
Event-based Pathology Data Prioritisation: A Study using Multi-variate
Time Series Classification
Jing Qi
1
, Girvan Burnside
2
, Paul Charnley
3
and Frans Coenen
1
1
Department of Computer Science, The University of Liverpool, Liverpool L69 3BX, U.K.
2
Department of Biostatistics, The University of Liverpool, Liverpool L69 3BX, U.K.
3
Wirral University Teaching Hospital NHS Foundation Trust, Arrowe Park Hospital, Wirral CH49 5PE, U.K.
Keywords:
Data Prioritisation, Time Series Classification, kNN, LSTM-RNN.
Abstract:
A particular challenge for any hospital is the large amount of pathology data that doctors are routinely pre-
sented with. Pathology result analysis is routine in hospital environments. Some form of machine learning
for pathology result prioritisation is therefore desirable. Patients typically have a history of pathology results,
and each pathology result may have several dimensions, hence time series analysis for prioritisation suggests
itself. However, because of the resource required, labelled prioritisation training data is typically not readily
available. Hence traditional supervised learning and/or ranking is not a realistic solution and some alternative
solution is required. The idea presented in this paper is to use the outcome event, what happened to a patient,
as a proxy for a ground truth prioritisation data set. This idea is explored using two approaches: kNN time
series classification and Long Short-Term Memory deep learning.
1 INTRODUCTION
The challenge of prioritising pathology time series
data using the tools and techniques of machine learn-
ing is that, in most cases, we do not have sufficient
amounts of training data, because of the clinical re-
source required to create such data, to support effec-
tive supervised learning. This means that some al-
ternative mechanism needs to be adopted. The fun-
damental idea presented in this paper is to use some
form of proxy for the training data set using meta-
knowledge about patients. More specifically using
meta-knowledge concerning the “final destination” of
patients, the outcome event for each patient, and use
this to build a outcome event classification system.
Three outcome events were considered: Emergency
Patient (EP), an In-Patient (IP) or an Out Patient (OP).
Then, given a new pathology result and the patient’s
pathology history, it would be possible to predict the
outcome event and then use this to prioritise the new
pathology result. For example if we predict the out-
come event for a patient to be EP, then the new pathol-
ogy result should be assigned high priority; however,
if we predict that the outcome event will be IP the new
pathology result should be assigned medium priority,
and otherwise low.
The hypothesis that this paper seeks to establish
is that there are patterns in patients’ historical lab
test results, which are markers as to where the pa-
tient “ended up” and which can hence be used for
prioritisation. To act as focus, the work presented is
directed at pathology lab test results related to renal
function, namely Urea and Electrolytes (U&E) tests.
This test provides an extra challenge in that it features
five components (tasks) each with an associated test
result value. In addition each task within a U&E test
has three values associated with it. Thus there are five
historical multi-variate time series per patient.
There are a number of multi-variate time series
classification algorithms that could be adopted to clas-
sify time series. Two are considered in this paper: (i)
k Nearest Neighbour (kNN) (Xing and Bei, 2019) and
(ii) Long short-term memory (LSTM). The first was
selected because it was the most frequency used al-
gorithm with respect to time series classification. A
value of k = 1 was adopted, as suggested in (Bagnall
et al., 2017). Dynamic Time Warping (DTW) was
used as the similarity measure.
The remainder of the paper is organised as fol-
lows. Section 2 presents previous work relating to the
work in this paper. An overview of the U&E appli-
cation domain is then given Section 3, followed by a
formalism in Section 4. The two proposed approaches
to event-based prioritisation, using kNN and LSTM,
are presented in Section 5. The evaluation of the pro-
posed approaches is then presented in Section 6. Fi-
nally, in Section 7, some conclusions and directions
for future work are considered.
Qi, J., Burnside, G., Charnley, P. and Coenen, F.
Event-based Pathology Data Prioritisation: A Study using Multi-variate Time Series Classification.
DOI: 10.5220/0010643900003064
In Proceedings of the 13th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2021) - Volume 1: KDIR, pages 121-128
ISBN: 978-989-758-533-3; ISSN: 2184-3228
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
121

2 PREVIOUS WORK
The prioritisation mechanism proposed in this paper
is founded on time series classification. Many time
series classification approaches have been proposed.
One of the most popular, and that used with respect to
the work presented in this paper, is k Nearest Neigh-
bour (kNN) classification. The fundamental idea of
kNN classification is to compare a previously unseen
time series, which we wish to label, with a “bank”
of time series whose labels are known, identify the k
most similar and use the labels from the k most simi-
lar to label the previously unseen time series. Usually
k = 1 is adopted because it avoids the need for any
conflict resolution.
