
Neuro-symbolic XAI for Computational Drug Repurposing
Martin Dranc
´
e
a
, Marina Boudin, Fleur Mougin
b
and Gayo Diallo
c
Inserm U1219, Bordeaux Population Health Research Center, Team ERIAS, University of Bordeaux, France
Keywords:
Artificial Intelligence, XAI, Drug Repurposing, Knowledge Graph, Bioinformatics.
Abstract:
Today in the health domain, the challenge is to build a more transparent artificial intelligence, less affected by
the opacity intrinsic to the mathematical concepts it uses. Among the fields which use AI techniques, is drug
development, and more specifically drug repurposing. DR involves finding a new indication for an existing
drug. The hypotheses generated by DR techniques must be validated. Therefore, the mechanism of generation
must be understood. In this paper, we describe the use of a state-of-the-art neuro-symbolic algorithm in order
to explain the process of link prediction in a knowledge graph-based computational drug repurposing. Link
prediction consists of generating hypotheses about the relationships between a known molecule and a given
target. More specifically, the implemented approach allows to understand how the organization of data in a
knowledge graph changes the quality of predictions.
1 INTRODUCTION
Today, it costs about 1.5 billion dollars and about fif-
teen years of research to develop a new drug (DiMasi
et al., 2016). These constraints are limiting factors that
do not encourage the pharmaceutical industry, which
is primarily concerned with cost-effectiveness. In par-
ticular, they do not invest in the development of new
drugs for so-called rare diseases. In this context, over
the last decade, drug repurposing (DR), which con-
sists of searching for a new indication for an exist-
ing drug (link between an existing molecule and a
given indication), has been attracting growing inter-
est. Advances in Artificial Intelligence (AI) methods
have made it possible to develop AI-based computa-
tional DR with very encouraging results (Ashburn and
Thor, 2004)(Stokes et al., 2020).
However, the use of “black-box” models, which
are completely opaque to the understanding of their
users (biomedical experts, clinicians, etc.), is currently
one of the main obstacles to the use of AI in the health
field. In this field indeed, AI should not be seen as
a decision maker per-se, but rather as a transparent
support to the users’ decision process. Recently, new
explainable AI (XAI) models, referred to as neuro-
symbolic models, have been developed. These latter
AI models are transparent by nature, i.e. users can
a
https://orcid.org/0000-0001-6365-531X
b
https://orcid.org/0000-0002-7436-3010
c
https://orcid.org/0000-0002-9799-9484
Table 1: Type and number of entities (nodes) in the
OREGANO KG.
Type Number
ATC Code 1,005
Drug 42,856
Gene 35,602
Effect 153
Target 254,289
Disease 8,997
Pathway 297
Activity 74
Phenotype 11,202
Indication 2,154
Side Effect 5,556
find and understand the mechanisms leading to a pre-
diction without having to modify, simulate or process
any information about the model’s operation itself.
Neuro-symbolic models combine symbolic and statis-
tical learning (Das et al., 2018). This combination al-
lows for robust predictions made by a neural network,
supported by the transparency provided by the use of
logical rules, understandable by humans. Among the
many advantages of using these neuro-symbolic mod-
els, a major benefit is the possibility of interaction be-
tween the model and users during the learning pro-
cess. Indeed, models that provide an explanation of
how they work can more easily be adjusted in the event
that some bias is detected.
220
Drancé, M., Boudin, M., Mougin, F. and Diallo, G.
Neuro-symbolic XAI for Computational Drug Repurposing.
DOI: 10.5220/0010714100003064
In Proceedings of the 13th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2021) - Volume 2: KEOD, pages 220-225
ISBN: 978-989-758-533-3; ISSN: 2184-3228
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All r ights reser ved

These models are now being applied to knowl-
edge graphs (KGs) in the context of DR. According
to (Paulheim, 2017), a KG has the following charac-
teristics: (i) it describes real-world entities and their
interactions within a graph, (ii) it defines classes and
relationships between these entities in the form of a
schema, (iii) it allows entities to be arbitrarily linked
together, and (iv) it can aggregate information from
different domains. These data structures are well
suited for describing biomedical information, as they
are usually derived from multiple databases and KGs
are perfect for maintaining the semantic relationship
between entities. Recently, as proved by (Himmel-
stein et al., 2017) and (Zhang et al., 2021), KG-based
DR has demonstrated interesting results through the
use of AI models and algorithms to predict new links
between existing entities in KGs. Unfortunately, the
lack of transparency of the models prevents the results
from being used more widely by experts in the field of
DR, including biochemists and clinicians.
