
Ingestion of a Data Lake into a NoSQL Data Warehouse:
The Case of Relational Databases
Fatma Abdelhedi
1
, Rym Jemmali
1,2
and Gilles Zurfluh
2
1
CBI
2
, Trimane, Paris, France
2
IRIT CNRS (UMR 5505), Toulouse University, Toulouse, France
Keywords: Data Lake, Data Warehouse, NoSQL, Big Data, Relational Database, MDA, QVT.
Abstract: The exponential growth of collected data, following the digital transformation of companies, has led to the
evolution of databases towards Big Data. Our work is part of this context and concerns more particularly the
mechanisms allowing to extract datasets from a Data Lake and to store them in a unique Data Warehouse.
This one will allow to realize, in a second time, decisional analyses facilitated by the functionalities offered
by the NoSQL systems (richness of the data structures, query language, access performances). This article
proposes an extraction mechanism applied only to relational databases of the Data Lake. This mechanism
relies on an automatic approach based on the Model Driven Architecture (MDA) which provides a set of
schema transformation rules, formalized with the Query/View/Transform (QVT) language. From the physical
schemas describing relational databases, we propose transformation rules that allow to generate a physical
model of a Data Warehouse stored on a document-oriented NoSQL system (OrientDB). This paper presents
the successive steps of the transformation process from the meta-modeling of the datasets to the application
of the rules and algorithms. We provide an experimentation using a case study related to the health care field.
1 INTRODUCTION
Due to the considerable increase of data generated by
human activities, Data Lakes have been created
within organizations, often spontaneously, by the
physical grouping of datasets related to the same
activity. A Data Lake (Nargesian, Zhu, Miller, Pu, &
Arocena, 2019) is a massive grouping of data
consisting of structured or unstructured datasets.
These datasets generally have the following
characteristics: (1) they can be stored on
heterogeneous systems, (2) each of them is exploited
independently of the others, (3) some of them can
contain raw data, i.e., data stored in their original
form and without being organized according to the
use that will be made of them, (4) the types and
formats of the data can vary. In practice, a Data Lake
can group together different datasets (Khine & Wang,
2018) such as relational databases, object databases,
Comma Separated Values (CSV) files, texts,
spreadsheet folders. The massive data contained in a
1
https://opusline.fr/health-data-hub-an-ambitious-french-
initiative-for-tomorrows-health/
Data Lake represents an essential reservoir of
knowledge for business decision makers. This data
can be organized according to a multidimensional
data model in order to support certain types of
decision processing (El Malki, Kopliku, Sabir, &
Teste, 2018). For example, in the French health
sector, a Data Lake has been created by the French
public health insurance company under the name of
“Espace Numérique de Santé” (ENS)
1
; it includes the
electronic health records of insured persons, access
authorization directories, health questionnaires, and
care planners. The case study presented in Section 2
is in the context of insurance companies' exploitation
of the ENS. It is this application that motivated our
work. Indeed, the heterogeneity of storage systems
combined with the diversity of content in the Data
Lake is a major obstacle to the use of data for
decision-making. To manipulate a Data Lake, a
solution consists in ingesting the data into a Data
Warehouse and then transforming it (grouping,
calculations, etc.). Ingestion is a process that consists
64
Abdelhedi, F., Jemmali, R. and Zurfluh, G.
Ingestion of a Data Lake into a NoSQL Data Warehouse: The Case of Relational Databases.
DOI: 10.5220/0010690600003064
In Proceedings of the 13th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2021) - Volume 3: KMIS, pages 64-72
ISBN: 978-989-758-533-3; ISSN: 2184-3228
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

in extracting data from various sources and then
transferring them to a repository where they can be
transformed and analyzed. For example, in (Meehan,
Tatbul, Aslantas, & Zdonik, s. d.) massive data from
various sources are ingested into a Data Warehouse
and exploited in the context of information retrieval
on the Web. Other works have introduced the concept
of polystore which preserves the initial data sources
(no ingestion) and allows querying them by creating
"data islands", each of which contains several
systems sharing a common query language. For
example, all relational databases are connected to the
"relational island", which is queried using standard
SQL. This solution, developed in particular in the
BiGDAWG (Duggan, Kepner, Elmore, & Madden,
2015) and ESTOCADA (Alotaibi et al., 2020)
projects, keeps the data in their native formats.