Time series classification using kNN entails two
challenges: (i) the data format for the input time se-
ries, and (ii) the nature of the similarity (distance)
measure to be used to establish similarity (Wang et al.,
2013). There are two popular time series formats:
(i) instance-based and (ii) feature-based. Using the
instance-based format the original time series format
is maintained. Using the feature-based representation,
properties of the time series are used (Wang et al.,
2008). For the work presented in this paper the in-
stance based format was used. There are a number of
similarity measure options including Euclidean, Man-
hattan and Minkowski distance measurement, but Dy-
namic Time Warping (DTW) is considered to be the
most effective with respect to the instance-based for-
mat, and offers the additional advantage that the time
series considered do not have to be of the same length
(Wang et al., 2013). For the work presented in this
paper DTW was adopted.
The recent success of deep learning offers a more
substantive way of processing time series than in the
case of traditional models. Among many deep learn-
ing techniques, Recurrent Neural Networks(RNNs)
are considered as an effective way of classifying time
series, because they allow for the processing of vari-
able length inputs and outputs by maintaining state
information across time steps. There are many ex-
amples in the literature where RNNs have been used
with respect to time series classification; see for ex-
ample (Choi et al., 2016; Esteban et al., 2015). Long
Short-Term Memory (LSTM) networks are a popular
form of RNNs. The advantage of RNNs in general,
and LSTMs in particular, is that they have shown to
be more accurate, with respect to time series classi-
fication, then kNN. However, kNN does not require
significant training or large amounts of training data
as in the case of RNNs (LSTMs). There are many
variations of LSTMs (Greff et al., 2016). In this pa-
per, the standard “vanilla” LSTM setup was used.
3 APPLICATION DOMAIN
The work presented in this paper is focused on the
Urea and Electrolytes (U&E) test; a commonly used
test to detect abnormalities of blood chemistry, pri-
marily kidney (renal) function and dehydration. A
U&E test is usually performed to confirm normal kid-
ney function or to exclude a serious imbalance of
biochemical salts in the bloodstream. The U&E test
data considered in this paper comprised, for each test,
measurement of levels of: (i) Sodium (So), (ii) Potas-
sium (Po), (iii) Urea (Ur), (vi) Creatinine (Cr), and
(v) Bicarbonate (Bi). The measurement of each is re-
ferred to as a “task”, thus we have five tasks per test.
In other words each U&E test results in five pathology
values. It is suggested that U&E pathology results can
be prioritised more precisely if the trend of the his-
torical records is taken into consideration, therefore
providing more efficient treatment for patients with a
potential risk of renal function conditions. Given a
new set of pathology values for a U&E test we wish
to determine the priority to be associated with this set
of values.
4 FORMALISM
In the context of the foregoing, the assumption is that
the training data comprises a set of pathology results,
D = {P
1
, P
2
, . . . }, where the class (event) label c for
each pathology record P
j
∈ D is known. As the focus
of the work is U&E test data, which comprises five
tasks (components), each record P
j
∈ D is of the form:
P
j
= hId, Date, Gender, T
So
, T
Po
, T
Ur
, T
Cr
, T
Bi
, ci (1)
Where T
so
to T
bi
are five multi-variate time series rep-
resenting, in sequence, pathology results for the five
tasks typically found in a U&E test: Sodium (So),
Potassium (Po), Urea (U r), Creatinine (Cr) and Bicar-
bonate (Bi); and c is the class label taken from a set of
classes C. Each time series T
i
has three dimensions:
(i) pathology result, (ii) normal low and (iii) normal
high. The normal low and high dimensions indicate
a “band” in which pathology results are expected to
fall. These values are less volatile than the pathology
result values themselves, but do change for each pa-
tient over time. Thus each times series T
i
comprises a
sequence of tuples, of the form hv, nl, nhi (pathology
result, normal low and normal high respectively).
To derive the class label for each record P
j
∈ D
reference was made to the outcome event(s) associ-
ated with each patient. For the evaluation presented
later in this paper, three outcome events were consid-
ered: (i) Emergency Patient (EP), an In-Patient (IP)
KDIR 2021 - 13th International Conference on Knowledge Discovery and Information Retrieval
122

or an Out Patient (OP) which were correlated with
the priority descriptors “high”, “medium” and “low”
respectively. Hence C = {high, medium, low}.
Given a new pathology result for a patient j,
comprised of five tuples, one per task, {V
So
n+1
,
V
Bu
n+1
,V
Ur
n+1
, v
Cr
n+1
, v
Bi
n+1
}, these will be incorpo-
rated into the patient record P
j
for the patient in ques-
tion by appending each new pathology tuple to the ap-
propriate time series T
i
to give {V
i
1
,V
i
2
. . .V
i
n
,V
i
n+1
}.
The patient record P
j
thus becomes the “query
record”, the record we wish to label.
5 MULTI-TIME SERIES
EVENT-BASED PATHOLOGY
DATA PRIORITISATION
The fundamental idea promoted in this paper is that
pathology results can be prioritised in terms of the
trend of a given patients’ pathology. In order to val-
idate this idea, two approaches were adopted, the
kNN-DTW approach and the LSTM-RNN approach.
Each is discussed in more detail below.