The current study is part of the OREGANO project
which aims to provide an end-to-end framework for
KG-based computational DR (Boudin, 2020). Specif-
ically, we describe the exploitation of a state-of-
the-art neuro-symbolic model and algorithm in the
OREGANO workflow to explain the results of the
link prediction process. Link prediction here consists
of generating hypotheses about the relationships be-
tween a known molecule and a given target. To do
this, we used PoLo (Policy-guided walks with Logi-
cal rules) (Liu et al., 2021), a neuro-symbolic model
achieving state-of-the-art results in KG link predic-
tion. It has already been used in the context of the
Hetionet KG (Himmelstein et al., 2017) to repurpose
drugs. In our work, we particularly investigated how
the structure of data in a KG changes the quality of
predictions.
2 MATERIALS AND METHODS
2.1 Data
The data that are used to develop the OREGANO KG
have been previously collected in the context of the
work described in (Boudin, 2020). They come from
various free and/or open source knowledge bases and
Linked Open Data. They have been then curated and
integrated into the OREGANO KG depicted in Fig-
ure 1. Currently, this graph has a total of 362,186
nodes of 11 different types and 802,949 relations of
19 different types. The detailed number of nodes per
data type can be found in Table 1 while the number of
relations (edges) is provided in Table 2.
Table 2: Type and number of relations (edges) in the
OREGANO KG.
Type Number
associated to 250,000
causes condition 8,584
decreases activity 2,446
decreases effect 102
decreases efficacy 106,558
has activity 5,565
has code 3,150
has effect 11,251
has indication 8,307
has phenotype 76,177
has side effect 129,330
has target 50,132
increases activity 6,636
increases effect 10,041
increases efficacy 34,071
is affecting 5,341
is substance that treats 60,889
participates in 24,919
2.2 Link Prediction
PoLo (Liu et al., 2021) is an algorithm that was pro-
posed in March 2021 to perform link prediction from
a KG. It is one of the only fully neuro-symbolic algo-
rithms that relies as much on mathematical methods as
on the use of logical rules from the information con-
tained in the KG on which it operates. Its operation
is inspired by MINERVA (Das et al., 2018), both of
which are based on the use of reinforcement learning
in order to optimize the movements of an agent in the
graph and thus make link prediction. From an initial
entity (here, a drug to be repurposed), an agent moves
in the graph from node to node. The decision of which
node to choose for the next move is determined by a
policy set by an long short-term memory (Hochreiter
and Schmidhuber, 1997). The transitions between the
nodes accumulate and form a path that represents a
chain of reasoning. These iterations are repeated a fi-
nite number of times, until the agent obtains a reward,
based on the path taken and the associated prediction.
This learning task is modeled as a Markov decision
process.