Our work aims at performing decisional
processing on a Data Lake. This problem is part of a
medical application in which massive data are stored
in a Data Lake that will be used by medical decision
makers. We have chosen to ingest the data from the
Data Lake into a Data Warehouse that will later be
reorganized for Big Data Analytics. This article deals
with the ingestion of relational databases and
voluntarily excludes other forms of datasets present
in the Data Lake. This choice is justified (1) by the
majority presence of relational databases in the ENS
used by our case study (cf. Section 2.1) and (2) by the
major difficulty of translating relational key type
links (attribute values) into pointer type links
(references in object-oriented models).
Our paper is organized as follows: in the
following Section 2, we present the medical
application that justifies our work's purpose. Section
3 describes the context of our study as well as our
research problem which aims at facilitating the
querying of data contained in a Data Lake by decision
makers. Section 4 describes the metamodels of the
databases used in our application. Section 5 presents
our contribution which consists in formalizing with
the Model Driven Architecture (MDA), the process
of transforming the databases of a Data Lake into a
NoSQL Data Warehouse. Section 6 describes an
experimentation of the proposed process based on our
medical application. Section 7 contrasts our proposal
with related works. Finally, Section 8 concludes this
paper and highlights possible directions for exploring
the continuity of the work.
2
https://www.trimane.fr
2 CONTEXT OF WORK
In this section, we present the case study that
motivated our work as well as the problem addressed
in this article.
2.1 Motivating Scenario
Our work is motivated by a project developed in the
health field for a group of private health insurance
companies. These insurance companies, stemming
from the social and solidarity economy, propose to
their customers a coverage of the medical expenses
which comes in complement of those refunded by a
public institution: the public health insurance fund.
To ensure the management of their clients, these
private health insurance companies are faced with a
significant increase in the volume of data processed.
This is all the truer since some of these companies act
as social security centers, i.e., clients transmit the
entire rebate file to a private insurance company,
which performs all the data processing related to the
file. In order to monitor these files, numerous
exchanges are necessary between the health insurance
fund, the private insurance companies and the health
professionals. Under the impetus of the State, the
health insurance fund has developed a digital health
platform (ENS). This allows the storage of the
medical data of each insured person. This data is
confidential, but the holder can authorize a health
professional or institution to consult or feed his ENS.
All accesses and actions carried out on the ENS by
non-holders are traced. Private health insurance
companies therefore have limited access to the ENS,
from which they can extract data in order to process
the files of their policyholders and, more broadly, to
carry out analyses of any kind (in compliance with
confidentiality rules).
For each insured person, the ENS contains
administrative data, the medical file (vitals, medical
imaging archives, reports, therapeutic follow-ups,
etc.), the history of rebates and questionnaires. When
the ENS is fully deployed at the national level, its
volume will be considerable since it concerns 67
million insured persons.
This project is being developed by Trimane
2
on
behalf of insurance companies.
In the context of this project, the ENS constitutes
a real Data Lake because of (1) the diversity of data
types, media, and formats (2) the volumes stored
which can reach several terabytes and (3) the raw
nature of the data. The objective of the project is to
Ingestion of a Data Lake into a NoSQL Data Warehouse: The Case of Relational Databases
65

study the mechanisms for extracting data from the
ENS and organizing it to facilitate analysis (Big Data
Analytics). For our application, the extracted data
comes mainly from relational databases present in the
ENS.
2.2 Problematic
Our work aims to develop a system allowing private
companies to create a Data Warehouse from a Data
Lake. This article deals more specifically with the
mechanisms of extraction and unification from
commonly used databases, we limit the framework of
our study as follows:
- The ENS Data Lake is the source of the data;
in this article, we voluntarily reduce its content
to relational databases. Indeed, this category
of datasets represents an important part of the
ENS data;
- The generated Data Warehouse is managed by
a document-oriented NoSQL database system.
