
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local
Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
Tabish Ashfaq, Nivedha Ramesh and Nawwaf Kharma
Department of Electrical and Computer Engineering, Concordia University, Montreal, Quebec, Canada
Keywords:
Neuroevolution, Evolutionary Algorithms, Convolutional Neural Networks, Deep Learning, Cartesian
Genetic Programming, Genetic Algorithms, Stochastic Local Search, Novelty Search, Simulated Annealing.
Abstract:
Convolutional neural networks (CNNs) are deep learning models that have been successfully applied to vari-
ous computer vision tasks. The design of CNN topologies often requires extensive domain knowledge and a
high degree of trial and error. In recent years, numerous Evolutionary Algorithms (EAs) have been proposed to
automate the design of CNNs. The search space of these EAs is very large and often deceptive, which entails
great computational cost. In this work, we investigate the design of CNNs using Cartesian Genetic Program-
ming (CGP), an EA variant. We then augment the basic CGP with methods for identifying potential/actual
local optima within the solution space (via Novelty Search), followed by further local optimization of each of
the optima (via Simulated Annealing). This hybrid EA methodology is evaluated using the MNIST data-set
for handwritten digit recognition. We demonstrate that the use of the proposed method results in considerable
reduction of computational effort, when compared to the basic CGP approach, while still returning competitive
results. Also, the CNNs designed by our method achieve competitive recognition results compared to other
neuroevolutionary methods.
1 INTRODUCTION
Image recognition is a widely studied application of
computer vision. In recent years Convolutional Neu-
ral Networks (CNNs) have proven to be especially ef-
fective at this task in a supervised learning context.
This has motivated the development of numerous evo-
lutionary algorithms (EAs) for the design of CNN ar-
chitectures and for the optimization of their hyperpa-
rameters (Baldominos et al., 2018; Xie and Yuille,
2017; McGhie et al., 2020).
The computational effort expended by such algo-
rithms in obtaining optimal solutions is considerable,
and thus, such an approach may not be a viable op-
tion when computational resources are constrained.
As with many applications of evolutionary comput-
ing, a large proportion of the computational effort is
expended in evaluating a population of individual so-
lutions, many of which may be highly unfit or simply
redundant. As such, if a candidate solution is deter-
mined (by some criteria) to be useless, it may be re-
moved from the population, which saves on compu-
tational time without degrading the effectiveness of
evolutionary search and optimization.
Therefore, we are motivated to investigate modi-
fications to the standard evolutionary approach which
may lead to a more efficient neural architecture search
procedure. This implies that potential solutions to the
problem should aim to reduce the number of wasted
cycles in the evolution process. This may be done
by identifying local optima within the solution space
and restricting the search to the neighbourhoods of
these local optima. For this purpose, we present a
method for approximate quantification of structural
diversity and for characterization of local neighbour-
hoods within a population of CNNs.
In this work, we investigate the use of a three-
stage evolutionary optimization approach. The first
stage in this approach aims to generate a diverse ini-
tial generation of CNNs using the Novelty Search al-
gorithm (Lehman and Stanley, 2011). The second
stage involves the evolution of CNN architectures us-
ing Cartesian Genetic Programming (CGP) (Miller
and Thomson, 2000). In the third stage, we select
the most diverse generation from the previous stage.
From this population, we sample the most optimal in-
dividuals in terms of both fitness and novelty, using
a multi-objective optimization algorithm. We attempt
Ashfaq, T., Ramesh, N. and Kharma, N.
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for Handwritten Digit Recognition.
DOI: 10.5220/0010648300003063
In Proceedings of the 13th International Joint Conference on Computational Intelligence (IJCCI 2021), pages 123-133
ISBN: 978-989-758-534-0; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
123

to exploit the local neighbourhoods of these individ-
uals using a stochastic local search (SLS) algorithm.
The SLS algorithm implemented for this paper is the
Simulated Annealing (SA) (Kirkpatrick et al., 1983)
algorithm.
The rest of this paper is organised as follows.
Section 2 provides a review of related work in neu-
roevolution. Section 3 describes the genetic encoding,
and our optimized evolutionary approach to design-
ing CNN architectures. We present the results of our
proposed methodology in Section 4. Finally, we end
with some concluding remarks and recommendations
for future work in Section 5.
2 RELATED WORK
Neuroevolution has been used to design various types
of neural network weights and topologies to address
a wide range of problems. In 2002, the highly
influential technique, Neuroevolution of Augment-
ing Topologies (NEAT) (Stanley and Miikkulainen,
2002), was developed and proved effective in evolv-
ing the architecture and weights of small neural net-
works. In 2009, this method was extended with the
introduction of the HyperNEAT algorithm (Stanley
et al., 2009), which allowed for the evolution of larger
scale neural networks through indirect encoding using
a Compositional Pattern Producing Network (CPNN).
However, this technique has been shown to be inef-
fective in performing image classification, and has re-
sulted in an error rate of 7.9% on the MNIST dataset
when used for feature learning with a CNN (Verbanc-
sics and Harguess, 2015).
In recent years, several works have been pub-
lished which attempt to automate the design of CNNs
through neuroevolution. Genetic CNN (Xie and
Yuille, 2017) is a Genetic Algorithm which uses a
fixed-length binary string representation. The au-
thors obtained competitive recognition results on the
MNIST dataset. However, they do not encode the
fully-connected part of the network, choosing to adapt
the fully-connected part of the basic LeNet architec-
ture (Lecun et al., 1998).
Baldominos et al. (Baldominos et al., 2018) stud-
ied the evolution of CNNs using both a GA with bi-
nary Gray encoding representation, as well as Gram-
matical Evolution which uses an integer-based en-
coding. They include both convolutional and fully-
connected components in their frameworks, in addi-
tion to parameterizing the connectivity pattern within
the networks, thereby allowing for the evolution of
recurrent structures. The authors also present a sim-
ilarity metric which they use to implement a niching
strategy. However, the metric presented in this paper
evaluates to zero for individuals with unequal num-
bers of convolutional and dense layers, and hence,
it does not provide a measure of similarity in many
cases.
Sun et al. (Sun et al., 2020) propose a GA
which encodes highly functional blocks in variable-
length chromosomes, and incorporates skip connec-
tions. Similarly, a CGP-based approach (Suganuma
et al., 2017) was published in 2017 which uses a func-
tion set of convolutional and residual units to reduce
the search space. In both of these cases, only the con-
volutional part of the network is encoded. In addi-
tion, these works do not attempt to measure structural
similarity between different neural networks. In this
work, we augment the CGP approach introduced by
Suganuma et al. by extending the function set and
incorporating transpose convolutions and fully con-
nected layers.
