑424. https://doi.org/10.3322/caac.21492
Hornbrook, M. C., Goshen, R., Choman, E., O’Keeffe-
Rosetti, M., Kinar, Y., Liles, E. G., & Rust, K. C.
(2017). Early Colorectal Cancer Detected by Machine
Learning Model Using Gender, Age, and Complete
Blood Count Data. Digestive Diseases and Sciences,
62(10), 2719‑2727. https://doi.org/10.1007/s10620-
017-4722-8
Jeon, J., Du, M., Schoen, R. E., Hoffmeister, M.,
Newcomb, P. A., Berndt, S. I., Caan, B., Campbell, P.
T., Chan, A. T., Chang-Claude, J., Giles, G. G., Gong,
J., Harrison, T. A., Huyghe, J. R., Jacobs, E. J., Li, L.,
Lin, Y., Le Marchand, L., Potter, J. D., … Hsu, L.
(2018). Determining Risk of Colorectal Cancer and
Starting Age of Screening Based on Lifestyle,
Environmental, and Genetic Factors.
Gastroenterology, 154(8), 2152-2164.e19.
https://doi.org/10.1053/j.gastro.2018.02.021
Kinar, Y., Kalkstein, N., Akiva, P., Levin, B., Half, E. E.,
Goldshtein, I., Chodick, G., & Shalev, V. (2016).
Development and validation of a predictive model for
detection of colorectal cancer in primary care by
analysis of complete blood counts : A binational
retrospective study. Journal of the American Medical
Informatics Association, 23(5), 879‑890.
https://doi.org/10.1093/jamia/ocv195
Ma, G. K., & Ladabaum, U. (2014). Personalizing
Colorectal Cancer Screening : A Systematic Review
of Models to Predict Risk of Colorectal Neoplasia.
Clinical Gastroenterology and Hepatology, 12(10),
1624-1634.e1.
https://doi.org/10.1016/j.cgh.2014.01.042
Nartowt, B., Hart, G. R., Muhammad, W., Liang, Y., &
Deng, J. (2019). A Model of Risk of Colorectal
Cancer Tested between Studies : Building Robust
Machine Learning Models for Colorectal Cancer Risk
Prediction. International Journal of Radiation
Oncology, Biology, Physics, 105(1), E132.
https://doi.org/10.1016/j.ijrobp.2019.06.2265
Schneider, J. L., Layefsky, E., Udaltsova, N., Levin, T.
R., & Corley, D. A. (2020). Validation of an
Algorithm to Identify Patients at Risk for Colorectal
Cancer Based on Laboratory Test and Demographic
Data in Diverse, Community-Based Population.
Clinical Gastroenterology and Hepatology, 18(12),
2734-2741.e6.
https://doi.org/10.1016/j.cgh.2020.04.054
The PRISMA 2020 statement : An updated guideline for
reporting systematic reviews | The EQUATOR
Network. (s. d.). Consulté 29 juillet 2021, à l’adresse
https://www.equator-network.org/reporting-
guidelines/prisma/
Thomas, M., Sakoda, L. C., Hoffmeister, M., Rosenthal,
E. A., Lee, J. K., van Duijnhoven, F. J. B., Platz, E.
A., Wu, A. H., Dampier, C. H., de la Chapelle, A.,
Wolk, A., Joshi, A. D., Burnett-Hartman, A., Gsur,
A., Lindblom, A., Castells, A., Win, A. K., Namjou,
B., Van Guelpen, B., … Hsu, L. (2020). Genome-
wide Modeling of Polygenic Risk Score in Colorectal
Cancer Risk. American Journal of Human Genetics,
107(3), 432‑444.
https://doi.org/10.1016/j.ajhg.2020.07.006
Usher-Smith, J. A., Walter, F. M., Emery, J. D., Win, A.
K., & Griffin, S. J. (2016). Risk Prediction Models for
Colorectal Cancer : A Systematic Review. Cancer
Prevention Research, 9(1), 13‑26.
https://doi.org/10.1158/1940-6207.CAPR-15-0274
Wang, Y.-H., Nguyen, P.-A., Islam, M. M., Li, Y.-C., &
Yang, H.-C. (2019). Development of Deep Learning
Algorithm for Detection of Colorectal Cancer in EHR
Data. Studies in Health Technology and Informatics,
264, 438‑441. https://doi.org/10.3233/SHTI190259
Machine Learning for Colorectal Cancer Risk Prediction: Systematic Review
511