This type of system offers (1) great flexibility
in reorganizing objects for analysis and (2)
good performance in accessing large volumes
of data (using MapReduce).
To achieve our goal, each database in the Data
Lake is extracted and converted into another model to
allow its storage in the Data Warehouse as shown in
Figure 1. We do not address here the problems related
to the selective extraction of data and their
aggregative transformation.
To test our proposals, we have developed a Data
Lake with multiple relational databases managed by
MySQL
3
and PostgreSQL
4
systems. These databases
contain respectively data describing the follow-up of
the insured and the processing of the files in a medical
center. The available metadata are limited to those
accessible on the storage systems (absence of
ontologies for example). The Data Warehouse, which
is supported by an OrientDB platform, must allow the
analysis of the care pathways of insured persons with
chronic pathologies. We chose the OrientDB
5
system
to store the Data Warehouse. Indeed, this document-
oriented NoSQL system allows to consider several
types of semantic links such as association,
composition and inheritance links; it is thus well
adapted to our case of study where the richness of the
links between objects constitutes an essential element
for decisional processes.
3
https://www.mysql.com
4
https://www.postgresql.org
3 RELATED WORKS
In this section, we present research work on
extracting data from a Data Lake and more
specifically data from several relational databases and
creating a NoSQL Data Warehouse. The advent of
Big Data has created several challenges for the
management of massive data; among these we find
the creation of architectures to ingest massive data
sources as well as the integration and transformation
of these massive data (Big Data) allowing their
subsequent query. In this sense, some works have
focused on the proposal of architectures (physical and
logical) allowing the use and the management of Data
Lakes. The article (Diamantini et al., 2018) proposes
an approach to structure the data of a Data Lake by
linking the data sources in the form of a graph
composed of keywords. Other works propose to
extract the data of a Data Lake from the metamodels
of the sources. The authors in (Candel, Ruiz, &
García-Molina, 2021) have proposed a metamodel
unifying NoSQL and relational databases. There are
several formalisms (Bruel et al., 2019) to express
model transformations such as the QVT standard, the
ATL language (Erraissi & Banane, 2020) which is a
non-standardized model transformation language
more or less inspired by the QVT standard of the
Object Management Group, etc.
Other works have studied only the transformation
of a relational database into a NoSQL database. Thus
in (Hanine, Bendarag, & Boutkhoum, 2015;
Stanescu, Brezovan, & Burdescu, 2016) the authors
developed a method to transfer data from relational
databases to MongoDB. This approach translates the
links between tables only by nesting documents. In
(Liyanaarachchi, Kasun, Nimesha, Lahiru, &
Karunasena, 2016), the authors present MigDB, an
application that converts a relational database
(MySQL) to a NoSQL one (MongoDB). This
conversion is done over several steps: transforming
tables into JSON files, then transmitting each JSON
file to a neural network. This network allows to
process the links at the JSON file level, either by
nesting or by referencing. This work considers all
association links (of different cardinalities) only. The
same is true in (Chickerur, Goudar, & Kinnerkar,
2015), where the authors propose a method for
transferring relational databases to MongoDB by
converting the tables into CSV files that are then
imported into MongoDB. However, the proposed
method simply converts tables into MongoDB
5
https://orientdb.com/docs/3.0.x/
KMIS 2021 - 13th International Conference on Knowledge Management and Information Systems
66

Figure 1: Architecture of the data extraction process from a Data Lake: RDBToNoSDW.
collections without supporting the various links
between tables.
4 OVERVIEW OF OUR
SOLUTION
Although a Data Lake can contain files of any format,
we focus in this article on the extraction of relational
databases and the feeding of a NoSQL Data
Warehouse. Several works have dealt with the
transfer of a relational database to a NoSQL database.
Thus, some works have proposed algorithms for
converting relational data to document-oriented
systems such as MongoDB (Mahmood, 2018);
however, these works transform relational links into
document nesting or DBRef links. However, these
NoSQL linkage solutions are not satisfactory with
respect to object systems
6
. Moreover, to our
knowledge, no study has been conducted to convert
several relational databases contained in a Data Lake
into a NoSQL Data Warehouse.