5.1 Event-based Data Prioritisation
using kNN
The kNN classification model uses a parameter k, the
number of best matches we are looking for. As al-
ready noted, k = 1 was used with respect to the eval-
uation reported later in this paper because this avoids
the need for a conflict resolution mechanism where
k > 1. As also noted earlier, Dynamic Time Warp-
ing (DTW) was used for similarity measurement be-
cause of its ability to operates with time series of
different length (Wang et al., 2013). The disadvan-
tage of DTW is its high computational complexity,
which is O(x× y) where x and y are the lengths of the
two time series under consideration. There are many
techniques available for reducing this time complex-
ity in the context of kNN classification. For the work
presented here “early-abandonment” (Rakthanmanon
et al., 2012) and LB-Keogh lower bounding (Vikram
et al., 2013) were used.
The traditional manner in which kNN is applied
in the context of time series analysis is to compare
a query time series with the time series in the kNN
bank. In the case of the U&E test data prioritisation
scenario considered here the process involved five
comparisons, once for each time series in the query
record P
j
, T
q
so
, T
q
po
, T
q
ur
, T
q
cr
and T
q
bi
. In addition,
although traditional kNN is applied to univariate time
series; in this case three-dimensional, multi-variate,
time series were used.
For each comparison five distance measures were
obtained. With respect to the proposed kNN, the five
tasks were considered independently and the final pri-
oritisation decided using a “High priority first and vot-
ing second” mechanism. Given the foregoing, the ap-
plication of kNN to label P
j
was as follows:
1. Calculate the LB-keogh overlap for the five com-
ponent time series separately and prune all records
in D where the overlap for any one time series was
greater than a threshold ε, to leave D
0
.
2. Apply DTW, with early-abandonment to each pair
hT
q
i
, T
j
∈ D
0
i where i indicates the U&E task.
3. Assign the class label c associated with the most
similar time series T
i
∈ D
0
to the time series T
q
i
of
a patient record P
j
.
4. Use the “High priority first and voting second”
mechanism to decide the final priority level for P
j
.
The intuitions underpinning the mechanism were:
(i) if any of the five time series T
q
i
is assigned as
high prioritisation label, the final label for a pa-
tient record P
j
should be high, (ii) else the final
label is the one that receives more than half of the
votes (given a “tiebreak” the higher level of the
two labels is assigned to the patient).
The choice of value for the lower bounding thresh-
old ε was of great importance as it affected the ef-
ficiency and the accuracy of the similarity search.
According to (Li et al., 2017), there is a threshold
value for ε whereby the time complexity for the lower
bounding is greater than simply using DTW distance
without lower bounding. The experiments presented
in (Li et al., 2017) demonstrated that this threshold
occurred when the value for ε prunes 90% of the time
series in D. For the evaluation presented in this paper
ε = 0.159 was used because, on average, this resulted
in 10% of the time series in D being retained.
5.2 Event-based Data Prioritisation
using LSTM-RNN
The event-based data prioritisation process founded
on LSTM commenced with the training of five LSTM
models one per task: LST M
so
, LST M
po
, LST M
ur
,
LST M
cr
and DLST M
bi
. Once we have the LSTMs
they can be used.
The overall architecture comprised three “layers”:
(i) the input layer, (ii) the model layer and (iii) the de-
cision layer. In the input layer the component time
series T
q
so
, D
q
po
, D
q
ur
, D
q
cr
and D
q
bi
are extracted
from the query record P
q
. Thus for each task we
have a multi-variate time series T
i
= {V
1
,V
2
, ...,V
m
},
Event-based Pathology Data Prioritisation: A Study using Multi-variate Time Series Classification
123

where V − J is a tuple of the form presented earlier,
and m ∈ [l
min
, l
max
]. Where necessary each time se-
ries T
i
is padded to the maximum length, l
max
using
the mean values for the pathology test values, normal
low and normal high values in T
i
. Each time series T
i
is then passed to the appropriate LSTM in the model
layer. Each LSTM comprised: (i) an input layer, (ii)
an LSTM layer with two layers of LSTM cells and
(iii) an output layer. The output layer included the
Logits and Softmax components.
The last layer is the architecture is the decision
layer where the final label is derived. After obtaining
all of the five outputs and predicted labels from the
five LSTM models, a decision logic module was used
to decide the final prioritisation level of the patient.
The Softmax function for normalising was as follows:
y
i
=
e
a
i
Σ
|C|
k=1
e
a
k
∀i ∈ 1...C (2)
Where: (i) |C| is the number of classes (three in this
case) and (ii) a
i
is the output of the LSTM layer. Fi-
nally the following “High priority first and voting sec-
ond” rule was applied to produce the end classifica-
tion: if any one of the five LSTMS produces a pre-
diction of “High” the final prediction is “high”, oth-
erwise average the five outputs produced by Softmax
function and then choose the class with the maximum
probability.