3 METHODS
We evolve two separate populations of CNNs. The
first of these populations, is optimized according to
the fitness criteria described in Section 3.2.2, begin-
ning with a randomly generated initial generation.
This population is henceforth referred to as Popula-
tion 1. In the second population, Population 2, as an
alternative to random initialization, we include a dis-
tinct initialization stage. In this stage, we begin with
a randomly generated population and optimize it for
novelty, using the Novelty Search algorithm (Lehman
and Stanley, 2011) along with the metric developed
in Section 3.3. This process yields the initial genera-
tion for the evolutionary algorithm. Lastly, we apply
a different survivor selection scheme during evolution
of each of the two populations, as described in Section
3.2.3.
The individual networks evolved in our algorithms
are each designed as a combination of two sub-
networks: a fully convolutional network followed by
a fully connected (dense) network.
3.1 Genetic Encoding
For the encoding of the fully convolutional sub-
network, we adapt a CGP based approach (Suganuma
et al., 2017) which uses the functional modules de-
scribed in Table 1 as the node functions. The fully-
connected sub-network is represented using a list of
node functions drawn from a function set of dense
layers. Thus, the genotype may be considered as the
combination of a CGP grid - where each element of
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
124

the grid contains a function gene as well as a connec-
tion gene - and a list of fully-connected node func-
tions.
The combination of these two encodings is ex-
pressed as a directed acyclic graph (DAG). The DAG
is an intermediate phenotype which may be adjusted
to ensure the validity of networks generated for a
given classification task. For the MNIST dataset, we
enforce the output layer of each network to be a dense
layer with 10 nodes and a softmax activation function.
Finally, the DAG representation is used to con-
struct the CNN, which is implemented as a Keras
(Chollet et al., 2015) model. In the remainder of this
paper, the CNN is referred to as the phenotype.
3.1.1 Representation of the Fully Convolutional
Sub-network
The fully-convolutional sub-network is represented
using a two-dimensional matrix, in which each ele-
ment of the matrix contains a string that describes the
node function gene as well as the indices of the input
nodes. Seven types of node functions are prepared
and included in the function set, namely, ConvBlock,
ResBlock, DeconvBlock, max pooling, average pool-
ing, concatenation, and summation.
The ConvBlock consists of standard convolution
processing with a stride of 1, followed by batch nor-
malization and a ReLU activation function. All con-
volutions are same convolutions, i.e. the input to the
convolution operation is padded with zeros, result-
ing in an output that matches the input in terms of
height and width of the activation map. Thus, the
height and width of an activation map is reduced only
through pooling layers. The function set contains
ConvBlocks of different numbers of output channels
and kernel sizes. ResBlocks are composed of Con-
vBlocks followed by a second convolution operation,
batch normalization, summation, and finally ReLU
activation. DeconvBlocks consists of a convolution
operation followed by batch normalization, ReLU ac-
tivation, and finally upsampling by a factor of 2.
The max pooling and average pooling functions
are applied over a window size of 2, using a stride
value of 2. The concatenation function takes two ac-
tivation maps as input, and concatenates them along
the feature axis. In the case where the two activa-
tion maps have unequal height and width, the larger
activation map is downsampled by max pooling prior
to concatenation. Finally, the summation function is
used to add the values of two activation maps. In cases
where direct summation is not possible due to incom-
patible numbers of channels in the activation maps,
the number of channels in the smaller of the two acti-
vation maps is increased by applying 1-by-1 convolu-
tion prior to summation.
3.1.2 Representation of the Fully-connected
Sub-network
The fully-connected sub-network is represented as a
list of DenseBlock node functions as described in Ta-
ble 1. This network is then appended to the output of
the fully convolutional sub-network. As mentioned
above, a softmax output layer with 10 nodes is ap-
pended to the end of the fully-connected network.
Figure 1 illustrates the genotype of the fittest solution
obtained by the proposed method.
3.2 Evolutionary Algorithm
Evolutionary strategies belong to the family of evo-
lutionary algorithms and are majorly applied to opti-
mization problems. The search paradigm inspired by
biological evolution involves applying mutation, re-
combination and selection operators to a population
of candidate solutions. In our work, we adopt the (20
+ λ) evolutionary strategy to evolve CNN architec-
tures. We use a value of λ = 16. An initial population
of 20 individuals is randomly generated subject to ini-
tialization constraints which ensure that the resulting
CNNs will fit in the available GPU memory.
3.2.1 Mutation Operators
Variation in the population is introduced through mu-
tation of function and connection genes – that is, a
given node may have its function swapped with a ran-
dom selection from the function set, or may have its
immediate neighbours swapped.
3.2.2 Fitness Evaluation
Each individual is trained on the MNIST training set
for 5 epochs, after which it is evaluated on the held-
out validation set. It should be noted that the test set
is not used for evaluation during evolution. Training
hyperparameters are kept constant for all individuals.
We use the Adam optimization algorithm (Kingma
and Ba, 2017) to minimize Categorical Cross-Entropy
loss. A learning rate of 1e-4 is used during training for
all individuals.