In our ingestion process, we have defined three
modules as shown in Figure 1. Our proposal is based
on Model Driven Architecture (MDA) which allows
to describe separately the functional specifications
and the implementation of an application on a
platform. Among the three models present in MDA
(CIM, PIM and PSM), we are located at the PSM
level where the logical schemas are described. We
also use the declarative language
Query/View/transform (QVT)
7
specified by the
Object Management Group OMG
8
which allows us to
describe the ingestion of data by model
transformations.
To use the MDA transformation mechanism, we
proposed two metamodels describing respectively the
source and target databases. From these metamodels,
6
http://www.odbms.org/odmg-standard/reading-room/od
mg-2-0-a-standard-for-object-storage/
7
https://www.omg.org/spec/QVT/1.2/PDF
we specified the transformation rules in QVT
language to ensure data ingestion.
5 DATABASES
METAMODELING
We present successively the metamodels of a source
Relational database and a target NoSQL database.
5.1 Relational Metamodel
The Data Lake, source of our process, can contain
several relational databases. A relational database
contains a set of tables made of a schema and an
extension. The schema of a table contains a sequence
of attribute names associated with atomic types (such
as the predefined types Integer, String, Date). The
extension is composed of a set of rows grouping
attribute values. Among the attributes of a table, we
distinguish the primary key whose values identify the
rows and the foreign keys which materialize the links.
Figure 2 represents the Ecore
9
metamodel of a
relational database.
5.2 Document-oriented NoSQL
Metamodel
The target of our process corresponds to the Data
Warehouse represented by a NoSQL database. A
document-oriented NoSQL database contains a set of
classes. Each class gathers objects that are identified
(by a reference) and composed of couples (attribute,
value); a value is defined by a basetype, multivalued
or structured. We distinguish a particular basic type,
the reference, whose values make it possible to link
the objects. These concepts are represented in Figure
3 according to the Ecore formalism.
8
The Object Management Group (OMG)
https://www.omg.org
9
https://download.eclipse.org/modeling/emf/emf/javadoc/
2.9.0/org/eclipse/emf/ecore/package-summary.html
Ingestion of a Data Lake into a NoSQL Data Warehouse: The Case of Relational Databases
67
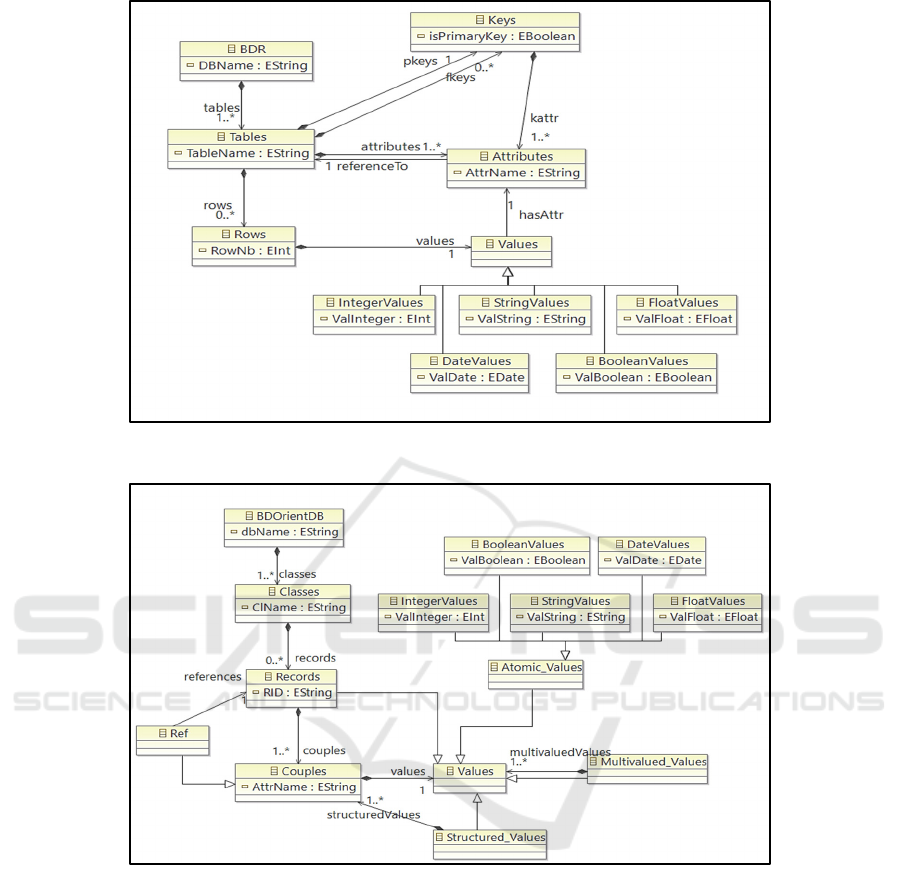
Figure 2: Metamodel of a relational database stored in the Data Lake.