The adopted individual LSTM architectures com-
prises 2 hidden layers and Logits plus Softmax func-
tion in the output layer, because multi-classes classi-
fication is being undertaken. For the LSTM to op-
erate five parameters needed to be tuned during the
training process. The parameters belong to two cate-
gories: (i) optimization parameters and (ii) model pa-
rameters. The optimization parameters were: Learn-
ing rate, batch size and number of epochs. The model
parameters were the number of hidden layers and the
number of hidden units. For the optimization, Adam
optimization was chosen due to its efficiency and the
nature of adaptive learning rate. For finding the op-
timal parameters, cross-entropy was used as the loss
function and the parameters tuned by observing the
loss and accuracy plots of the training and validation
data.
6 EVALUATION
This section presents the evaluation of the proposed
multi-time series event-based pathology data priori-
tising approach using kNN and LSTM as described
above. The metrics used were accuracy, precision and
recall. In all cases the evaluation was conducted using
a desktop machine with a 3.2 GHz Quad-Core Intel
Core i5 processor and 24 GB of RAM. For the LSTM
a GPU laptop was used fitted with a NVIDA GeForce
RTX 2060 unit. Five-fold cross validation was used
through-out. For the evaluation U&E pathology data
provided by the Wirral Teaching Hospital in Mersey-
side in the UK was used. This was used to create
three data sets: (i) D
f emale
, (ii) D
male
and (iii) D
all
(where D
all
= D
f emale
∪ D
male
). An overview of the
U&E evaluation data sets is given in Sub-section 6.1.
The objectives of the evaluation were:
1. To identify the optimum parameter settings in the
context of LSTM approach.
2. To compare the operation of the kNN and LSTM
approaches in terms of effectiveness.
3. To compare the operation of the kNN and LSTM
approaches in terms of efficiency (runtime).
The results with respect to each of the above are dis-
cussed in sub-sections 6.2 and 6.3 respectively.
6.1 Evaluation Dataset
The Wirral Teaching Hospital U&E pathology test
data comprised four data tables. The first three
were event data tables: (i) Emergency Patient (EP),
(ii) Inpatient(IP) and Outpatient (OP); comprised of
180,865, 226,634 and 955,318 records respectively
and corresponding to High, Medium and Low prior-
ity. The fourth was a Laboratory (Lab) data table,
comprised of 532,801 records, holding the pathology
results; this included results for patients in the event
data tables and patients that had never visited the hos-
pital, but were treated at their local surgery. The data
sets contain patient records over a two year span. The
LAB dataset was the primary dataset used for the
evaluation reported here. The event data sets were
used for generating outcome event labels (classes) for
the time series held in the LAB dataset.
Some statistics concerning the data set are given
in Table 1. From the table it can be observed that
there is a significant imbalance between the number
of records associated with each class, this is not an
issue when using kNN with k = 1, but it is an issue
when using LSTMs, as highly imbalanced data may
pose bias towards the majority class. An oversam-
pling technique was adopted to address this issue with
respect to the RNN training.
Each record in the LAB table, R
i
, representing a
pathology result for a single task in a U&E test, was
of the form:
R
i
= hID, Task, Date,Value,Unit, Max, Min, Genderi
(3)
KDIR 2021 - 13th International Conference on Knowledge Discovery and Information Retrieval
124
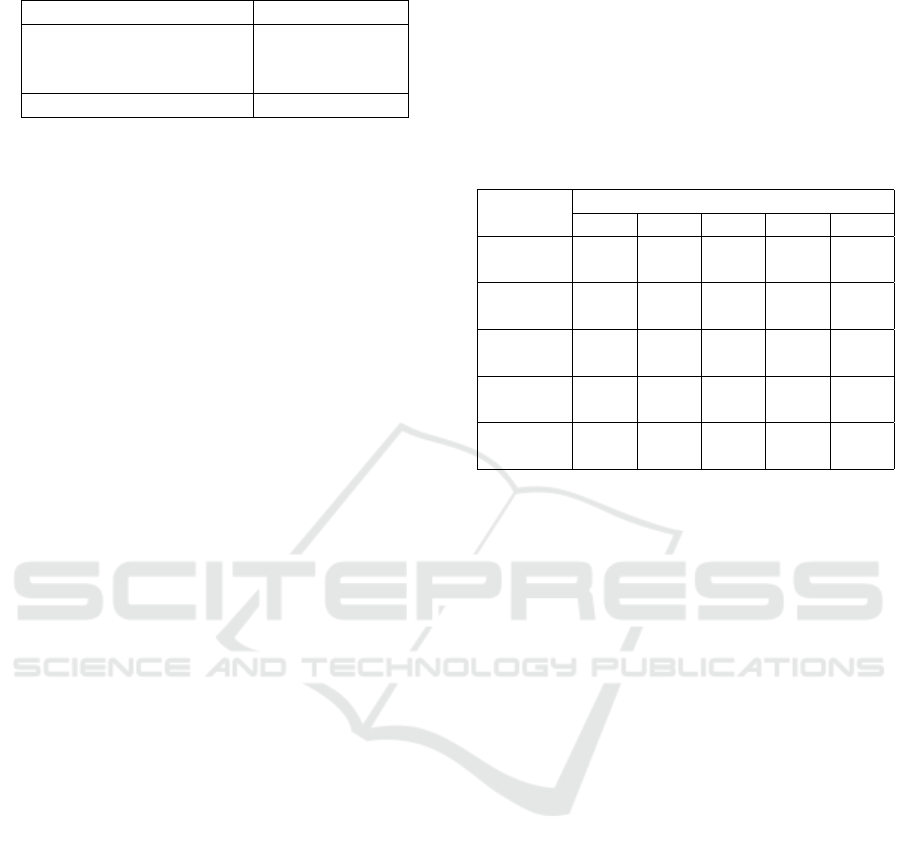
Table 1: U&E Data set statistics.