The partially trained networks are then evaluated
using the F
1
score, defined in (1), where TP: true pos-
itive, FP: false positive, FN: false negative:
F
1
=
T P
(T P +
1
2
(FP + FN))
(1)
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
125
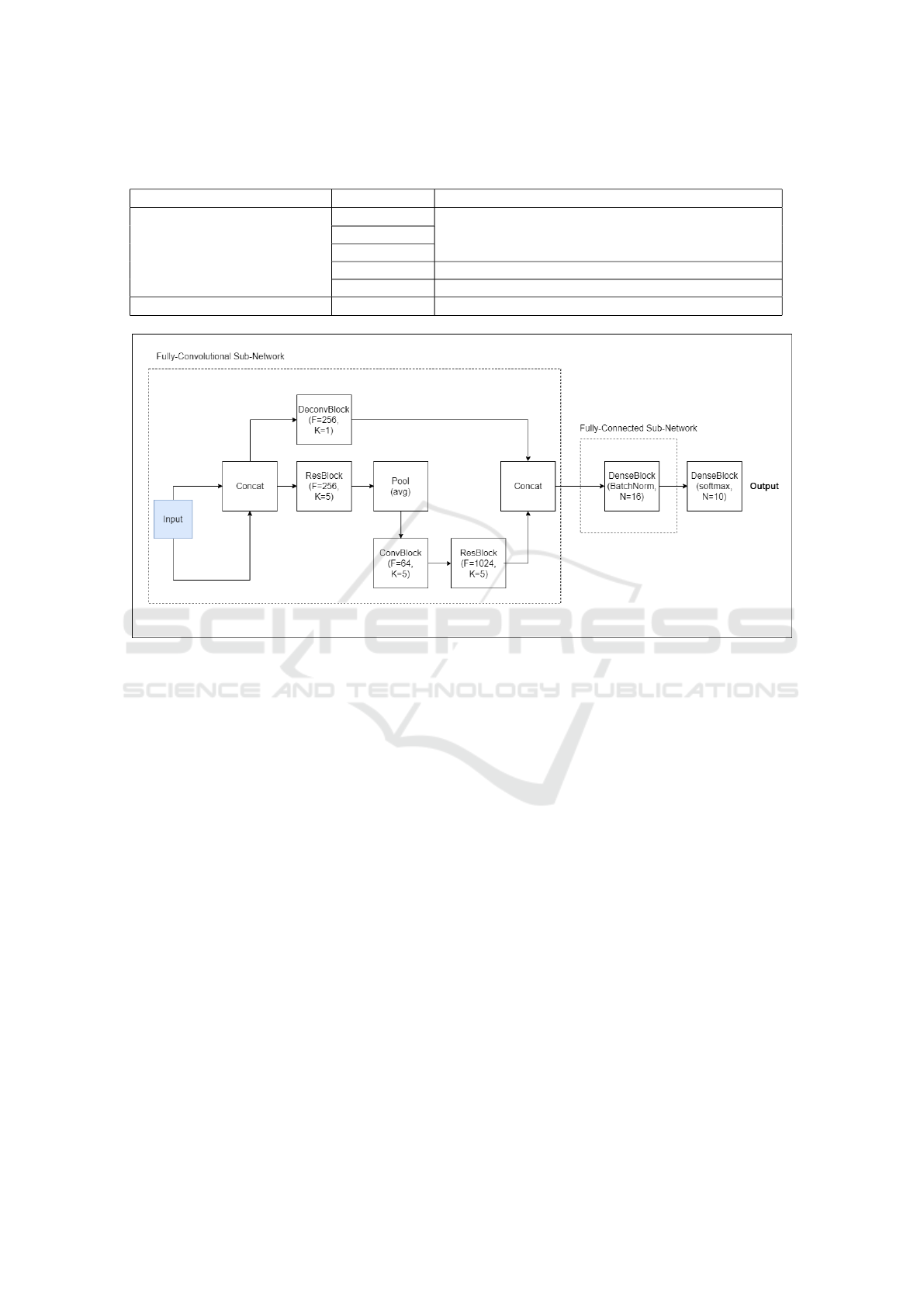
Table 1: Node functions present in the CGP function set. F: number of filters (output channels), K: kernel size, N: number
of units in a dense layer.
Sub-Network Node Type Variation
Fully-Convolutional Network
ConvBlock
F = {1, 2, 4, 8, 16, 32, 64, 128, 256, 512, 1024}
K = {1, 3, 5}
ResBlock
DeconvBlock
Pooling Average pooling, Max pooling
MergeBlock Summation, Concatenation
Fully-Connected Network DenseBlock N = {8, 16, 32, 64, 128, 256, 512, 1024, 2048, 4096}
Figure 1: Genotype of the best performing CNN architecture obtained by the proposed method. The figure demonstrates how
the active path of the CGP genotype is expressed, resulting in an encoding that describes the CNN in terms of the highly
functional modules enumerate in Table 1.
3.2.3 Parent Selection and Survivor Selection
For parent selection, we use tournament selection
(Miller and Goldberg, 1995) from the current gen-
eration with a window size of 5. We select 20% of
the population without replacement. For survivor se-
lection, we use tournament selection from the pool
of parents and offspring of the current generation,
without replacement. A window size of 5 is used.
We favour individuals with smaller model size, where
model size is defined as the number of trainable pa-
rameters of a CNN. Thus, when comparing two indi-
viduals, the lesser fit of the two may be selected if: (1)
its fitness lies within a tolerance of 0.05 of the fitter
individual, and (2) it has fewer trainable parameters
than the fitter individual. In the case of the second
population, survivor selection is realised in a similar
manner: the first clause is the same as above. The
second clause is amended to favour individuals with a
higher novelty score: the lesser fit of two individuals
may be selected if it has fewer trainable parameters,
or has a higher novelty score, than the fitter individ-
ual.
3.2.4 Offspring Generation
Offspring are generated from parents using the afore-
mentioned mutation operators. In the case that a child
is produced via a mutation that does not alter the ac-
tive path (the longest list of connected nodes) its fit-
ness is set equal to that of its parent.
3.2.5 Termination Criteria
Evolution is terminated after 200 generations, result-
ing in 3204 fitness evaluations.
