
Substitution of the Fittest: A Novel Approach for Mitigating
Disengagement in Coevolutionary Genetic Algorithms
Hugo Alcaraz-Herrera
a
and John Cartlidge
b
University of Bristol, Bristol, U.K.
Keywords:
Coevolution, Disengagement, Genetic Algorithms.
Abstract:
We propose substitution of the fittest (SF), a novel technique designed to counteract the problem of disengage-
ment in two-population competitive coevolutionary genetic algorithms. The approach presented is domain-
independent and requires no calibration. In a minimal domain, we perform a controlled evaluation of the
ability to maintain engagement and the capacity to discover optimal solutions. Results demonstrate that the
solution discovery performance of SF is comparable with other techniques in the literature, while SF also
offers benefits including a greater ability to maintain engagement and a much simpler mechanism.
1 INTRODUCTION
While attempting the problem of designing optimal
sorting networks using a genetic algorithm (GA),
Hillis decided that rather than use randomly gener-
ated input lists to evaluate sorting networks, he would
instead co-evolve a population of input lists that are
evaluated on their ability to not be sorted (Hillis,
1990). By coupling the evolution of input lists with
the evolution of the networks to sort those lists, Hillis
attempted to create an “arms race” dynamic such
that input lists consistently challenge networks that
are sorting them. As networks improve their abil-
ity to sort, so lists become more difficult to sort, etc.
This coevolutionary approach significantly improved
results and generated wide interest amongst evolu-
tionary computation (EC) practitioners. In particular,
coevolution offers EC the ability to tackle domains
where an evaluation function is unknown or diffi-
cult to operationally define; and through self-learning,
coevolutionary systems offer potential for the “holy
grail” of open-ended evolutionary progress.
However, it soon emerged that coevolution can
suffer from some “pathologies” that cause the system
to behave in an unwanted manner, and prevent contin-
ual progress towards some desired goal. For instance,
coevolving populations may continually cycle with no
overall progress; populations may progress in an un-
intended and unwanted direction; or populations may
a
https://orcid.org/0000-0002-9991-662X
b
https://orcid.org/0000-0002-3143-6355
disengage and stop progressing entirely (Watson and
Pollack, 2001). These pathologies have been studied
in depth and a variety of techniques have been intro-
duced as remedy (Popovici et al., 2012). However,
there is still much to be understood, and no panacea
has been discovered.
Contribution: We propose substitution of the fittest
(SF), a novel domain-independent method designed
to tackle the problem of disengagement in two-
population competitive coevolutionary systems. We
explore and evaluate SF in the deliberately simple
“greater than” domain, specifically designed for elab-
orating the dynamics of coevolution (Watson and Pol-
lack, 2001). We compare performance and system
dynamics against autonomous virulence adaptation
(AVA), a technique that has been shown to reduce the
likelihood of disengagement and improve optimisa-
tion in various domains (Cartlidge and Ait-Boudaoud,
2011). Initial results suggest that SF has some bene-
fits over AVA. We evaluate and discuss the reasons
why, and present avenues for future investigation.
2 BACKGROUND
Coevolutionary genetic algorithms with two distinct
populations are often described using terminology
that follows the biological literature. As such, and fol-
lowing Hillis’ original formulation, the populations
are often named as “hosts” and “parasites” (Hillis,
1990). In such cases, the host population tends to
Alcaraz-Herrera, H. and Cartlidge, J.
Substitution of the Fittest: A Novel Approach for Mitigating Disengagement in Coevolutionary Genetic Algorithms.
DOI: 10.5220/0010661200003063
In Proceedings of the 13th International Joint Conference on Computational Intelligence (IJCCI 2021), pages 59-67
ISBN: 978-989-758-534-0; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
59

denote the population of candidate “solutions” that
we are interested in optimising (e.g., the sorting net-
works), while the parasite population tends to denote
the population of test “problems” for the solution pop-
ulation to solve (e.g., the lists to sort); i.e., the hosts
are the models and the parasites are the training set;
or alternatively the hosts are the learners and the para-
sites are the teachers. Throughout this paper, we tend
to use the host-parasite terminology to distinguish co-
evolving populations. However, while this terminol-
ogy is meaningful in asymmetric systems where one
population (the model) is of most interest, it should
be noted that in symmetric systems, such as games of
self-play where both coevolving populations are mod-
els with the same encoding scheme, the two popula-
tions become interchangeable and the names host and
parasite have less meaning.
In an ideal scenario, two-population competitive
coevolution will result in an arms race such that both
populations continually evolve beneficial adaptions
capable of outperforming competitors. As a result,
there is continual system progress towards some de-
sired optimum goal. However, this ideal scenario
rarely materialises. In practice, coevolutionary sys-
tems tend to exhibit pathologies that restrict progress
(Watson and Pollack, 2001). These include cycling,
where populations evolve through repeated trajecto-
ries like players in an endless game of rock-paper-
scissors; and while short-term evolution exhibits con-
tinual progress, there is no long-term global progress
(Cartlidge and Bullock, 2004b). Alternatively, popu-
lations may start to overspecialise on sub-dimensions
of the game, such that evolved solutions are brittle
and do not generalise (Cartlidge and Bullock, 2003).
Furthermore, one population may begin to dominate
the other to such an extent that populations disen-
gage and evolutionary progress fails altogether, with
populations left to drift aimlessly (Cartlidge and Bul-
lock, 2004a). The likelihood of suffering from these
pathologies can be exacerbated by the problem do-
main. Cycling is more likely when the problem ex-
hibits intransitivity; overspecialisation is more likely
in multi-objective problems; and disengagement is
more likely if the problem has an asymmetric bias that
favours one population (Watson and Pollack, 2001).
Numerous techniques have been proposed for mit-
igating the pathologies that prevent continual coevo-
lutionary progress (for detailed reviews, see (Popovici
et al., 2012; Miguel Antonio and Coello Coello,
2018)). We can roughly group these approaches into
three broad categories; although in practice many
techniques straddle more than one category.