Event (Priority) Num. Patients
Emergency Patient (High) 255
In Patient (Medium) 123
Out Patient (Low) 3,356
Total 3,734
Where: (i) ID is the unique code for the patient, (ii)
Task is the name of the task (Sodium, Potassium,
Urea, Creatinine and Bicarbonate). (iii) Date is the
date the test was conducted, (iv) Value is the pathol-
ogy value for the task, (v) Unit is the units for the
Value, and (vi) Max and Min are the bounds for the
anticipated normal range for the Value (for the patient
and task in question, not the same for all patients).
Some data cleaning was first undertaken, removing
patients with missing or non-numeric task values and
feature scaling to benefit the faster convergence of the
LSTM.
The time series data set D = {P
1
, P
2
, . . . },
where each P
j
patient record was of the form
hID, TestDate, Gender, T
So
, T
Po
, T
Ur
, T
Cr
, T
Bi
, ci (see
Section 4), required five time series equating to the
five tasks included in the U&E test data (Sodium,
Potassium, Urea, Creatinine and Bicarbonate). The
time series were constructed by processing the data
for each patient up until an outcome event. The
values, up to and including the event value, were then
used to construct the relevant time series. If a patient
appeared in more than one event data set, for example
a patient was an “out patient” and then became an “in
patient” and finally became an “emergency patient”,
then the time series prior to the“emergency event”
was used, because the pattern of the “emergency
patient” indicates the highest priority. Also there
were a small number of patients (less than 1% of
the total data set) who did not appear in any of the
event data sets, in other words the patients remained
with their general practitioners. This group of patient
records was removed from the training data. Time
series comprised of less than three time stamps
were also removed. Each P
j
patient record was then
labelled according to the priority indicated by the
event value time stamp.
The final data set, D
all
, comprised 3,734 time se-
ries; 255 high priority, 123 medium priority and 3,356
low priority, covering all five tasks. Thus there were
747 records (3,734/5) in each fold of the cross valida-
tion. The records in each fold were stratified so that
there was an equal distribution of classes in each fold.
The D
f emale
data set was comprised of 1,960 time se-
ries; 136 high priority, 55 medium priority and 1,769
low priority. The D
male
, comprised of 1,774 time se-
ries; 119 high priority, 68 medium priority and 1,587
low priority. All three data sets were used for the eval-
uation. The reason for exploring the distinction be-
tween genders was because it had been suggested that
there maybe gender differences for the prioritisations
being investigated (Halbesma et al., 2008; Tomlinson
and Clase, 2019).
Table 2: LSTM Parameter Settings for the five LSTMs (one
per task).
Para.