3.3 Diversity Measures and Novelty
Search
In order to study the effect of structural diversity on
the efficiency of the evolutionary algorithm in obtain-
ing optimal solutions, a second population is gen-
erated using the Novelty Search algorithm (Lehman
and Stanley, 2011). The resulting population is then
used as the initial population for the evolutionary al-
gorithm, following the procedure described in the pre-
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
126
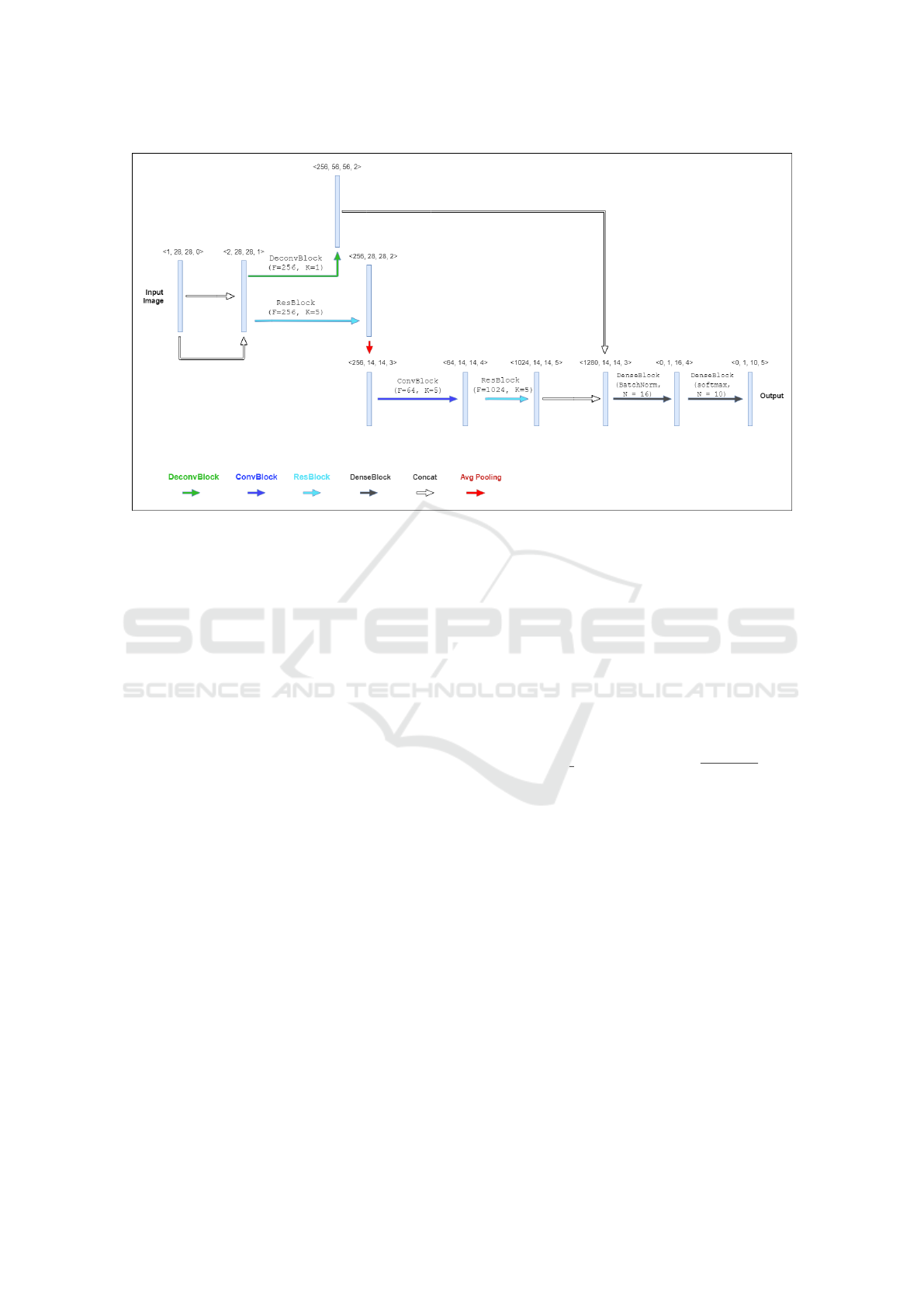
Figure 2: Phenotype of the best performing CNN architecture obtained by the proposed method, with the vector representation
of each layer indicated. The figure illustrates the functions of each of the modules represented in the genotype illustrated in
Figure 1. The coloured arrows indicate the various functions: DeconvBlock (green), ConvBlock (blue), ResBlock (cyan),
DenseBlock (dark gray), Concatenation (white), and Average Pooling (red).
vious section. This is the aforementioned Population
2.
We develop a novelty metric based on cosine
similarity as defined below. This metric is used in
our Novelty Search algorithm to explore the solution
space.
3.3.1 Vector Representation of the CNN
The structural information in the phenotype of can-
didate solutions is represented using sets of 4-
dimensional vectors, where each vector represents a
successive layer of the CNN. This is illustrated in Fig-
ure 2. A single layer, v
i
, in a CNN architecture can
be represented as in (2).
v
i
=
c
i
x
i
y
i
d
i
(2)
Where c
i
, x
i
and y
i
describe the size of the layer’s
activation in terms of the numbers of channels, the
width, and the height, respectively. The parameter d
i
represents the depth of the layer, i.e., the minimum
number of layers between this layer and the input
layer.
This representation allows us to obtain an approx-
imate measure of the structural similarity between so-
lutions using vector similarity measures such as co-
sine similarity.
3.3.2 Cosine Similarity and K Nearest
Neighbours Computation
The cosine similarity between two vectors is defined
as in (3).
cosine similarity(v
1
,v
2
) =
v
1
· v
2
(|v
1
||v
2
|)
(3)
The similarity between two individuals is com-
puted as described by Algorithm 1. The novelty of
an individual is then calculated using the normalized
sum of its similarity with its k nearest neighbours
(Cover and Hart, 1967), with k=11. This value is then
scaled by a factor of 100, and the result is subtracted
from 100. Thus, novelty scores lie within the range of
0-100.
3.4 Local Optimization
The proposed method involves the use of EAs for ex-
ploration and the use of an SLS algorithm for the local
optimization of near-optima. In order to demonstrate
this procedure, we select from Populations 1 and 2 the
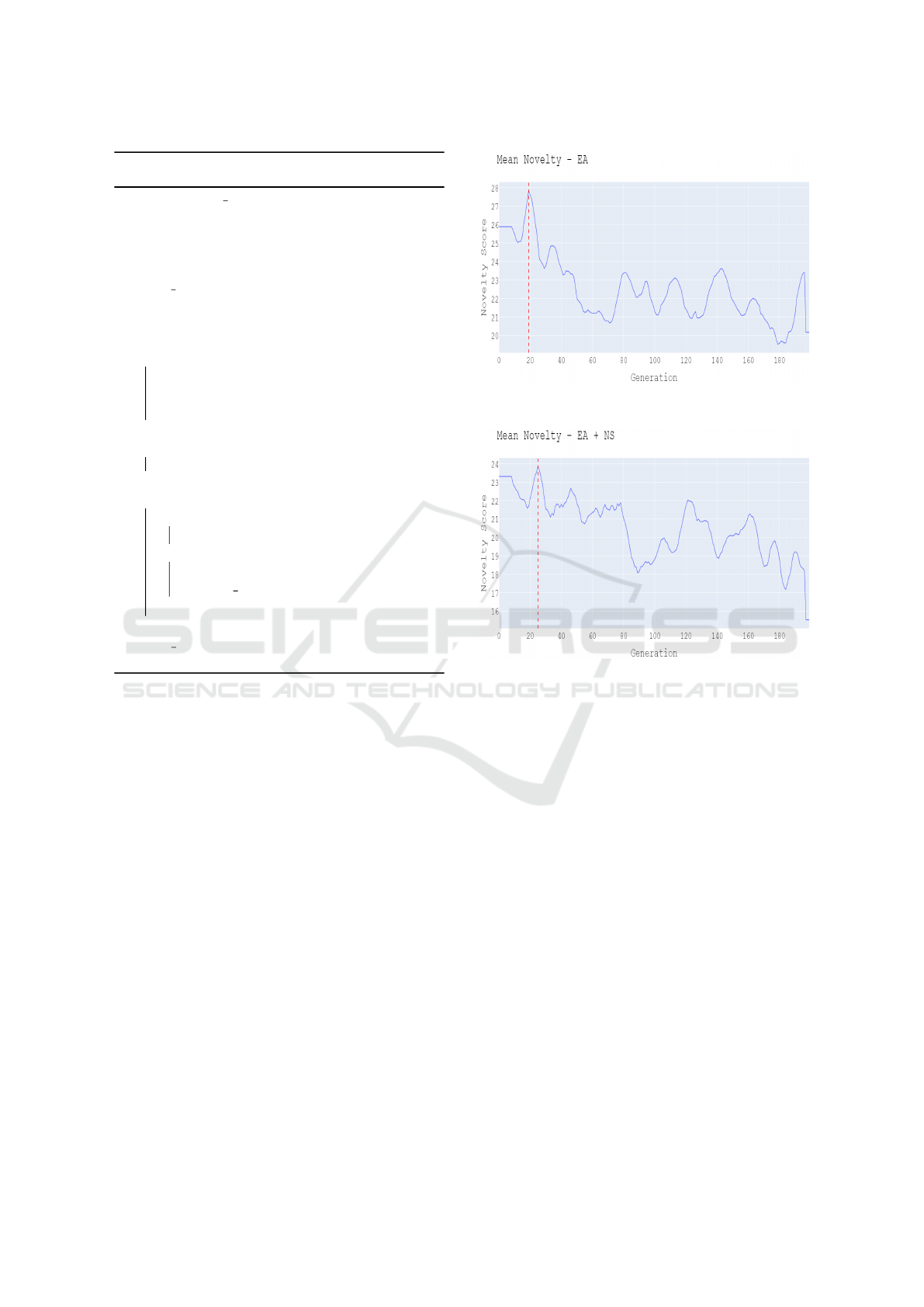
generations with highest mean novelty, as illustrated
in Figure 3. The novelty score of an individual is as
defined in Section 3.3.2. Thus, every architecture in
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
127
Algorithm 1: Computing similarity between two individu-
als.
Result: norm sum
V1: vector set representation of individual 1
V2: vector set representation of individual 2
similarity: array of length max(length(V1),
length(V2))
norm sum: normalized element-wise sum of
similarity array
V1 = V1[1:length(V1)-1];
V2 = V2[1:length(V2)-1];
if length(V2) >length(V1) then
temp ← V 1;
V 1 ← V2;
V 2 ← temp;
end
for i ← 0 to length(V1) - length(V2) do
V2.append([0,0,0,0])
end
for i ← 0 to length(V1) do
if norm(V2) = 0 then
similarity[i] ← 0;
else
similarity[i] ←
cosine similarity(V 1[i],V 2[i]);
end
end
norm sum ←
sum(similarity)/length(similarity)
each of these sampled generations contains an evalu-
ation score (F
1
Score) and a novelty score. Based on
these two metrics, we select individuals which are de-
termined to be located near/at local optima. We then
perform an extensive search of their local neighbor-