Figure 3: The Data Warehouse metamodel: a document-oriented NoSQL database.
6 OUR DATA PROCESSING
SOLUTION
This involves transferring relational databases from
the Data Lake to a NoSQL database corresponding to
the Data Warehouse. To carry out this ingestion
process, we have defined three modules that will
successively ensure (1) the transformation of
relational data into NoSQL data (CreateDW module),
(2) the conversion of relational links (foreign keys)
into references (ConvertLinks module) and (3) the
merging of tables containing objects of the same
semantics (MergeClasses module).
6.1 CreateDW Module
This module transforms each relational database of
the Data Lake into a unique NoSQL database
according to the MDA approach. The NoSQL
warehouse being unique, it will contain the data
coming from the different relational databases of the
Data Lake. The application of a set of transformation
rules defined on the metamodels of Section 4,
generates a set of classes in a NoSQL database. We
informally present the rules that have been expressed
in the QVT language.
KMIS 2021 - 13th International Conference on Knowledge Management and Information Systems
68

Rule 1: Each table in a relational database is
transformed into a class in the NoSQL database. To
avoid synonymy, the name of the class is prefixed by
the name of the original database.
Rule 2: Each row of a table, associated with its
schema, is transformed into a record in the
corresponding target class; the record then contains a
set of couples (attribute, value). The primary key is
stored as any attribute. At this stage, the foreign keys
are also stored with their relational values; they will
be converted into references by the ConvertLinks
module.
These two rules, which we formalized in QVT
language, are applied for each relational database of
the Data Lake and feed the NoSQL database; we will
present their syntax in Figure 4 of the
experimentation section. In parallel with the
application of these transformation rules, an
algorithmic processing allows to record metadata;
these metadata match each relational primary key
with the Record ID (RID) of the corresponding record
in the NoSQL database.
6.2 ConvertLinks Module
In the standard object-oriented systems
10
, links are
materialized by references. Since this principle is
used in NoSQL systems, it is necessary to convert
relational foreign keys that have been transferred to
the Data Warehouse into references.
The mechanism we have developed in
ConvertLinks is not based on the expression of MDA
rules but corresponds to an algorithmic process. In the
NoSQL database, all records of a class are
systematically marked with identifiers (RID for
Record ID). During the transfer of data into the
records, the relational primary and foreign keys were
transferred in the form of pairs (attribute, value).
Thus, thanks to the metadata recorded by the previous
CreateDW module, the values of the foreign keys are
converted into RID.
6.3 MergeClasses Module
Data ingestion from the Data Lake was performed by
transferring data from different relational databases to
the NoSQL database (in the previous processes).
10
http://www.odbms.org/wp-
content/uploads/2013/11/001.04-Ullman-CS145-ODL-
OQL-Fall-2004.pdf
However, it is frequent that tables with the same
semantics from different relational databases are
transferred; these tables are called "equivalent". For
example, the DB1-Insured and DB2-Patients tables
from the DB1 and DB2 databases contain data on the
insured; some properties in these tables are redundant
and others are complementary. Two tables are
considered "equivalent" if they contain data related to
the same entity (same primary key). The CreateDW
module has allowed us to create a class in the NoSQL
database for each table. It is therefore useful to group
the data contained in "equivalent" tables into a single
class in the NoSQL database. To achieve this
grouping, we relied on an ontology establishing the
correspondences between the terms of the relational
databases contained in the Data Lake. This ontology
is provided by data administrators bringing their
business expertise. These administrators, after
consultation, provided the business rules that apply to
the tables considered as semantically equivalent.