First, there are archive methods, which are de-
signed to preserve potentially valuable adaptations
from being “lost” during the evolutionary process.
The first coevolutionary archiving technique is the
Hall of Fame (HoF) (Rosin and Belew, 1997). Ev-
ery generation, the elite member of each population
is stored in the HoF archive. Then, individuals in
the current population are evaluated against current
competitors and also against members of the HoF.
This ensures that later generations are evaluated on
their capacity to beat earlier generations as well as
their contemporaries. However, as the archive grows
each generation, simple archiving methods like the
HoF can become unwieldy over time. To counter
this, more sophisticated and efficient archiving meth-
ods have been introduced to simultaneously minimise
archive size while maximising archive “usefulness”.
An efficient example is the Layered Pareto Coevo-
lutionary Archive (LAPCA), which only stores in-
dividuals that are non-dominated and unique; while
the archive itself is pruned over time to keep the size
within manageable bounds (de Jong, 2007). More
recent variations on Pareto archiving approaches in-
clude rIPCA, which has been applied to the problem
of network security through the coevolution of adver-
sarial network attack and defence dynamics (Garcia
et al., 2017). Pareto dominance has also been em-
ployed for selection without the use of an archive,
for example the Population-based Pareto Hill Climber
(Bari et al., 2018); and Pareto fronts have been incor-
porated into an “extended elitism” framework, where
offspring are selected only if they Pareto dominate
parents when evaluated against the same opponents
(Akinola and Wineberg, 2020).
A second popular class of approaches attempt
to maintain a diverse set of evolutionary challenges
through the use of spatial embedding and multiple
populations. Spatially embedded algorithms – where
populations exist on an n-dimensional plane and in-
dividuals only interact with other individuals in the
local neighbourhood – have been shown to succeed
where other non-spatial coevolutionary approaches
fail. Explanations for how spatial models can help
combat disengagement through challenge diversity
have been explored in several works (Wiegand and
Sarma, 2004; Williams and Mitchell, 2005). Chal-
lenge diversity can also be maintained through the
use of multiple genetically-distinct populations (i.e.,
with no interbreeding or migration). Examples in-
clude the friendly competitor, where two model pop-
ulations (one “friendly” and one “hostile”) are coe-
volved against one test population (Ficici and Pollack,
1998). Tests are rewarded if they are both easy to be
defeated by a friendly model and hard to be beaten
by a hostile model; thereby ensuring pressure on tests
to evolve at a challenge-level consistent with the abil-
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
60

ity of models. Recently, a new method incorporat-
ing the periodic spawning of sub-populations, and
then re-integration of individuals that perform well
across multiple sub-populations back into the main
population has been shown to encourage continual
progress in predator-prey robot coevolution (Simione
and Nolfi, 2021).
Finally, there are approaches that focus on adapt-
ing the mechanism for selection such that individuals
are not selected in direct proportion to the number of
competitions that they win; i.e., selection favours in-
dividuals that are not unbeatable. An early endeavour
in this area is the phantom parasite, which marginally
reduces the fitness of an unbeatable competitor, while
all other fitness values remain unchanged (Rosin,
1997). Later, the Φ function was introduced for
the density classification task to coevolve cellular au-
tomata rules that classify the density of an initial con-
dition (Pagie and Mitchel, 2002). The Φ function
translates all fitness values such that individuals are
rewarded most highly for being equally difficult and
easy to classify (i.e., by being classified correctly half
of the time); while individuals that are always classi-
fied or always unclassified are punished with low fit-
ness. However, while Φ worked well, it was limited
by being domain-specific.
More generally applicable is the reduced vir-
ulence technique (Cartlidge and Bullock, 2002;
Cartlidge and Bullock, 2004a). Inspired by the be-
haviour of biological host-parasite systems, where the
virulence of pathogens evolves over time, reduced vir-
ulence is the first domain-independent technique with
tunable parameters that can be configured. After gen-
erating a parasite score through competition, reduced
virulence applies the following non-linear function to
generate a fitness for selection:
f (x
i
,υ) =
2x
i
υ
−
x
2
i
υ
2
(1)
where 0 ≤ x
i
≤ 1 is the relative (or subjective) apti-
tude of individual i and 0.5 ≤ υ ≤ 1 represents the
virulence of the parasite population. When υ = 1,
equation (1) preserves the original ranking of para-
sites (i.e., the ranking of competitive score, x) and
is equivalent to the canonical method of rewarding
parasites for all victories over hosts. When υ = 0.5,
equation (1) rewards maximum fitness to parasites
that win exactly half of all competitions. Therefore,
in domains where there is a bias in favour of one
population (the “parasites”), setting a value of υ < 1
for the advantaged population reduces the bias dif-
ferential in order to preserve coevolutionary engage-
ment. Reduced virulence demonstrated improved per-
formance, but is limited by requiring υ to be deter-
mined in advance. In many domains, bias may be
difficult to determine and may change over time. To
tackle this problem, reduced virulence has been in-
corporated into a human-in-the-loop system enabling
a human controller to steer coevolution during run-
time by observing the system behaviour and altering
the value of υ in real time (Bullock et al., 2002).
Later, autonomous virulence adaptation (AVA) –
a machine learning approach that automatically up-
dates υ during coevolution – was proposed (Cartlidge
and Ait-Boudaoud, 2011). Each generation t, AVA
updates υ using:
υ
t+1
= υ
t
+ ∆
t
(2)
∆
t
= µ∆
t−1
+ α(1 − µ)(τ − X
t
) (3)
where 0 ≤ α,µ,τ ≤ 1 are learning rate, momentum,
and target value, respectively; and X
t
is the nor-
malised mean subjective score of the population.
1
Rigorous calibration of AVA settings demonstrated
that values α = 0.0125, µ = 0.3, and τ = 0.56 can
be applied successfully in a number of diverse do-
mains. In particular, it was shown that AVA can co-
evolve high performing sorting networks and maze
navigation agents with much greater computational
efficiency than archive techniques such as LAPCA
(Cartlidge and Ait-Boudaoud, 2011).