2.3 Logical Rules
PoLo uses meta-paths, in the form of Horn rules, to
evaluate the gain provided to the agent according to
its predictions. In order to work, PoLo needs a set
of meta-paths that are considered reliable and that de-
scribe possible paths between the entities to be re-
Neuro-symbolic XAI for Computational Drug Repurposing
221
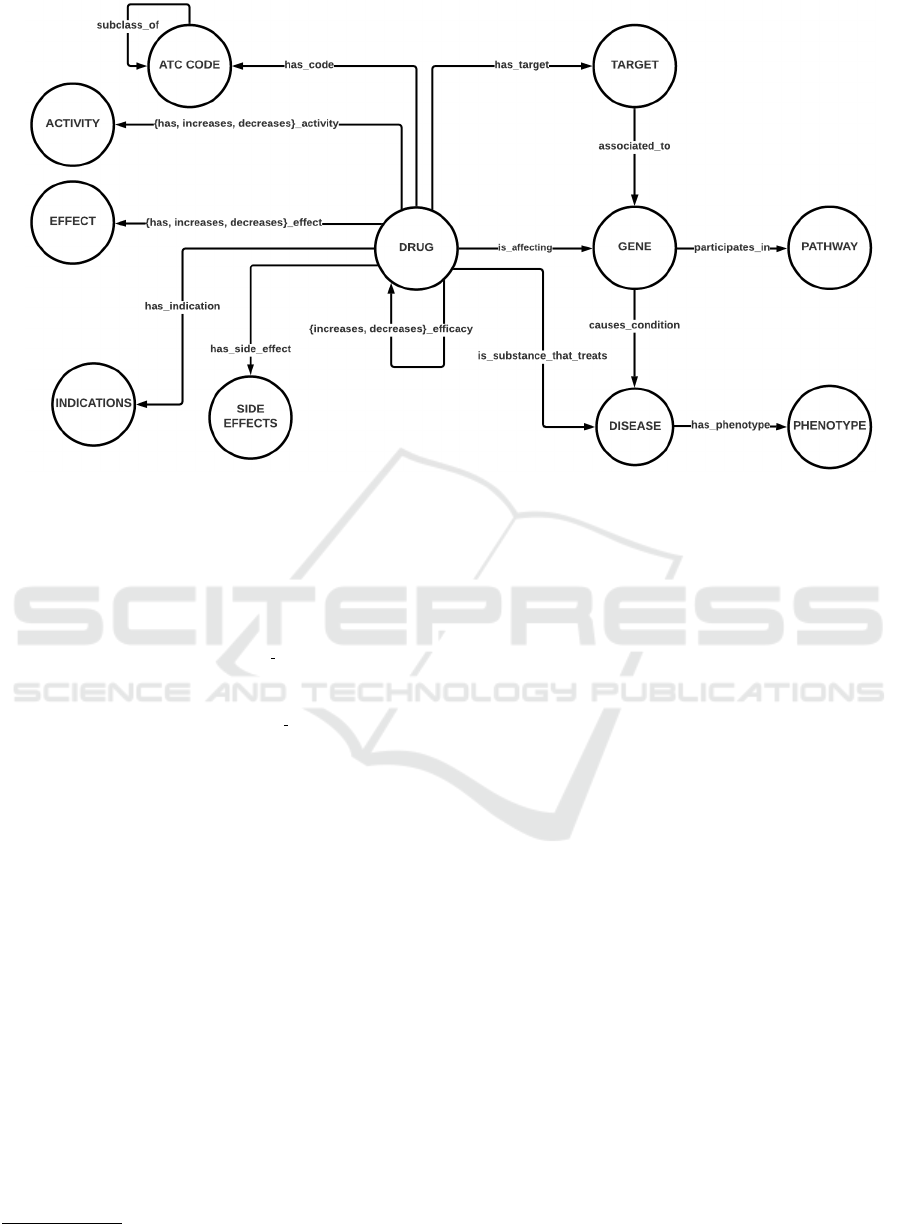
Figure 1: Schema of the OREGANO KG.
purposed, in this case between a drug and a disease.
These meta-paths must be “scored” according to their
reliability, a very reliable path having a score close to
1 and a very unreliable path a score close to 0. An
example of a meta-path is described as follows:
Aspirin
treats
−−−→ Headache
caused by
−−−−−−→ Dehydration
and becomes :
Drug
treats
−−−→ Symptom
caused by
−−−−−−→ Cause
As mentioned in (Liu et al., 2021), AnyBURL (Any-
time Bottom-Up Rule Learning algorithm) (Meilicke
et al., 2019) was applied to the OREGANO KG to
identify a set of rules intrinsic to the data. These rules
were transformed into meta-paths that were used by
PoLo during its learning phase. Based on these meta-
paths, PoLo evaluates the reward to be given to the
agent responsible for the link prediction.
2.4 Discovery Patterns
In addition to PoLo’s explainable logic rules, discov-
ery patterns have been used as proposed in (Himmel-
stein et al., 2017) and (Zhang et al., 2021). Discovery
patterns based on the exploration of semantic links be-
tween data allow the discovery of mechanistic links
between entities in a KG, but also to strengthen the
explanation of the prediction provided by the AI al-
gorithm. To provide a visual exploration, we relied
on Neo4j
1
, together with its query language Cypher.