Task
Bo Cr Po So Ur
Learning
0.01 0.01 0.01 0.01 0.01
Rate
Batch
512 128 256 512 512
Size
Epochs 1000 1000 1000 1000 1000
Hidden
2 2 2 2 2
Layer
Hidden
32 32 16 32 32
Units
6.2 Parameter Settings for LSTM
The general way for finding the best parameters for
deep neural network models is to analyse the learning
curve and accuracy plot of the training and validation
data. The most popular learning curve used for this
purpose is loss over time. Loss measures the model
error, in other words, “how bad the performance of
the model is”. Thus, the lower the loss is, the bet-
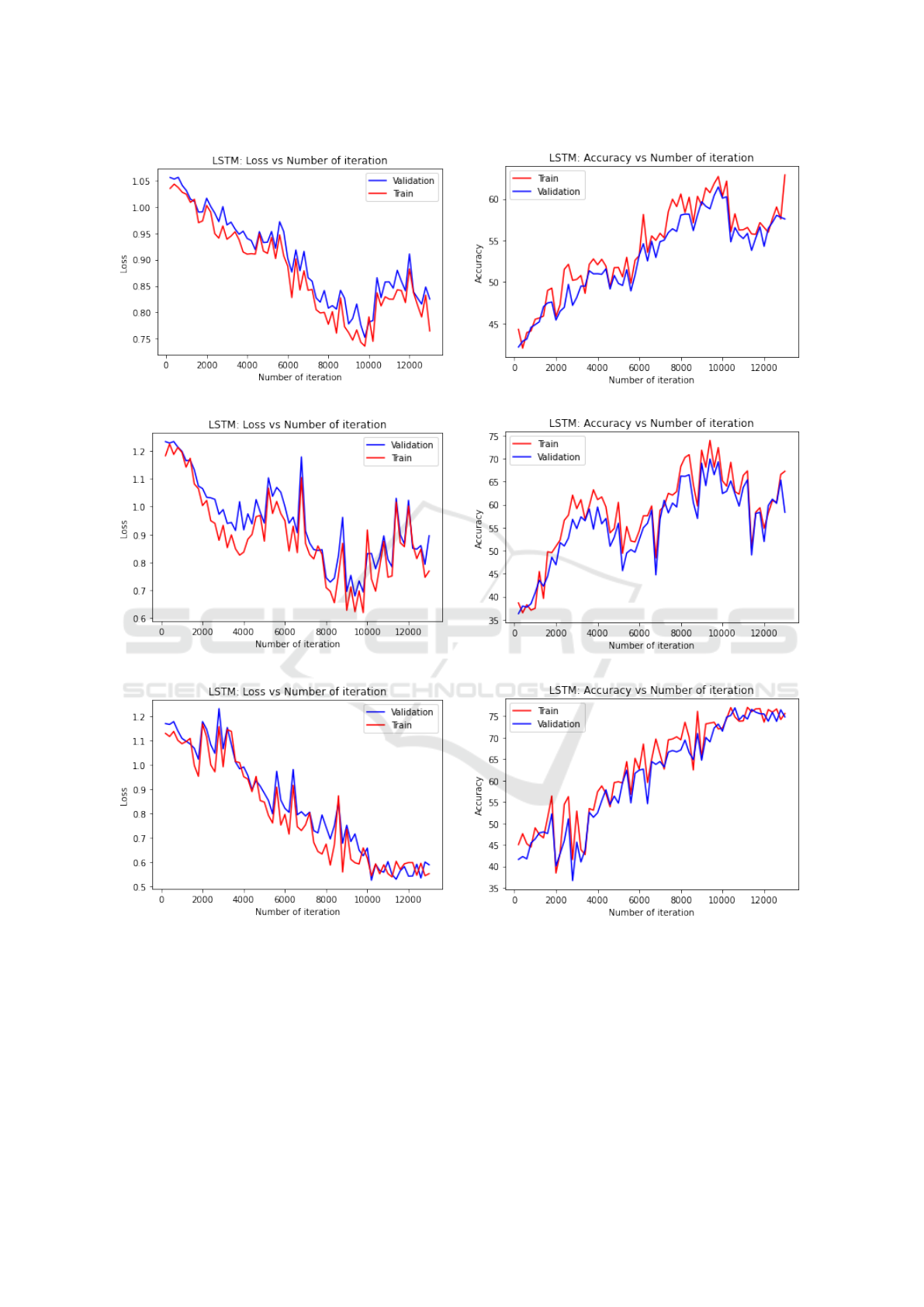
ter the model performance. Figure 1 shows the aver-
age loss and accuracy plots for each of the three data
sets considered. For each graph in Figure 1 the x-axis
gives the number of times the weights in the network
were updated, and the y-axis the loss value. From the
figures, we can observe that oscillations appear in all
of the loss and accuracy plots and that convergence is
not obvious. Possible reasons for this include: (i) the
oversampling solution for dealing with the class im-
balanced problem meant that there were insufficient
sequences for the LSTM to learn from; and (ii) that
the event-based mechanism used as the proxy ground
truth of the data set may not be entirely representative.
The final best settings for the parameters are given in
Table 2.
6.3 Comparison of Approaches
The average accuracy, precision and recall results for
each fold of the five-cross validation, for the kNN and
LSTM approaches, are given in Tables 3 and 4. Note
that the results are the average results of the three
Event-based Pathology Data Prioritisation: A Study using Multi-variate Time Series Classification
125
(a) Loss (D
all
)
(b) Accuracy (D
all
)
(c) Loss (D
f emale
)
(d) Accuracy (D
f emale
)
(c) Loss (D
male
)
(d) Accuracy (D
male
)
Figure 1: Loss and Accuracy curves for the LSTM generation process.
evaluation data sets. The results of the precision and
recall for the class high are highlighted. The overall
average (Ave) and standard deviation (SD) are given
in the last two rows. Note that the SD values are low,
indicating that there is little variation across the folds.
From the table it can be seen that the RNN approach
consistently outperformed the kNN approach. A gen-
eral observation is that the precision and recall values
might be argued to be on the low side, possibly in-
dicating either: (i) that the hypothesised event-based
prioritisation approach, is not as good a predictor of
priority, as anticipated, (ii) the irregular nature (dis-
tribution of time stamps) of the time series, which
was not considered, may have an adverse effect. For
the LSTM-RNN models, the way that the class im-
balanced problem was dealt with may also have ad-
KDIR 2021 - 13th International Conference on Knowledge Discovery and Information Retrieval
126

Table 3: Average Precision and Recall (Three data set) of kNN.