3 SUBSTITUTION OF THE
FITTEST
We introduce substitution of the fittest (SF), a novel
technique designed to combat disengagement that
is domain-independent and requires no calibration.
Disengagement tends to occur when one population
“breaks clear” of the competing population such that
all individuals in the leading population outperform
all individuals in the trailing population. Therefore,
in simple terms, SF is designed to apply a “brake” to
the population evolving more quickly; while for the
population trying to keep pace, SF applies an “accel-
eration”. Consequently, the advantage of the leading
population over the trailing population is reduced. In
this way, SF is designed to keep populations engaged.
Unlike standard evolutionary algorithms, where
individuals are evaluated using an “absolute” fitness
function that is exogenous to the evolutionary pro-
cess, competitive coevolutionary GAs utilise a “rel-
ative” (or “subjective”) fitness evaluation, where fit-
ness ψ
i
of an individual i is endogenously assigned
1
For the initial t < 5 generations, to avoid immediate
disengagement in cases of extreme bias differential, equa-
tion (3) is replaced by ∆
t
= (0.5 − X
t
)/t; so virulence can
immediately adapt to high (υ = 1) or low (υ = 0.5) values.
Substitution of the Fittest: A Novel Approach for Mitigating Disengagement in Coevolutionary Genetic Algorithms
61

based on performance against other evolving individ-
uals. Usually, score ψ
i
is simply the proportion of
“victories” that i secures across a series of competi-
tions against evolving opponents. These competitive
interactions between coevolving populations describe
a coupled system that has potential to develop into
an arms-race of continual progress. However, when
the populations decouple, i.e., when disengagement
occurs, all information regarding the relative differ-
ences in performance of individuals is lost, such that
∀i, j : ψ
i
= ψ
j
. This is problematic and causes the
coevolving populations to drift.
The current state of a population can be measured
by the population mean subjective aptitude:
σ
pop
=
∑
n
i=1
ψ
i
n
(4)
where n is the number of individuals in the population
and 0 ≤ σ ≤ 1. Then, σ values for each population
can be used to measure the level of disengagement,
defined as:
δ = |σ
popA
− σ
popB
| (5)
where 0 ≤ δ ≤ 1. When populations have similar σ,
then disengagement δ has a low value close to zero.
When populations are fully disengaged, i.e., when
σ
popA
= 1 and σ
popB
= 0, or when σ
popA
= 0 and
σ
popB
= 1, then δ = 1. During the coevolutionary pro-
cess, δ
t
is calculated and stored each generation t. If
δ
t+1
≤ δ
t
, disengagement did not increase; otherwise,
disengagement did increase and so SF is triggered.
When SF is triggered, we first calculate the num-
ber of individuals to be substituted, κ, defined as:
κ = nδ
1
δ
(6)
where n is the number of individuals in the popula-
tion and the result is rounded up to the nearest inte-
ger. The value of κ increases non-linearly as a func-
tion of δ. As populations approach disengagement (δ
near 1), the number of substitutions tends to n. It is
important to point out that if κ >
n
2
, then effectively
only n − κ substitutions occur. For instance, given a
population with n = 6 parasites whose aptitudes are
[0.8,0.6,0.4,0.2,0.1,0.0] and κ = 4, after the substitu-
tion, their aptitudes will be [0.0,0.1,0.2,0.4,0.1,0.0].
In this example, half the population was not modified
and the two fittest individuals were substituted by the
two worst individuals. Furthermore, if δ = 1, popu-
lations would not be modified as the number of indi-
viduals to be substituted is the same as the population
size (i.e., κ = n). On the contrary, when δ < 0.3, κ
tends to 0. During these times, populations are suffi-
ciently engaged and substitutions are not necessary.
The next step in the SF process consists of com-
paring σ
popA
and σ
popB
and then substituting κ indi-
viduals in each population using the following rules:
• Population with lowest σ: Rank all individuals by
subjective aptitude ψ
i
. Then, the κ individuals
with the highest ψ
i
replace the κ individuals with
the lowest ψ
i
. Finally, the subjective aptitude of
every individual is increased by:
ψ
0
i
= min(ψ
i
+ δ,1) (7)
taking minimum value to ensure 0 ≤ ψ
0
i
≤ 1.
• Population with highest σ: Rank all individuals by
ψ
i
. Then κ individuals with the lowest ψ
i
replace
the κ individuals with the highest ψ
i
; following
replacement, the subjective aptitude of every indi-
vidual is decreased using:
ψ
0
i
= max(ψ
i
− δ,0) (8)
taking maximum value to ensure 0 ≤ ψ
0
i
≤ 1.
As described, SF affects each population in a dif-
ferent manner. For the population that evolves more
quickly, the proportion of individuals to be randomly
selected is increased. For instance, if the highest
ranked individual whose ψ < δ, then the κ individ-
uals would be selected at random because they would
have ψ
0
= 0. On the other hand, for the population
which evolves slower than the other, the effect is the
opposite, i.e., if the lowest ranked individual whose
ψ + δ ≥ 1, then all κ individuals would have ψ
0
= 1
and hence those individuals would have high proba-
bility of being selected.
Following SF, selection is performed and genetic
operators are applied as usual. In the minimal exper-
iments we present in the following sections, we use
tournament selection and apply mutation, i.e., popu-
lations are asexual and recombination is not used.
4 EXPERIMENTAL METHOD
4.1 The “Greater Than” Game
The greater than game (Watson and Pollack, 2001)
was introduced as a minimal (and analytically
tractable) substrate capable of demonstrating some of
the pathological dynamics of coevolution; in partic-
ular disengagement. The game consists of maximis-
ing scalar values through a comparison-based func-
tion where given two scalar values, α and γ, the
function operates as score(α,γ) = 1 if α > γ, 0 oth-
erwise. Here, we use a slightly modified “greater
than or equals” game that rewards draws; such that
score(α,γ) = 1 if α > γ; 0.5 if α = γ; 0 otherwise.