1
https://neo4j.com/
It is a system for building and manipulating data in
the form of a graph-oriented database (GDB). These
GDBs have the advantage of preserving the structure
of the KG, by storing the information contained in the
nodes but also in the edges. Thus, the modeling of
complex data and queries are facilitated. From the
OREGANO project data, a GDB has been built on
Neo4j. The purpose of this GDB is to bring an ad-
ditional explanation and visualization, in order to in-
validate or validate the predictions made by PoLo. The
advantage of this method is that it offers the possibility
to build precise queries, allowing access to informa-
tion or links that had not been spotted by AnyBURL
or PoLo, but that do exist in the data.
2.5 Impact of the Data in the KG
As with any machine learning task, it is important to
consider the format of the data. Much of the current
research attempts to improve the results of link predic-
tion by improving the performance of the algorithms.
However, in the areas of AI that have grown in recent
years, there has been an evolution of algorithms along
with data processing methods. These methods ensure
optimal learning, for example by modifying or aug-
menting the data. For KGs, as far as we know, little or
no work has been done to determine the best way to
structure the data in KG and its possible impact on the
predictions. A first approach studied here concerns the
importance of the ratio between the entities of the KG.
Let r be the relationship of interest linking two types
KEOD 2021 - 13th International Conference on Knowledge Engineering and Ontology Development
222

of entities T 1 and T 2, what is the optimal number of
objects of types T 1 and T 2 in the graph to achieve a
good prediction? To answer this question in the con-
text of DR, several types of link prediction have been
tested. First, AnyBURL and PoLo were used to pre-
dict the following types of links:
• associated to between a protein and a gene;
• causes condition between a gene and a disease;
• has phenotype between a disease and a phenotype;
• has target between a drug and a protein;
• is affecting between a drug and a gene;
• is substance that treats between a drug and a dis-
ease;
• participates in between a gene and a pathway.
In a second approach, the relation between the amount
of training data and the quality of the prediction was
tested. For this purpose, the link prediction task is re-
run focusing exclusively on the links has target and
is substance that treats, but changing the number of
triples of these types. For each type of relation,
the first experiments have been launched with 5000
triples, then 5000 triples are added per additional ex-
periment, until all the triples are reached.
3 RESULTS
Table 3 shows the Mean Reciprocal Rank (MRR)
score for each PoLo link prediction based on the
different relation types. These results show that
the quality of predictions cannot be solely related
to the quality of the extracted rules or their num-
ber. Indeed, the best predictions are reached for the
is substance that treats relation for which 630 rules
are available with the best confidence index equal
to 0.70. However, for the is affecting relation, the
MRR is close to 0 while many rules are also avail-
able and have a satisfactory confidence index. In con-
trast, the predictions for the relation causes condition
are ranked third in terms of MRR, with only 112 Horn
rules available and a confidence index three times
lower than that of is affecting. Predictions involving
participates in achieve an MRR of 0.3 based on only
68 rules.
Regarding the impact of the amount of training
data, the results are presented in Figure 2. These
results show that, for both relations, increasing the
amount of training data has rather a negative impact
on the quality of predictions. Furthermore, we can
clearly see the correlation between the MRR and the
number of times a rule given by AnyBURL produces
the correct result.
As a final result for the DR problem, best
results were found when predicting the relation
is substance that treats and can be found in Table 4.
These results, which are slightly better than with Het-
ionet, are only partial results, the other (major) part
coming from the explanations provided by PoLo for
each of the predictions made. From this generated file,
the following results emerged:
• 682 drugs were identified as candidates for being
repurposed;
• these candidates impact a total of 447 diseases;
• out of 630 Horn rules provided by AnyBURL to
PoLo, only 10 of them were effectively used for
predictions;
• on these 10 rules, the most frequent one has been
used 600 000 times, the second only 21 000 times.