By exploiting this ontology
11
, the MergeClasses
module creates new classes in the NoSQL database.
In the presence of several equivalent tables, the
module groups the data in a single class. This
treatment is not limited to a union operation between
records; in fact, distinct records concerning the same
entity can have complementary attributes that will be
combined in a single record.
7 IMPLEMENTATION AND
TECHNICAL ENVIRONMENT
In this section, we describe the techniques used to
implement the RDBToNoSDW process of Figure 1.
The first module (CreateDW) implements
transformation rules, we used a technical
environment adapted to modeling, metamodeling and
model transformation. We used the Eclipse Modeling
Framework (EMF)
12
model transformation
environment with the Ecore metamodeling language
to create and manipulate metamodels. Ecore is based
on XMI to instantiate the models. Algorithmic
processing has been coded in Java. In fact, EMF is
compatible with Java. Figure 4 shows the complete
architecture of our RDBToNoSDW process.
11
The principles of exploitation of the ontology are not
detailed in this article.
12
https://www.eclipse.org/modeling/emf
Ingestion of a Data Lake into a NoSQL Data Warehouse: The Case of Relational Databases
69
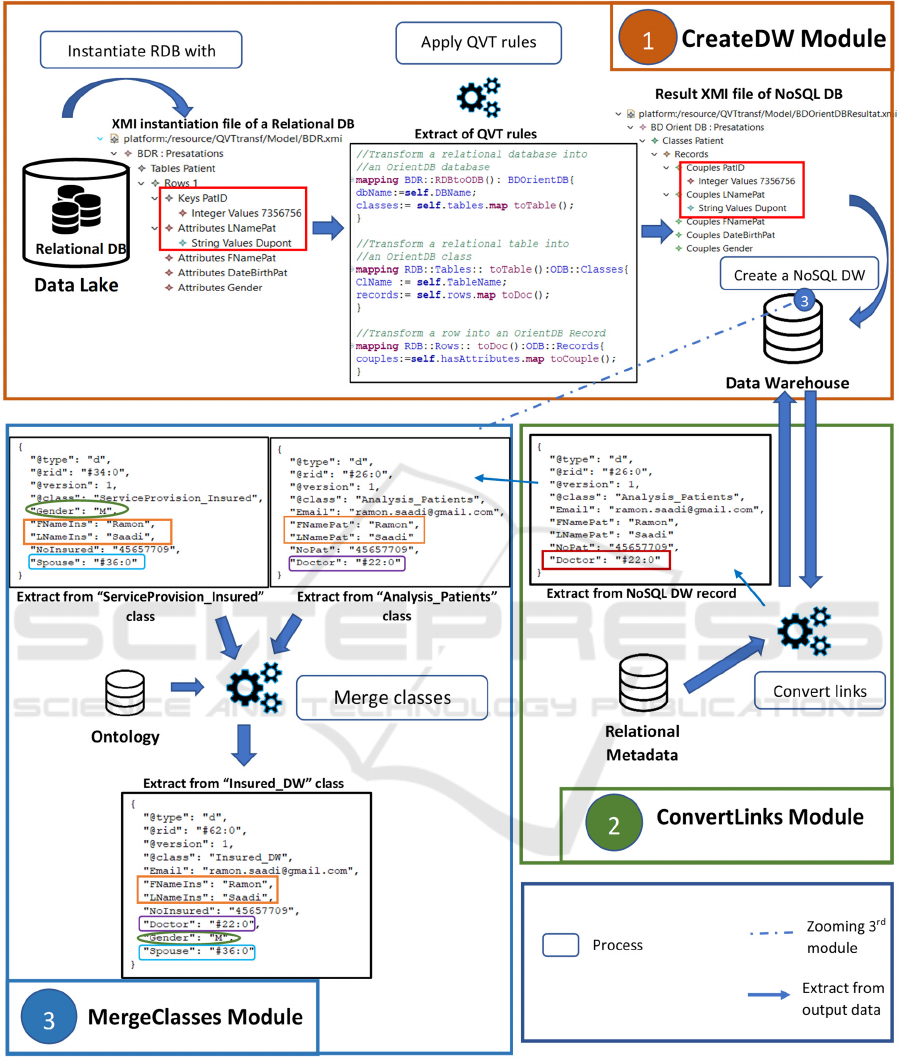
Figure 4: Data flow and processing in the RDBToNoSDW process.