The coevolutionary set-up consists of two isolated
populations, each with n individuals. Each individual
is represented by a binary string with l = 100 bits and
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
62
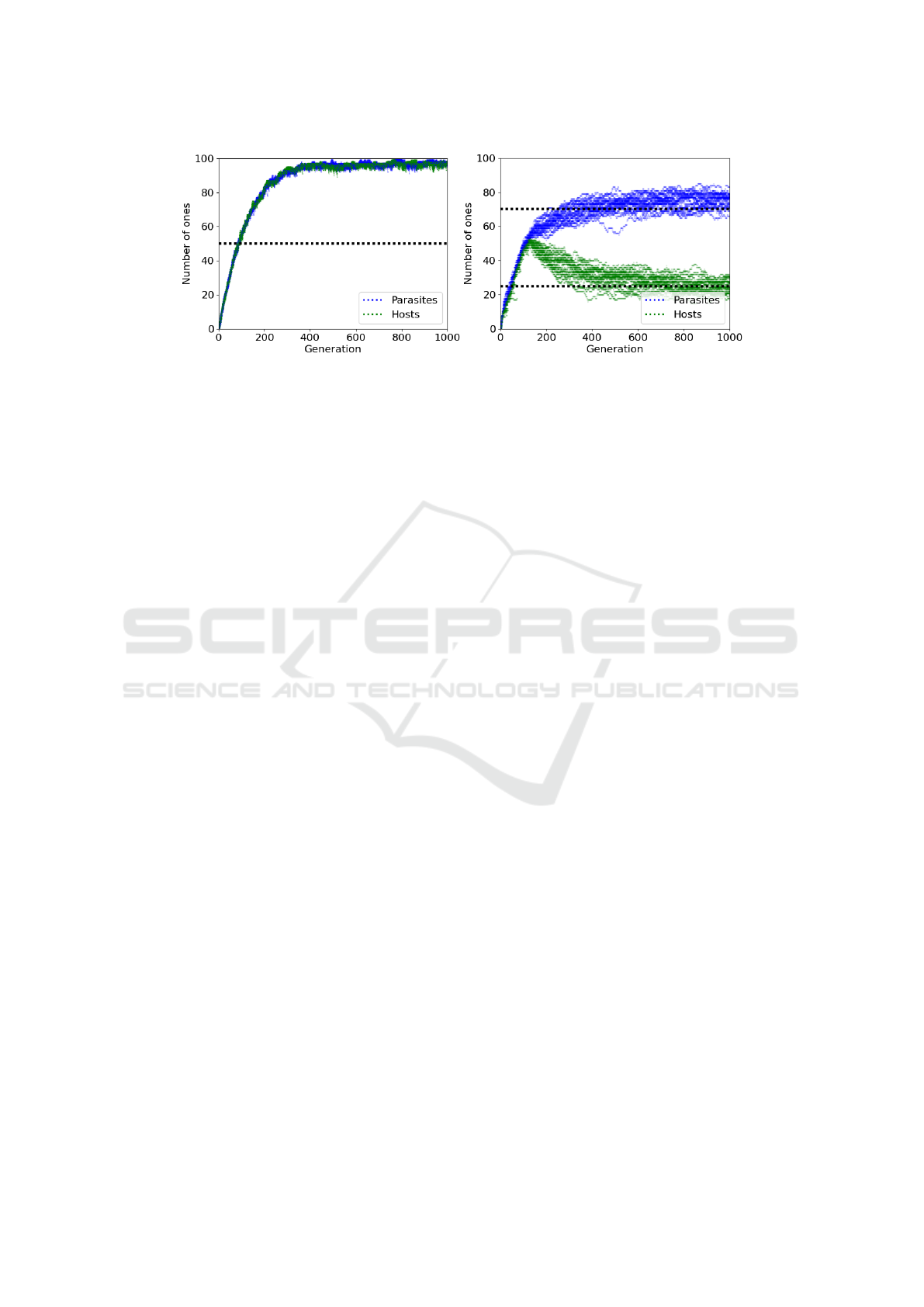
Figure 1: Coevolution: (left) equal bias β
h
= β
p
= 0.5; and (right) differential bias β
h
= 0.25, β
p
= 0.75.
the “objective” purpose of the coevolutionary system
is to evolve individuals with bit-strings containing all
ones (i.e., scalar values of l = 100). Every genera-
tion, to generate a subjective aptitude score, each in-
dividual plays the greater than game against a sam-
ple of S opponents. Tournament selection is used to
select individuals for reproduction, and the only ge-
netic operator is mutation (i.e., reproduction is asex-
ual). Mutation has a bias controlled by parameter β,
where 0 ≤ β ≤ 1. For each bit, there is a probability
m of mutation occurring. When it occurs, the bit is
assigned a new value at random, with probability β of
assigning a 1, and probability 1 − β of assigning a 0;
i.e., when β = 0.5, mutation has an equal chance of
assigning the bit to 1 or 0; when β = 0 mutation will
always assign the bit to 0; and when β = 1 mutation
will always assign the bit to 1. This bias parameter β
allows the simple game to emulate the intrinsic asym-
metry of real and more complex domains, where it is
often easier for one population to outperform another.
Under mutation bias alone, i.e., when populations are
disengaged and left to drift under the absence of se-
lection pressure, we expect the population to tend to-
wards having β×l ones. Therefore, for a bias β = 0.5,
we would expect the population to drift towards scalar
values of 50.
In our two-population competitive set up, we label
the populations as as hosts and parasites. Each popu-
lation has an independent bias value β, which controls
the problem difficulty for each population. We use
β
h
to label the bias value of the host population and
β
p
to label the bias value of the parasite population.