4 DISCUSSION
The use of PoLo to repurpose drugs through link
prediction has produced a significant amount of re-
sults. These results reached an accuracy not previ-
ously achieved from the OREGANO KG using other
methods such as TransE (Bordes et al., 2013). More-
over, the use of a neuro-symbolic model offers reliable
explanations for the origin of each prediction. These
results need to be analyzed by DR experts in order to
get the best out of them. It is with this in mind that
these results are currently being further investigated
by the clinical genetics unit of the Bordeaux Univer-
sity Hospital. The aim of this collaboration is to give
access to the results to clinicians who will be able to
evaluate and select those they consider most interest-
ing to explore in their research. The possible explana-
tion for each repurposing will come from logical rules
used by PoLo but also from the post-hoc mechanistic
explanation provided by the visualization capability of
the Neo4j graph database.
Regarding the fact that providing more training
data does not help to obtain better predictions, two im-
portant points should be highlighted:
• The quality and number of rules produced by Any-
BURL are not systematically correlated with the
quality of a prediction;
• The increase in the number of triples in the training
data does not necessarily mean an improvement in
performance.
This last point is particularly interesting since it is
counter-intuitive. We observe that, in both cases, the
MRR drops when the number of rules producing a cor-
rect prediction decreases, which is normal if we con-
sider that the PoLo agent’s gain is directly related to
Neuro-symbolic XAI for Computational Drug Repurposing
223
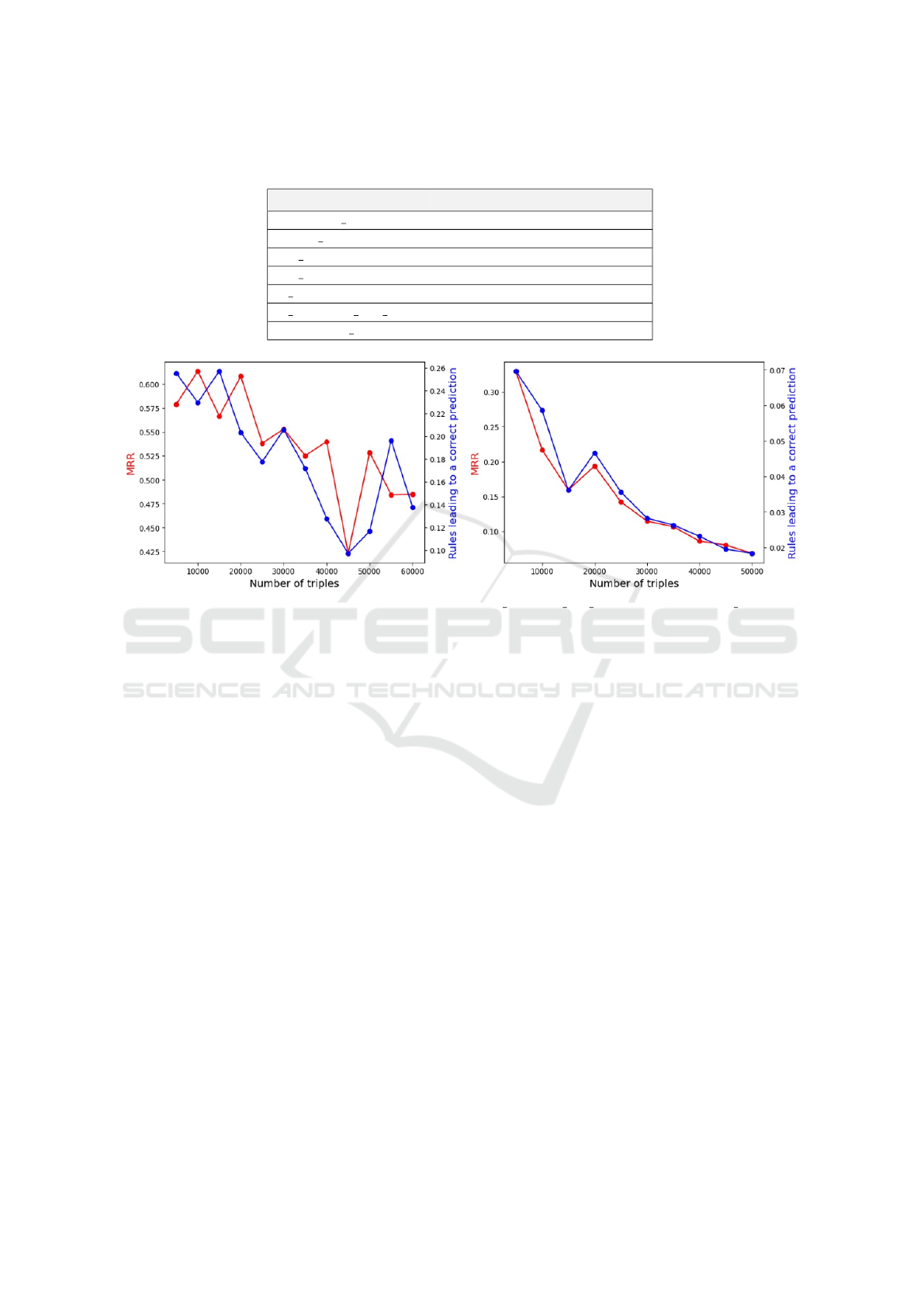
Table 3: MRR (Mean Reciprocal Rank) produced by PoLo for each type of relation, based on the best quality and number of
rules extracted by AnyBURL.