hood to discover optimal architectures.
Here, we distinguish two other populations: the
group of individuals selected for local optimization
from Population 1 is henceforth referred to as Popu-
lation 3. Similarly, the group of individuals selected
from Population 2 will be referred to as Population 4.
3.4.1 Identification of Local Optima
As every architecture has an evaluation metric and
a novelty metric, the sampling of local optima can
be treated as a multi-objective optimization problem.
Among the many existing evolutionary algorithms for
multi-objective optimization, we use the NSGA-II al-
gorithm. NSGA-II has evolved over the last few years
with many new variants (D’Souza et al., 2010) which
have reduced its time-complexity or/and improved its
convergence to the true Pareto Optimal front.
The algorithm proceeds by performing a non-
dominated sorting of the CNN structures based on
(a) Variation of mean novelty over different generations of
the evolutionary algorithm for Population 1.
(b) Variation of mean novelty over different generations of
the evolutionary algorithm for Population 2, which is
optimized for fitness and novelty.
Figure 3: Mean novelty vs time: evolutionary algorithm
(top) and evolutionary algorithm with Novelty Search (bot-
tom). In each case, the vertical line indicates the generation
selected for local optimization.
their multi-objective fitness values, and ranking the
solutions into different non dominated fronts as in
Figure 4. Thereafter, we sample the solutions by
selecting those individuals belonging to the highest
ranked front first, followed by those of the next front
and so on, until we have obtained at least 4 individ-
uals. In case there is space only for part of a front
in the new population, we use a crowded-distance op-
erator to determine the individuals that are from the
least crowded regions within that front. Details of the
crowded-distance operator can be obtained from (Deb
et al., 2002).
The solutions sampled from Population 1 comprise
rank 1 and rank 2 solutions, while the four solutions
sampled from Population 2 comprise rank 1 solutions
only. These constitute the Population 3 and 4, respec-
tively.
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
128

(a) Pareto Optimal Front for Population 1.
(b) Pareto Optimal Fronts for Population 2.
Figure 4: The generation with the highest mean novelty
score that is sampled from Population 1 and Population 2
is fed into the NSGA II algorithm. The solutions in pink
belong to rank 1, solutions in blue belong to rank 2 and so-
lutions in yellow belong to further lower ranks.
3.4.2 Simulated Annealing
The solutions found by NSGA II are fed into a local
search optimizer. A local search algorithm explores
the neighborhood of a candidate by comparing it with
neighboring solutions. This search is guided by a cost
function that measures the quality of a given solution.
In our work, we have chosen Simulated Annealing
as the local optimization algorithm. Simulated An-
nealing is a heuristic method which was inspired by
the physical annealing of metals (Kirkpatrick et al.,
1983). The search algorithm iteratively compares a
given solution to another neighboring solution in the
local neighborhood of the current solution. The cost
function reflects the difference between the fitness of
the current solutions and that of a neighbour. If the
cost is positive (neighbour is fitter), the algorithm se-
lects the neighboring solution as the (working) opti-
mal one for the next iteration. However, if the cost
is negative, the algorithm selects either solution (cur-
rent or neighbour), on a probabilistic basis described
by (4).
P = exp(
cost
T
) (4)
The algorithm starts with an initial ’high temper-
ature’, when the probability of accepting bad solu-
tions is higher. This ensures that the algorithm does
not get stuck in a local optimum. The temperature
is iteratively reduced in line with a cooling schedule.