When bias differential is high, i.e., when the value of
β
p
is much larger than the value of β
h
(or vice versa),
disengagement becomes more likely as the game is
much easier for the parasites (alternatively, the hosts)
to succeed. In more complex domains, there is often
asymmetry in problem difficulty for coevolving pop-
ulations. By varying β
p
and β
h
, we are able to control
the asymmetry of problems in the simple greater than
game.
Our experimental set up is detailed as follows. We
coevolve two isolated populations, each with 25 indi-
viduals (n = 25). The length of the binary array (an
individual) is 100 (l = 100) and each bit is initialised
to 0. For generating a competitive score, we use a
sample size of 5 (S = 5). Tournament size for selec-
tion is 2. The probability of mutation per bit is 0.005
(m = 0.005). Finally, each evolutionary run lasts for
1000 generations.
4.2 Disengagement
The effect of disengagement can most clearly be elab-
orated by visualising what happens when it occurs.
Figure 1 (left) presents the coevolution of two popu-
lations, each with equal bias β
p
= β
h
= 0.5. We see
that populations remain engaged throughout the evo-
lutionary run. This engagement provides a continual
gradient for selection and encourages an “arms race”
of increasing performance. As a result, both popula-
tions reach optimal performance of 100 ones. This is
far higher than 50 ones (dotted line) that both popu-
lations would be expected to reach if drifting through
space under mutation alone, i.e., when selection pres-
sure is removed.
In contrast, Figure 1 (right) demonstrates the
pathology of disengagement. Here, there is a differen-
tial bias in favour of parasites, such that β
p
= 0.75 and
β
h
= 0.25. Initially, both populations remain engaged
and selection drives evolutionary progress, with both
populations reaching approximately 50 ones by gen-
eration 150. However, the impact of differential bias
in favour of parasites then leads to a disengagement
event such that all parasites have more ones than their
competing hosts; resulting in a subjective score of
zero for all hosts and a subjective score of one for
all parasites. At this point selection pressure is re-
moved as all individuals have an equal (and there-
fore random) chance of selection, leaving the popu-
lations to drift under mutation alone. As expected,
the parasite population drifts to the parasite mutation
Substitution of the Fittest: A Novel Approach for Mitigating Disengagement in Coevolutionary Genetic Algorithms
63
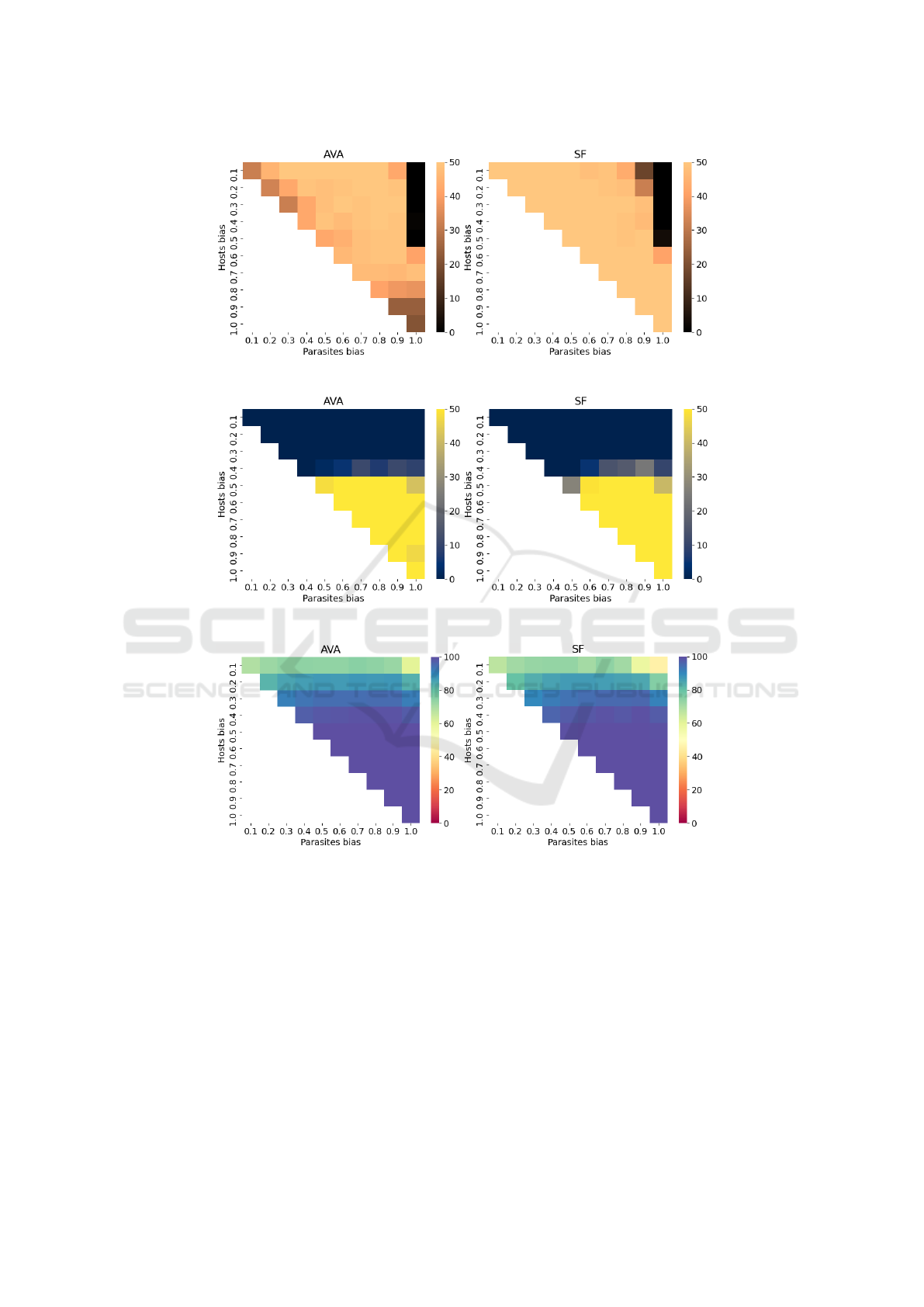
Figure 2: Number of runs with no disengagement; AVA (left) and SF (right) across all bias levels (50 trials).