Relation Number Best Score MRR
associated to 21 0.43 0.0003
causes condition 112 0.14 0.15
has phenotype 96 0.11 0.07
has target 489 0.163 0.07
is affecting 399 0.42 0.004
is substance that treats 630 0.70 0.49
participates in 68 0.92 0.3
Figure 2: Impact of the number of relations in the training data for is substance that treats on the left and has target on the
right.
the path taken for the prediction. These results there-
fore suggest that a surplus of training data makes the
use of the rules proposed by AnyBURL rarer, leading
to less frequent rewards and thus to a decrease in per-
formance of PoLo predictions. Another explanation
for this decrease in performance could be how more
training data brings more noise to the system. PoLo
uses training triples to determine what are the diseases
for which drugs can be repurposed. The more training
data, the more different diseases can be proposed as
a result. Based on this observation, a useful step that
could be added would be to filter the training data to
keep only those that contain diseases of interest.
The disparity of the metrics according to the type
of relation chosen demonstrates the importance of the
structure of the data in the prediction task. Today,
AI research in the field of drug repositioning focuses
mainly on improving prediction algorithms, without
really paying attention to the data and its quality.
These new models are systematically tested on the
same evaluation datasets, without ever reconsidering
their structure. However, it is necessary to under-
stand how the organization of the data within a KG
changes the quality of the predictions, not least be-
cause biomedical data in general are different from the
“test” datasets typically used in the literature. In addi-
tion to these considerations around data, it is important
to remember that the goal of more transparent AI is to
be trusted by clinicians and other healthcare experts.
With this in mind, algorithms should be proposed that
allow for greater interaction with clinicians, in order to
offer greater flexibility in the choice of how they oper-
ate, as here with the logical rules to be followed. Cur-
rently, the set of “good” logical rules to follow is given
to PoLo using AnyBURL. AnyBURL is a powerful
tool for extracting the main logical rules and meta-
paths that exist in a KG, but these rules are ranked
based on their ability to describe the data, not on their
relevance from a medical perspective. Since PoLo is
trained using these rules in the reinforcement learning
process, it will be important to filter these rules to keep
only those that are medically plausible. Another way
to do this is to have the clinicians construct the rules
themselves, based on their a priori knowledge. The
fact that PoLo only used 10 of the 630 proposed rules
and only one of them most of the time (600 000 times)
shows that these rules are not efficient in repurposing
drugs. This is also a problem when considering the use
of reinforcement learning based on these rules, as the
agent should be rewarded when it uses different med-
ically valid rules and not when it uses only the most
common one among the data.
KEOD 2021 - 13th International Conference on Knowledge Engineering and Ontology Development
224

Table 4: Comparison of the metrics obtained for drug repositioning through the is substance that treats relation on the
OREGANO KG with the results obtained on Hetionet.