The most popular cooling schedule is the geometric
cooling schedule, which defines a parameter α with
a value between 0 and 1. At the conclusion of every
iteration, the temperature is updated in line with (5).
T = α ∗ T (5)
As the algorithm progresses the temperature de-
creases exponentially according to the cooling sched-
ule, making the algorithm more greedy. Thus the
probability of choosing solutions with lower fitness
decreases with increasing iterations. Simulated an-
nealing terminates after a fixed number of iterations
and yields the most optimal solutions from the local
neighborhood of the initial candidate solutions. We
have chosen an initial temperature of 1000, with α =
0.91. The algorithm terminates after 183 iterations
when the final value of temperature is 0.000032155.
3.4.3 Estimation of Local Neighborhood
To define the neighborhood function we use the co-
sine similarity metric defined in (3). We compute
the cosine distance between the generated neighbor,
produced by mutating a candidate solution, and every
other individual in the population. The mutated in-
dividual is then assigned to the neighborhood of the
candidate solution with which it has the highest simi-
larity.
3.5 Experimental Setting
The MNIST (LeCun and Cortes, 2010) data-set is
a set of 28x28 pixel gray-scale images of handwrit-
ten digits. It consists of 60,000 training images and
10,000 test images. In our experiments, we split the
training images into a training set of 50,000 images
and a held-out validation set of 10,000 images. Image
pixel values are normalized by dividing each value by
255.
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
129

All experiments are conducted on machines with
the following features: two Intel E5-2650 v4 Broad-
well 2.2GHz CPUs with 24 CPU cores, four NVIDIA
P100 Pascal 16 GB GPUs, and 128 GB RAM.
4 RESULTS
Using the methods described in the preceding section,
we develop four distinct populations of CNNs:
1. Population 1: the population obtained using the
standard evolutionary algorithm.
2. Population 2: the population obtained using the
evolutionary algorithm with Novelty Search ini-
tialization.
3. Population 3: the population obtained from the
standard evolutionary algorithm and subsequent
local optimization via Simulated Annealing.
4. Population 4: the population obtained from the
evolutionary algorithm with Novelty Search ini-
tialization and subsequent local optimization via
Simulated Annealing.
From each of these populations, the fittest four in-
dividuals are selected and trained completely using
the 50,000 images from the MNIST data-set reserved
for training. The held-out validation data-set is used
to prevent over-fitting by applying the early stopping
regularization method (Caruana et al., 2000). Each
network is trained for up to 100 epochs, with an early
stopping patience parameter of 20 epochs.
The trained networks are then evaluated on the
previously unseen 10,000 test images. The resulting
recognition error is reported in Table 2 along with the
number of trainable parameters. The number of fit-
ness evaluations completed before obtaining each so-
lution is also reported. The minimum error obtained
by any individual across all populations is 0.48%, and
was found in population 4.
4.1 Recognition Error
From the results over the four populations, we can ob-
serve that individuals from each population obtained
competitive recognition errors. However, the large
discrepancy in performance between the fittest and
fourth fittest individuals obtained by the EA alone
(Population 1) suggests a failure by the EA in exploit-
ing the many peaks of the fitness landscape, simulta-
neously. The variance in recognition error of the four
best individuals obtained by the EA alone is 0.0169.
When novelty is included as an objective in Pop-
ulation 2, we observe a slight reduction in perfor-
mance in terms of fitness value and recognition error.
The four best individuals in this case exhibit approxi-
mately equal performance. Since they have high nov-
elty scores, it is reasonable to infer that they belong
to distinct regions within the solution space. In this
case, the variance in recognition error is 0.0013.
Populations 3 and 4 are obtained through local op-
timization. Recall that during local optimization, each
individual is constrained from entering the neighbour-
hood of the solution space occupied by any of the
other individuals in the population. An inspection of
the recognition errors reveals that some individuals in
Population 3 have become constrained to sub-optimal
regions of the solution space, when compared to indi-
viduals in Population 4. In addition, the variances in
recognition error exhibited by these two populations
are 0.0172 and 0.0025, respectively.
From the discussion above, it appears that the in-
clusion of novelty as a search objective results in a
reduction in the variance of recognition error, by an
order of magnitude. This suggests that, by enforcing
exploratory behaviour, our method discovers highly-
fit solutions that come from different parts of the so-
lution space- resulting in a highly diverse and highly
fit set of solutions.
4.2 Number of Fitness Evaluations
Here, we conduct a comparison of the number of fit-
ness evaluations required to obtain optimal solutions
in each of the four populations. Similarly to our dis-
cussion on the variance of recognition errors, we con-
sider the mean and standard deviation of the number
of fitness evaluations required to obtain the four best
solutions for each of the four populations. For pop-
ulations 1 through 4, these values are 1655 ± 1260,
2986 ± 204, 905 ± 111, and 1030 ± 46 fitness evalu-
ations, respectively. The average time taken for each
fitness evaluation was found to be 3.12 minutes per
evaluation.
The populations obtained through local optimiza-
tion via Simulated Annealing appear to require fewer
fitness evaluations in order to obtain optimal solu-
tions, compared to those obtained through the evolu-
tionary approach (p < .05). This is further illustrated
by Figure 5, which demonstrates that the evolution-
ary algorithms incur a large number of wasted cycles
in between discoveries of optimal solutions. This is
in contrast to Figure 6, which illustrates the ability of
the Simulated Annealing algorithm to rapidly climb
towards local optima. This is especially salient after
approximately 600 fitness evaluations, when the tem-
perature of the cooling schedule has reduced, and the
algorithm has become more deterministic.
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
130

Table 2: Summary of the four fittest individuals obtained from each population. Individuals are sorted by test error. Popula-
tions 1 through 4 are as described in the preceding sections, and reiterated in Section 4.
Population Individual #
# Trainable
Parameters
# Fitness
Evaluations
Fitness
Score
Test
Error (%)
Population 1
1 5.493 M 2676 0.992 0.57
2 0.968 M 3104 0.987 0.69
3 9.175 M 136 0.990 0.71
4 103.364 M 704 0.989 0.93
Population 2
1 850.491 M 2746 0.993 0.64
2 1.018 M 3158 0.987 0.72
3 0.232 M 3217 0.989 0.72
4 7.358 M 2821 0.985 0.73
Population 3
1 7.400 M 1004 0.994 0.67
2 14.488 M 964 0.993 0.52
3 32.219 M 716 0.992 0.83
4 19.118 M 936 0.992 0.84
Population 4
1 34.011 M 1072 0.994 0.48
2 46.063 M 1024 0.993 0.54
3 7.550 M 956 0.992 0.57
4 8.863 M 1068 0.992 0.62
Lastly, the lower variance exhibited by popula-
tions 3 and 4 suggests that the local optimization ap-
proach will reliably obtain optimal solutions within a
narrow range of number of fitness evaluations.