Figure 3: Number of runs where hosts reached optimum; AVA (left) and SF (right) across all bias levels (50 trials).
Figure 4: Mean number of ones of best host; AVA (left) and SF (right) across all bias levels (50 trials).
bias (dotted line) of 75, while hosts degrade to the
host mutation bias of 25. The high bias differential
between populations (β
p
− β
h
= 0.5) not only causes
the initial disengagement event, but ensures that post-
disengagement populations drift through different re-
gions of genotype/phenotype space and will never re-
engage through chance alone.
In general, the greater the bias differential be-
tween populations, the greater the likelihood of dis-
engagement occurring. As shown, disengagement
severely hinders coevolutionary progress.
5 SF VS AVA: A COMPARISON
To measure the performance of SF, we perform a
thorough comparison against AVA (Cartlidge and Ait-
Boudaoud, 2011), which has been previously shown
to dramatically reduce the effects of disengagement
in the greater than game and also in several more
complex and realistic domains, including design-
ing minimal-length sorting networks and discovering
classifier systems for maze navigation. To understand
how the two approaches are likely to perform in more
complex domains, where population asymmetries are
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
64

more likely, we trial both SF and AVA in simulations
where mutation bias is varied across all possible levels
(β
p
≥ β
h
) in the range [0.1, 1.0]. The mutation bias
was configured in favour of parasites (except when
β
p
= β
h
); although this decision is arbitrary and re-
sults where bias is in favour of hosts would yield sym-
metrically similar results. For each bias scenario, we
performed 50 experimental trials. To analyse perfor-
mance of SF and AVA, we utilise three metrics: (i) the
reliability of the technique to maintain population en-
gagement; (ii) the capacity to discover optimal hosts
containing all ones; and (iii) the mean number of ones
that hosts reach before disengagement occurs. The
following sections describe our findings.
5.1 Maintaining Engagement
Since the main objective of AVA and SF is to main-
tain engagement during coevolution, regardless of
bias fluctuations that may arise, it is fundamental to
study their response under diverse bias levels. Fig-
ure 2 presents a heatmap showing the number of runs
where AVA and SF maintained population engage-
ment during the full coevolutionary process (regard-
less of whether an optimal host is found).
Overall, we see that SF is able to maintain en-
gagement more successfully than AVA across a di-
verse range of bias differentials. For 38 of the 50 bias
pairings, SF maintains engagement for the full coevo-
lutionary run of 1000 generations across all 50 trials.
For AVA, however, this number is only 12. Moreover,
SF maintains engagement for the full coevolutionary
run in at least 40 out of 50 trials across 49 bias pair-
ings, while for AVA this number is 42.
Relative to SF, results suggest that AVA tends to
struggle in scenarios (i) where the bias of both popu-
lations are either the same (symmetrical systems) or
similar; and (ii) where parasite bias is very high (e.g.,
β
p
= 1.0) and there is a large bias differential between
parasites and hosts. In comparison, although SF also
fails in scenarios where parasites have very high bias,
SF is capable of maintaining engagement where AVA
is not.
Regarding the result obtained by AVA, in the orig-
inal research (Cartlidge and Ait-Boudaoud, 2011), it
was calibrated to only handle bias levels in the range
[0.5, 1.0]. Moreover, in the original experiments the
number of generations is 750 whereas in these exper-
iments, the number of generations is 1000. The du-
ration of experimental trials is a key factor inasmuch
AVA, in a number of bias scenarios, tends to allow
disengagement after optimal hosts are found. For in-
stance, when β
h
= 0.5, β
p
= 1.0, populations tend to
first reach the optimum, but then later, around genera-
tions 850-900, the populations disengage. This unex-
pected behaviour suggests that AVA parameters may
require recalibration to maintain engagement over
long time periods when there is a high bias differ-
ential. It also demonstrates an advantage of SF over
AVA, as SF has no parameter settings to calibrate.
5.2 Reaching the Optimum
Another essential aspect to analyse is the capability
to reach the optimal zone. Figure 3 presents the num-
ber of runs where hosts (more precisely, at least one
host) reached the optimal, regardless of whether or
not populations disengage after this point. We see a
similar pattern for both SF and AVA. AVA reached
the optimal zone at least 40 times under 21 bias lev-
els, where as SF was capable of reaching 40 times or
more under 20 bias levels. Furthermore, both AVA
and SF reached the optimal zone a maximum of 50
times (i.e., every time) under 18 bias scenarios.
As expected, in multiple bias scenarios hosts were
not capable of reaching the optimum when hosts have
a very low mutation bias (β
h
< 0.5).
5.3 General Performance
Figure 4 shows the mean maximum number of ones
of the best host across all bias configurations, regard-
less of whether or not populations disengage or hosts
reach the optimum. Again, performance of SF and
AVA is similar. AVA reaches at least 90 ones under
36 bias scenarios, whereas SF reaches 90 ones in 35
scenarios. Furthermore, AVA and SF both reach 100
ones under 18 possible bias scenarios.
Results suggest that AVA and SF tend to behave
similarly across most bias levels. However, when
there is a significant bias differential (e.g., β
h
=
0.1,β
p
= 0.9), AVA enables populations to reach a
greater performance (closer to the optimum) than SF.
5.4 Coevolutionary Dynamics
Figures 5 and 6 present example runs to highlight the
effects of AVA (left) and SF (right) during the coevo-
lutionary process. When bias differential is relatively
large (Figure 5; β
h
= 0.3 and β
b
= 0.7) both AVA
and SF maintain engagement throughout. However,
when bias differential is very large (Figure 6; β
h
= 0.2
and β
b
= 1.0), disengagement occurs under both tech-
niques, but tends to occur earlier (around generation
410 for AVA, compared with generation 550 for SF).