Data MRR Hits@1 Hits@3 Hits@10
OREGANO 0.4980 0.3898 0.5551 0.7300
HETIONET 0.4300 0.3370 0.4700 0.6410
5 CONCLUSION
In this paper, we have described a neuro-symbolic
XAI solution applied to the DR task using a KG. The
study has been implemented and evaluated in the con-
text of the OREGANO project using a state-of-the-art
algorithm which enables the explainability. Results
equivalent to those of the state of the art were obtained,
as well as several ways to provide an explanation for
each prediction, i.e. intrinsic to the model’s opera-
tion, but also post-hoc in a mechanistic way using fea-
tures of a graph oriented database. The challenges sur-
rounding this work are numerous. First, it is important
to better understand how the organization of data in a
KG affects the prediction task. This will be partic-
ularly important for the application of these methods
to DR for rare diseases, where data is by definition
less abundant. Also, more flexible methods must be
thought of, allowing biochemists and other clinicians
to easily participate in the learning process, bringing
their knowledge to the sum of data available for DR.
REFERENCES
Ashburn, T. and Thor, K. (2004). Drug repositioning: Identi-
fying and developing new uses for existing drugs. Na-
ture reviews Drug discovery, 3:673–83.
Bordes, A., Usunier, N., Garc
´
ıa-Dur
´
an, A., Weston, J., and
Yakhnenko, O. (2013). Translating embeddings for
modeling multi-relational data. In Advances in Neu-
ral Information Processing Systems 26.
Boudin, M. (2020). Computational approaches for drug
repositioning: Towards a holistic perspective based on
knowledge graphs. Proceedings of the 29th ACM In-
ternational Conference on Information & Knowledge
Management, page 3225–3228.
Das, R., Dhuliawala, S., Zaheer, M., Vilnis, L., Durugkar,
I., Krishnamurthy, A., Smola, A., and McCallum, A.
(2018). Go for a walk and arrive at the answer: Rea-
soning over paths in knowledge bases using reinforce-
ment learning. In International Conference on Learn-
ing Representations.
DiMasi, J. A., Grabowski, H. G., and Hansen, R. W. (2016).
Innovation in the pharmaceutical industry: New es-
timates of r&d costs. Journal of Health Economics,
47:20–33.
Himmelstein, D. S., Lizee, A., Hessler, C., Brueggeman, L.,
Chen, S. L., Hadley, D., Green, A., Khankhanian, P.,
and Baranzini, S. E. (2017). Systematic integration of
biomedical knowledge prioritizes drugs for repurpos-
ing. eLife, 6:e26726.
Hochreiter, S. and Schmidhuber, J. (1997). Long Short-
Term Memory. Neural Computation, 9(8):1735–1780.
Liu, Y., Hildebrandt, M., Joblin, M., Ringsquandl, M., Rais-
souni, R., and Tresp, V. (2021). Neural multi-hop
reasoning with logical rules on biomedical knowledge
graphs. In Verborgh, R., Hose, K., Paulheim, H.,
Champin, P.-A., Maleshkova, M., Corcho, O., Ris-
toski, P., and Alam, M., editors, The Semantic Web,
pages 375–391, Cham. Springer International Publish-
ing.
Meilicke, C., Chekol, M. W., Ruffinelli, D., and Stucken-
schmidt, H. (2019). Anytime bottom-up rule learning
for knowledge graph completion. In IJCAI.
Paulheim, H. (2017). Knowledge graph refinement: A sur-
vey of approaches and evaluation methods. Semantic
Web, 8:489–508.
Stokes, J. M., Yang, K., Swanson, K., Jin, W., Cubillos-
Ruiz, A., Donghia, N. M., MacNair, C. R., French, S.,
Carfrae, L. A., Bloom-Ackermann, Z., Tran, V. M.,
Chiappino-Pepe, A., Badran, A. H., Andrews, I. W.,
Chory, E. J., Church, G. M., Brown, E. D., Jaakkola,
T. S., Barzilay, R., and Collins, J. J. (2020). A
deep learning approach to antibiotic discovery. Cell,
180(4):688–702.e13.
Zhang, R., Hristovski, D., Schutte, D., Kastrin, A., Fiszman,
M., and Kilicoglu, H. (2021). Drug repurposing for
covid-19 via knowledge graph completion. Journal of
Biomedical Informatics, 115:103696.
Neuro-symbolic XAI for Computational Drug Repurposing
225