Fold Num. Acc. Pre. High Pre. Medium Pre. Low Rec. High Rec. Medium Rec. Low
1 0.585 0.414 0.400 0.545 0.637 0.577 0.666
2 0.632 0.534 0.688 0.578 0.678 0.467 0.714
3 0.576 0.412 0.541 0.674 0.588 0.535 0.647
4 0.523 0.598 0.541 0.634 0.712 0.4688 0.505
5 0.566 0.444 0.384 0.598 0.541 0.487 0.785
Ave 0.576 0.480 0.510 0.605 0.631 0.507 0.663
SD 0.039 0.082 0.124 0.050 0.068 0.047 0.103
Table 4: Average Precision and Recall (Three data set) of RNN.
Fold Num. Acc. Pre. High Pre. Medium Pre. Low Rec. High Rec. Medium Rec. Low
1 0.671 0.578 0.374 0.711 0.811 0.641 0.412
2 0.642 0.475 0.552 0.735 0.758 0.468 0.577
3 0.622 0.553 0.577 0.708 0.669 0.547 0.703
4 0.608 0.615 0.714 0.699 0.712 0.563 0.697
5 0.645 0.466 0.766 0.596 0.699 0.476 0.778
Ave 0.638 0.538 0.597 0.690 0.730 0.539 0.633
SD 0.024 0.065 0.120 0.054 0.056 0.071 0.143
Table 5: Average Cross-Validation Precision and Recall of All Models.
Models Acc. Pre. High Pre. Medium Pre. Low Rec. High Rec. Medium Rec. Low
LSTM-RNN-G 0.612 0.575 0.551 0.689 0.788 0.587 0.633
LSTM-RNN-F 0.645 0.541 0.415 0.711 0.678 0.615 0.612
LSTM-RNN-M 0.657 0.648 0.825 0.670 0.724 0.415 0.654
KNN-G 0.597 0.421 0.512 0.852 0.695 0.546 0.745
KNN-F 0.565 0.387 0.673 0.678 0.645 0.498 0.698
KNN-M 0.566 0.632 0.345 0.285 0.553 0.477 0.546
Ave 0.607 0.534 0.554 0.648 0.681 0.523 0.648
versely affected performance.
Table 5 gives the overall average performance re-
sults when the three data sets are considered in isola-
tion. From the table it is interesting to see that for the
gender LSTM-RNN models, the accuracy is slightly
better than the general LSTM-RNN model, whilst this
does not feature with respect to kNN models applied
to the different data sets. Thus there is still no ob-
vious evidence to demonstrate whether the prioritisa-
tion pattern from the data is related to gender, more
investigation is needed here.
Figure 2, presents the runtimes for the kNN and
LSTM models with respect different sizes of input
data from 500 to 5, 000 increasing in steps of 500
and using one fold of the five-cross validation. From
the figure it can be seen that when using kNN with
DTW is considerably less efficient than when using
the LSTM model. An improvement can be made by
changing the representation approach of the time se-
ries to optimise the data structure, so as to enable a
more efficient implementation of kNN and DTW. For
the training time of a single task LSTM in a single
epoch we can see from the figure that the time effi-
ciency is considerably higher than in case of the kNN
model. We can also observe that the run time line is
not linear in the case of the LSTM, as the run time is
also influenced by other parameters from the hidden
layers.