Interestingly, in Figure 6 we see that SF induces
different population dynamics with respect to AVA.
Under SF, in the early generations, parasites exhibit
Substitution of the Fittest: A Novel Approach for Mitigating Disengagement in Coevolutionary Genetic Algorithms
65
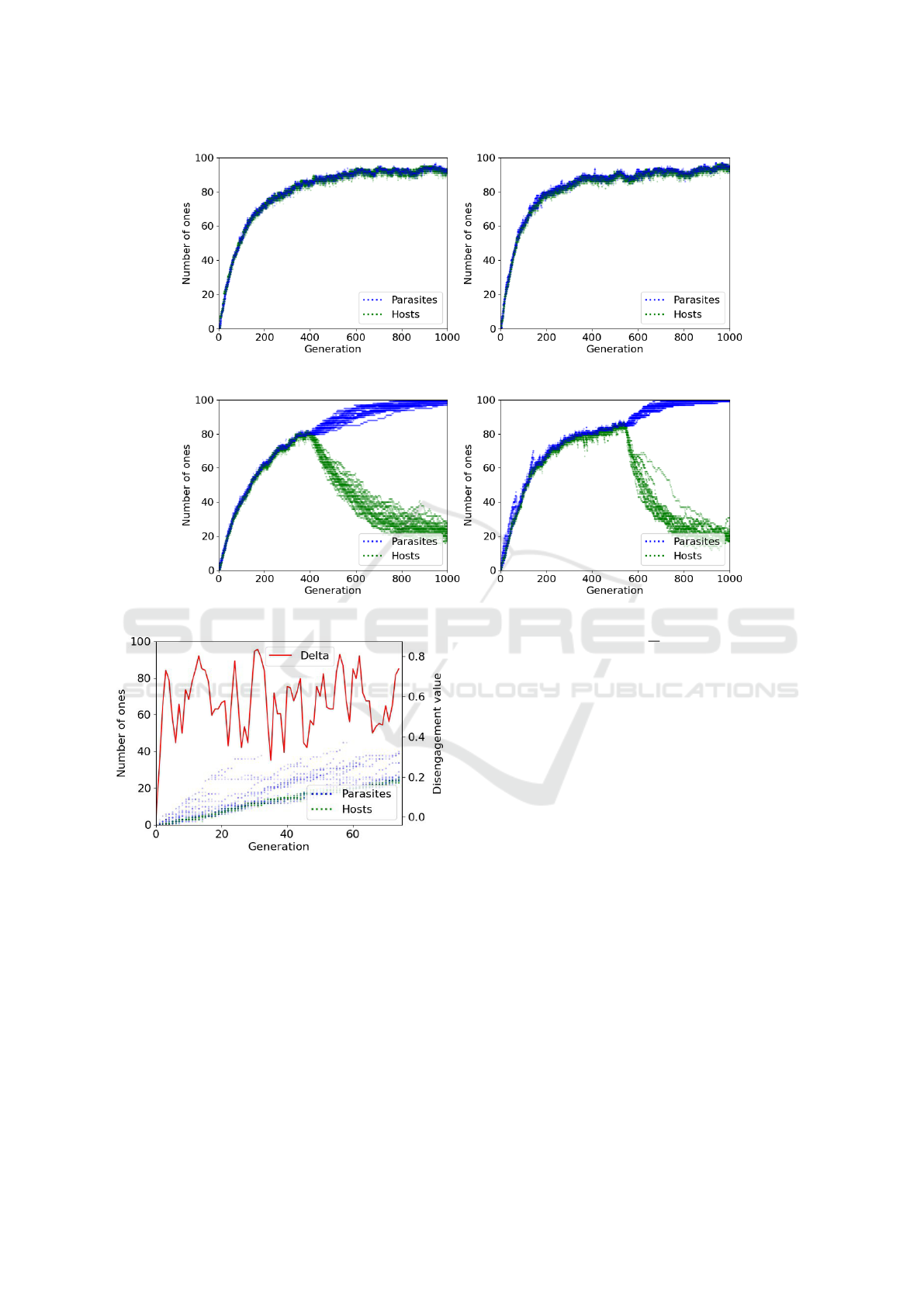
Figure 5: Example coevolutionary runs under AVA (left) and SF (right) with β
h
= 0.3, β
p
= 0.7.
Figure 6: Example coevolutionary runs under AVA (left) and SF (right) with β
h
= 0.2, β
p
= 1.0.
Figure 7: Initial generations of one run (β
h
= 0.1, β
p
= 1.0)
using SF, showing absolute fitness of parasites and hosts
(left axis) and δ value of disengagement (right axis).
lots of variation, with some outliers drifting far from
the engaged populations. Then, around generations
90 and 170, these outlier lineages suddenly disappear.
A similar “cull” effect is not observed with AVA.
To further investigate the cull effect in SF and its
direct relation with disengagement value δ, Figure 7
presents the initial generations of one example run
with very high bias differential (β
h
= 0.1, β
p
= 1.0).
Around generation 30, we see that there is a single
outlier parasite containing around 40 ones. As a con-
sequence, δ ≈ 0.81 and κ = 25(0.81
1/0.81
) = 19 (see
Equation 6). Since 19 >
25
2
, the effective substitu-
tions are 6 (see example given in Section 3). There-
fore, the 6 fittest parasites, including the outlier, are
substituted by the 6 worst parasites. Consequently,
the subsequent generations do not present outliers and
δ (and therefore κ) decreases. Other outlier lineages
later begin to emerge and the process repeats. In con-
trast, AVA tends to keep both populations more tightly
coupled throughout.