4.3 Comparison to Related Works
The proposed method obtains competitive perfor-
mance in terms of recognition error on the MNIST
test data-set. However, the trained networks obtained
by our method do not rival the state-of-the-art. During
training, it was observed that the early stopping tech-
nique halted training well before 100 epochs. Further
improvements may be obtained by including other
regularization techniques, such as the inclusion of
dropout layers (Srivastava et al., 2014). A compari-
son to related works is provided in Table 3.
5 CONCLUSIONS AND FUTURE
WORK
In this paper, we have investigated the use of a three-
stage CGP-based hybrid evolutionary algorithm (EA)
for the training of CNNs, used for handwritten digit
recognition. The well-known MNIST data-set is used
to assess the performance of the trained CNNs.
The first stage is meant to generate a population
of diverse CNNs, and for that reason we propose a
method for measuring structural similarity between
CNN architectures. The second stage increases the
fitness of the population (while not sacrificing diver-
Table 3: Comparison of the recognition error (%) on the
MNIST test dataset to related works.
Model MNIST (%)
psoCNN
(Fernandes Junior and Yen, 2019)
0.32
GeNet
(Xie and Yuille, 2017)
0.36
HyperNEAT
with CNN
(Verbancsics and Harguess, 2015)
7.90
evoCNN
(Sun et al., 2019)
1.18
IPPSO
(Wang et al., 2018)
1.13
(Baldominos et al., 2018)
0.37
Our method 0.48
sity) by means of artificial evolution. Most critically,
we demonstrate how the final stage of the proposed
methodology (simulated annealing) is able to opti-
mize the potential locally optimal individuals found
in previous stages, and do so more efficiently than a
non-hybridized EA.
The results of our experiments suggest that the
proposed method reduces, and reliably, the compu-
tational effort needed for obtaining optimal solutions,
compared to a standard evolutionary approach. We
quantify computational effort in terms of the number
of fitness evaluations required to obtain optimal so-
lutions. The number of fitness evaluations required
by the proposed method was found to be significantly
fewer than the number required by the standard EA (p
< .05).
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
131
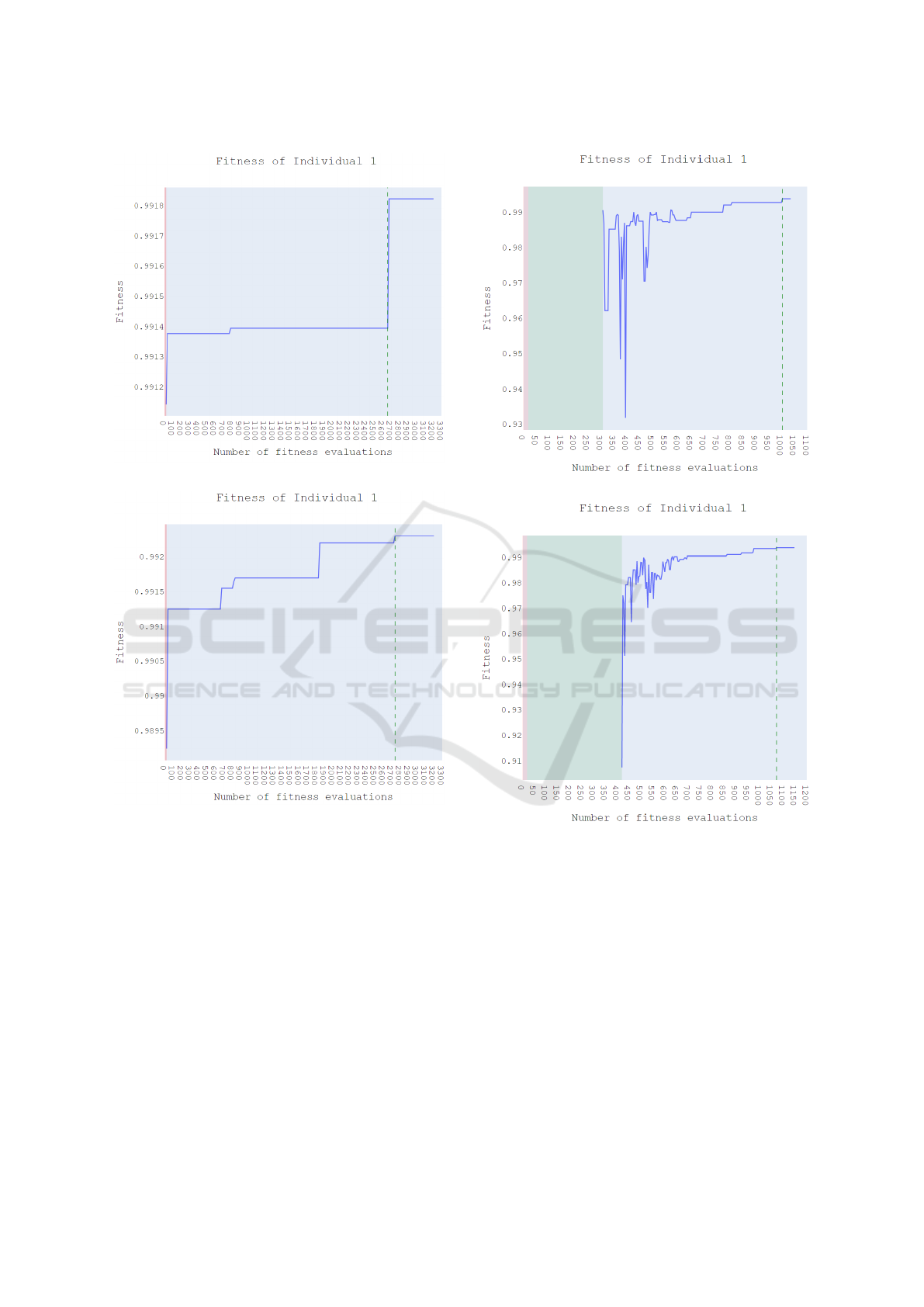
(a) Fitness of Individual 1 from Population 1.
(b) Fitness of Individual 1 from Population 2.
Figure 5: The above figures indicate the variation of fitness
over number of fitness evaluations during the evolutionary
process for Populations 1 and 2. The best fitness for (5a) is
obtained after 2676 evaluations and that for (5b) is obtained
after 2746 evaluations. The shaded red region represents the
number of fitness evaluations during the initialization of the
population.