To experiment our process, we stored several
relational databases in the Data Lake; these are test
databases directly inspired by the medical case study
(Section 2.1). Figure 5 shows two extracts of the
schemas of two databases: the "Service Provision"
database and the "Analysis" database. They have been
implemented with the PostgreSQL RDBMS and the
MySQL RDBMS respectively.
KMIS 2021 - 13th International Conference on Knowledge Management and Information Systems
70

a) The « Service Provision » database
b
) The « Analysis » database
Figure 5: Extracts from the relational schemas of a two Data Lake databases.
Figure 6: Extract from the list of Data Warehouse classes stored in OrientDB.
Figure 7: Extract from the "Insured_DW" class.
The CreateDW module (1) of our process (Figure
4) generates a unique NoSQL database from the two
Data Lake’s databases. To do so, it uses a relational
metamodel and a NoSQL metamodel as represented
with Ecore (Figures 2 and 3) as well as the
instantiation with the XMI language of the two
relational databases. The transformation rules have
been translated with the QVT language and apply to
all relational databases independently of the RDBMS.
The ConvertLinks module (2) allows converting
foreign keys into RID links. For example, in Figure 5,
the fields "Affiliation" and "Doctor" in the table
"Patient" will be converted. Finally, the
MergeClasses module (3) groups the records of the
classes considered as "equivalent" based on the
ontology provided by the experts.
The result of the treatments carried out by the 3
modules described above is represented by Figure 6.
Figure 6 represents the list of classes (corresponding
to the relational database tables).
Figure 7 shows an extract of the new class
"Insured_DW" containing the merged records of the
two classes "ServiceProvision_Patient" and
"Analysis_Insured".
8 DISCUSSION
Various articles have studied the extraction and
management of data from a Data Lake. Thus, some
works have focused on proposing solutions to
structure heterogeneous data coming from different
Ingestion of a Data Lake into a NoSQL Data Warehouse: The Case of Relational Databases
71

databases in a Data Lake. The paper (Diamantini et
al., 2018) proposes an approach to structure the data
of a Data Lake by linking the data sources in the form
of a graph composed of keywords. Other works
propose a metamodel unifying several data source
metamodels thus allowing the unification of data
from multiple sources. The authors in (Candel, Ruiz,
& García-Molina, 2021) proposed a metamodel
unifying the logical schemas of the four most
common types of NoSQL systems and relational
systems. Our solution is based on extracting data from
a data lake and loading it into a data warehouse. To
do so, we first proposed two metamodels representing
the physical models of each database: the first
metamodel concerns relational databases as source,
the second metamodel concerns document-oriented
NoSQL databases as target database of our solution.
We used EMF as our metamodeling tools. EMF
allowed us to formalize the transformation rules from
a source metamodel to the target metamodel. We
relied on the QVT standard to express our
transformation rules. The solution we propose allows
the interrogation of data contained in a Data Lake
thanks to the creation of a Data Warehouse.
9 CONCLUSION AND
PERSPECTIVES
This paper proposed a process to ingest data from a
Data Lake to a Data Warehouse; this one is made of
a unique NoSQL database and the Data Lake contains
several databases. We have limited the content of the
Data Lake to relational databases. Three modules
ensure the ingestion of the data. The CreateDW
module transforms each relational database into a
unique NoSQL database by applying MDA rules.
This mechanism will be used and extended to
transform other types of databases in the Data Lake.