6 CONCLUSIONS
This research has introduced SF as an alternative tech-
nique to mitigate disengagement in competitive co-
evolutionary genetic algorithms. Using a minimal
problem domain to enable exposition, we compared
the performance of SF with AVA, a technique in the
literature that has been shown to combat disengage-
ment in a variety of domains. Experimental results
suggest that, in general, SF has similar performance to
AVA in terms of discovery of optimal solutions. How-
ever, SF is also shown to have better performance than
AVA in terms of consistently maintaining engagement
across a wide variety of bias differentials, i.e., where
there is a large inherent advantage in favour of one co-
evolving population. The mechanism of SF is deliber-
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
66

ately designed to be simple and domain independent,
requiring no domain knowledge or specific calibra-
tion. This makes SF more easy to implement than
other techniques and offers the possibility of being
more generally applicable.
However, one of the potential weaknesses ob-
served in SF is the highly-fluctuating behaviour in-
duced in populations (i.e., the “cull” effect), which
might lead to sudden disengagement in other more re-
alistic domains. Thus, we believe that SF deserves
further exploration; although it has shown suitable
performance in a simple domain, experiments in more
complex domains such as maze navigation or sort-
ing networks (Cartlidge and Ait-Boudaoud, 2011) are
necessary to demonstrate its reliability. Furthermore,
a robust comparison against other state-of-art tech-
niques will be performed. Finally, we intend to ex-
plore the effects that SF has on other coevolutionary
pathologies, such as overspecialisation and cycling.
ACKNOWLEDGEMENTS
Hugo Alcaraz-Herrera’s PhD is supported by The
Mexican Council of Science and Technology (Con-
sejo Nacional de Ciencia y Tecnolog
´
ıa - CONACyT).
John Cartlidge is sponsored by Refinitiv.
REFERENCES
Akinola, A. and Wineberg, M. (2020). Using implicit multi-
objectives properties to mitigate against forgetfulness
in coevolutionary algorithms. In Genetic and Evo-
lutionary Computation Conference, GECCO, pages
769–777.
Bari, A. G., Gaspar, A., Wiegand, R. P., and Bucci, A.
(2018). Selection methods to relax strict acceptance
condition in test-based coevolution. In Congress on
Evolutionary Computation, CEC, pages 1–8.
Bullock, S., Cartlidge, J., and Thompson, M. (2002).
Prospects for computational steering of evolutionary
computation. In Workshop Proceedings 8th Int. Conf.
on Artificial Life, ALIFE, pages 131–137.
Cartlidge, J. and Ait-Boudaoud, D. (2011). Autonomous
virulence adaptation improves coevolutionary opti-
mization. IEEE Transactions on Evolutionary Com-
putation, 15(2):215–229.
Cartlidge, J. and Bullock, S. (2002). Learning lessons from
the common cold: How reducing parasite virulence
improves coevolutionary optimization. In Congress
on Evolutionary Computation, CEC, pages 1420–
1425 vol.2.
Cartlidge, J. and Bullock, S. (2003). Caring versus shar-
ing: How to maintain engagement and diversity in
coevolving populations. In Banzhaf, W., Ziegler, J.,
Christaller, T., Dittrich, P., and Kim, J. T., editors, Ad-
vances in Artificial Life, pages 299–308, Berlin, Hei-
delberg. Springer Berlin Heidelberg.
Cartlidge, J. and Bullock, S. (2004a). Combating coevolu-
tionary disengagement by reducing parasite virulence.
Evolutionary Computation, 12(2):193–222.
Cartlidge, J. and Bullock, S. (2004b). Unpicking tartan
CIAO plots: Understanding irregular coevolutionary
cycling. Adaptive Behavior, 12(2):69–92.
de Jong, E. D. (2007). A monotonic archive for pareto-
coevolution. Evolutionary Computation, 15(1):61–93.
Ficici, S. G. and Pollack, J. B. (1998). Challenges in
coevolutionary learning: Arms-race dynamics, open-
endedness, and mediocre stable states. In Interna-
tional Conference on Artificial Life, ALIFE, pages
238–247.
Garcia, D., Lugo, A. E., Hemberg, E., and O’Reilly, U.-
M. (2017). Investigating coevolutionary archive based
genetic algorithms on cyber defense networks. In
Genetic and Evolutionary Computation Conference
Companion, GECCO, pages 1455–1462.
Hillis, W. D. (1990). Co-evolving parasites improve simu-
lated evolution as an optimization procedure. Phys. D,
42(1–3):228–234.
Miguel Antonio, L. and Coello Coello, C. A. (2018). Co-
evolutionary multiobjective evolutionary algorithms:
Survey of the state-of-the-art. IEEE Transactions on
Evolutionary Computation, 22(6):851–865.
Pagie, L. and Mitchel, M. (2002). A comparison of evo-
lutionary and coevolutionary search. International
Journal of Computational Intelligence and Applica-
tion, 2(1):53–59.
Popovici, E., Bucci, A., Wiegand, R. P., and De Jong, E. D.
(2012). Coevolutionary principles. In Handbook of
Natural Computing, pages 987–1033. Springer Berlin
Heidelberg, Berlin, Heidelberg.
Rosin, C. D. (1997). Coevolutionary Search Among Adver-
saries. PhD thesis, Department of Computer Science,
University of California, San Diego, California.
Rosin, C. D. and Belew, R. K. (1997). New methods for
competitive coevolution. Evolutionary Computation,
5(1):1–29.
Simione, L. and Nolfi, S. (2021). Long-term progress and
behavior complexification in competitive coevolution.
Artificial Life, 26(4):409–430.
Watson, R. A. and Pollack, J. B. (2001). Coevolutionary
dynamics in a minimal substrate. In Genetic and Evo-
lutionary Computation Conference, GECCO, pages
702–709.
Wiegand, R. P. and Sarma, J. (2004). Spatial embedding
and loss of gradient in cooperative coevolutionary al-
gorithms. In Parallel Problem Solving from Nature,
PPSN, pages 912–921.
Williams, N. and Mitchell, M. (2005). Investigating the suc-
cess of spatial coevolution. In Genetic and Evolution-
ary Computation Conference, GECCO, pages 523–
530.
Substitution of the Fittest: A Novel Approach for Mitigating Disengagement in Coevolutionary Genetic Algorithms
67