In addition, we have observed that including nov-
elty as an objective has the effect of reducing the vari-
ance of recognition errors by an order of magnitude
compared to the pure evolutionary approach. The
proposed method involves optimizing individual solu-
tions within a constrained local neighbourhood. Thus,
we infer from this result that the proposed method ef-
fectively obtains optimal solutions belonging to dif-
ferent regions of the solution space.
(a) Fitness of Individual 1 from Population 3.
(b) Fitness of Individual 1 from Population 4.
Figure 6: The above figures indicate the variation of fitness
over the local optimization process for Populations 3 and
4. The best fitness for (6a) is obtained after 1004 evalua-
tions and that for (6b) is obtained after 1072 evaluations,
during the optimization phase. The shaded red region rep-
resents the number of fitness evaluations during the initial-
ization and the green region represents the number of fitness
evaluations during the evolutionary process until sampling
is done.
The best recognition error obtained by our method
on the MNIST test data-set was found to be 0.48%. In
comparison to related works, this reflects competitive
performance. However, some peer competitors have
obtained recognition error as low as 0.32%. In order
to improve performance of the CNNs evolved using
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
132

our CGP framework, we recommend the investigation
of regularization techniques, such as dropout. Also,
the evolution of recurrent structures within the CNNs
may be explored for improved performance.
Future work may explore measures of structural
similarity which act on the DAG representation of
CGP genotype, such as Graph Edit Distance (Sanfe-
liu and Fu, 1983). In addition, the method may be ap-
plied to other CNN tasks such as image segmentation.
Lastly, an investigation of alternative SLS algorithms
may yield further improvements in terms of reduced
computational cost.
ACKNOWLEDGEMENTS
We acknowledge the support of the Natural Sci-
ences and Engineering Research Council of Canada
(NSERC).
REFERENCES
Baldominos, A., Saez, Y., and Isasi, P. (2018). Evolution-
ary convolutional neural networks: An application to
handwriting recognition. Neurocomputing, 283:38–
52.
Caruana, R., Lawrence, S., and Giles, L. (2000). Overfit-
ting in neural nets: Backpropagation, conjugate gra-
dient, and early stopping. NIPS’00, page 381–387,
Cambridge, MA, USA. MIT Press.
Chollet, F. et al. (2015). Keras. https://keras.io.
Cover, T. and Hart, P. (1967). Nearest neighbor pattern clas-
sification. IEEE Transactions on Information Theory,
13(1):21–27.
Deb, K., Pratap, A., Agarwal, S., and Meyarivan, T. (2002).
A fast and elitist multiobjective genetic algorithm:
Nsga-ii. IEEE Transactions on Evolutionary Compu-
tation, 6(2):182–197.
D’Souza, R., Sekaran, C., and Kandasamy, A. (2010). Im-
proved nsga-ii based on a novel ranking scheme. Jour-
nal of Computing, 2.
Fernandes Junior, F. E. and Yen, G. G. (2019). Particle
swarm optimization of deep neural networks architec-
tures for image classification. Swarm and Evolution-
ary Computation, 49:62–74.
Kingma, D. P. and Ba, J. (2017). Adam: A method for
stochastic optimization.
Kirkpatrick, S., Gelatt, C. D., and Vecchi, M. P. (1983).
Optimization by simulated annealing. Science,
220(4598):671–680.
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
LeCun, Y. and Cortes, C. (2010). MNIST handwritten digit
database.
Lehman, J. and Stanley, K. O. (2011). Novelty search and
the problem with objectives. In Riolo, R., Vladislavl-
eva, E., and Moore, J. H., editors, Genetic Program-
ming Theory and Practice IX, pages 37–56. Springer
New York, New York, NY.
McGhie, A., Xue, B., and Zhang, M. (2020). Gpcnn:
evolving convolutional neural networks using genetic
programming. In 2020 IEEE Symposium Series on
Computational Intelligence (SSCI), pages 2684–2691,
Canberra, ACT, Australia. IEEE.
Miller, B. and Goldberg, D. (1995). Genetic algorithms,
tournament selection, and the effects of noise. Com-
plex Syst., 9.
Miller, J. F. and Thomson, P. (2000). Cartesian genetic pro-
gramming.
Sanfeliu, A. and Fu, K.-S. (1983). A distance measure be-
tween attributed relational graphs for pattern recogni-
tion. IEEE Transactions on Systems, Man, and Cyber-
netics, SMC-13(3):353–362.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I.,
and Salakhutdinov, R. (2014). Dropout: a simple way
to prevent neural networks from overfitting. The jour-
nal of machine learning research, 15(1):1929–1958.
Stanley, K. O., D’Ambrosio, D. B., and Gauci, J. (2009). A
Hypercube-Based Encoding for Evolving Large-Scale
Neural Networks. Artificial Life, 15(2):185–212.
Stanley, K. O. and Miikkulainen, R. (2002). Evolving Neu-
ral Networks through Augmenting Topologies. Evo-
lutionary Computation, 10(2):99–127.
Suganuma, M., Shirakawa, S., and Nagao, T. (2017). A
genetic programming approach to designing convolu-
tional neural network architectures. In Proceedings
of the Genetic and Evolutionary Computation Confer-
ence, pages 497–504, Berlin Germany. ACM.
Sun, Y., Xue, B., Zhang, M., Yen, G. G., and Lv, J.
(2020). Automatically designing cnn architectures
using the genetic algorithm for image classification.
IEEE Transactions on Cybernetics, 50(9):3840–3854.
Sun, Y., Yen, G. G., and Yi, Z. (2019). Evolving unsuper-
vised deep neural networks for learning meaningful
representations. IEEE Transactions on Evolutionary
Computation, 23(1):89–103.
Verbancsics, P. and Harguess, J. (2015). Image classifica-
tion using generative neuro evolution for deep learn-
ing. In 2015 IEEE Winter Conference on Applications
of Computer Vision, pages 488–493.
Wang, B., Sun, Y., Xue, B., and Zhang, M. (2018). Evolving
deep convolutional neural networks by variable-length
particle swarm optimization for image classification.
In 2018 IEEE Congress on Evolutionary Computation
(CEC), pages 1–8.
Xie, L. and Yuille, A. (2017). Genetic cnn. In 2017 IEEE
International Conference on Computer Vision (ICCV),
pages 1388–1397, Venice. IEEE.
A Hybrid Evolutionary Algorithm, Utilizing Novelty Search and Local Optimization, Used to Design Convolutional Neural Networks for
Handwritten Digit Recognition
133