The ConvertLinks module translates relational links
(keys) into references in accordance with the
principles of object databases supported by the
OrientDB system. Finally, the MergeClasses module
merges semantically equivalent classes from different
Data Lake databases; this merge is based on an
ontology provided by business experts.
Currently, we are continuing our work on the
ingestion of other types of data sources from a Data
Lake. Indeed, the Data Lake of our medical case study
contains various database types.
REFERENCES
Alotaibi, R., Cautis, B., Deutsch, A., Latrache, M.,
Manolescu, I., & Yang, Y. (2020). ESTOCADA :
Towards scalable polystore systems. Proceedings of the
VLDB Endowment, 13(12), 2949‑2952.
Bruel, J., Combemale, B., Guerra, E., Jézéquel, J., Kienzle,
J., Lara, J., Mussbacher, G., and al. (2019). Comparing
and classifying model transformation reuse approaches
across metamodels. Software and Systems Modeling.
Candel, C. J. F., Ruiz, D. S., & García-Molina, J. J. (2021).
A Unified Metamodel for NoSQL and Relational
Databases. ArXiv:2105.06494 [cs].
Chickerur, S., Goudar, A., & Kinnerkar, A. (2015).
Comparison of Relational Database with Document-
Oriented Database (MongoDB) for Big Data
Applications. 2015 8th International Conference on
Advanced Software Engineering Its Applications
(ASEA) (p. 41‑47).
Diamantini, C., Lo Giudice, P., Musarella, L., Potena, D.,
Storti, E., & Ursino, D. (2018). A New Metadata Model
to Uniformly Handle Heterogeneous Data Lake
Sources: ADBIS 2018 Short Papers and Workshops,
AI*QA, BIGPMED, CSACDB, M2U, BigDataMAPS,
ISTREND, DC, Budapest, Hungary, September, 2-5,
2018, Proceedings (p. 165‑177).
Duggan, J., Kepner, J., Elmore, A. J., & Madden, S. (2015).
The BigDAWG Polystore System. SIGMOD Record,
44(2), 6.
El Malki, M., Kopliku, A., Sabir, E., & Teste, O. (2018).
Benchmarking Big Data OLAP NoSQL Databases. In
N. Boudriga, M.-S. Alouini, S. Rekhis, E. Sabir, & S.
Pollin (Éds.), Ubiquitous Networking, Lecture Notes in
Computer Science (Vol. 11277, p. 82‑94). Cham:
Springer International Publishing.
Erraissi, A., & Banane, M. (2020). Managing Big Data
using Model Driven Engineering: From Big Data Meta-
model to Cloudera PSM meta-model (p. 1235‑1239).
Hanine, M., Bendarag, A., & Boutkhoum, O. (2015). Data
Migration Methodology from Relational to NoSQL
Databases, 9(12), 6.
Khine, P. P., & Wang, Z. S. (2018). Data lake : A new
ideology in big data era. ITM Web of Conferences, 17.
Liyanaarachchi, G., Kasun, L., Nimesha, M., Lahiru, K., &
Karunasena, A. (2016). MigDB - relational to NoSQL
mapper. 2016 IEEE International Conference on
Information and Automation for Sustainability (ICIAfS)
(p. 1‑6).
Mahmood, A. A. (2018). Automated Algorithm for Data
Migration from Relational to NoSQL Databases. Al-
Nahrain Journal for Engineering Sciences, 21(1), 60.
Meehan, J., Tatbul, N., Aslantas, C., & Zdonik, S. (s. d.).
Data Ingestion for the Connected World, 11.
Nargesian, F., Zhu, E., Miller, R. J., Pu, K. Q., & Arocena,
P. C. (2019). Data lake management: Challenges and
opportunities. Proceedings of the VLDB Endowment,
12(12), 1986‑1989.
Stanescu, L., Brezovan, M., & Burdescu, D. D. Federated
Conference on Computer Science and Information
Systems (2016). Automatic Mapping of MySQL
Databases to NoSQL MongoDB (p. 837‑840).
KMIS 2021 - 13th International Conference on Knowledge Management and Information Systems
72