7 CONCLUSION
In this paper, a mechanism for event-based pathol-
ogy data prioritisation has been proposed for multi-
variate time series pathology result data. The mo-
tivation was the large amount of pathology data re-
ceived by hospital departments which necessitates
some form of prioritisation. The challenge was that
there is no ground-truth prioritisation data available,
because of the resource required to create this. Two
approaches were explored, one using the kNN with
DTW as a distance measurement, and one using an
LSTM mechanism. The fundamental idea underpin-
ning the event-based prioritisation is to classify newly
Event-based Pathology Data Prioritisation: A Study using Multi-variate Time Series Classification
127
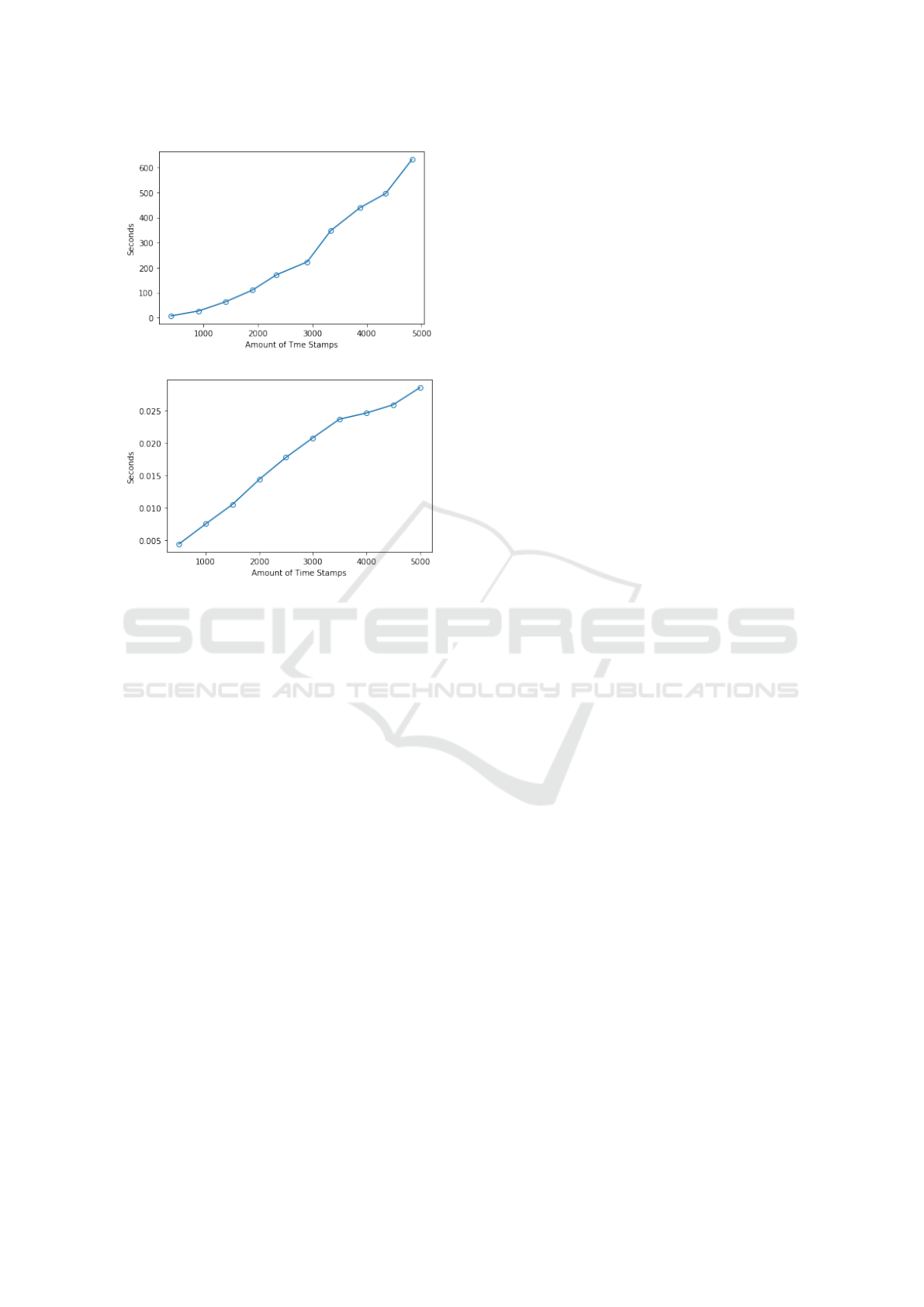
(a) Run time of kNN
(b) Run time of single task LSTM-RNN
Figure 2: Run time with different data size, (a) kNN model,
(b) LSTM-RNN model.
generated pathology data in terms of the anticipated
outcome event and then to use this outcome event
as a prioritisation marker. The proposed mechanism
was evaluated using U&E laboratory test data. The
results demonstrated that the LSTM mechanism pro-
duced the best recall and precision of 0.788 and 0.648
respectively. A criticism of the proposed RNN ap-
proach is that the process of running five LSTMs
separately is time consuming and complicated, meth-
ods using a stacked deep learning network ensemble
might be more preferable. Another criticism is that
the classification was conducted using crisp bound-
aries which may not be the most appropriate.
REFERENCES
Bagnall, A., Lines, J., Bostrom, A., Large, J., and Keogh,
E. (2017). The great time series classification bake
off: a review and experimental evaluation of recent
algorithmic advances. Data Mining and Knowledge
Discovery, 31(3):606–660.
Choi, E., Bahadori, M. T., Kulas, J. A., Schuetz, A., Stew-
art, W. F., and Sun, J. (2016). Retain: An interpretable
predictive model for healthcare using reverse time at-
tention mechanism. arXiv preprint arXiv:1608.05745.
Esteban, C., Schmidt, D., Krompaß, D., and Tresp, V.
(2015). Predicting sequences of clinical events by us-
ing a personalized temporal latent embedding model.
In 2015 International Conference on Healthcare In-
formatics, pages 130–139. IEEE.
Greff, K., Srivastava, R. K., Koutn
´
ık, J., Steunebrink, B. R.,
and Schmidhuber, J. (2016). Lstm: A search space
odyssey. IEEE transactions on neural networks and
learning systems, 28(10):2222–2232.
Halbesma, N., Brantsma, A. H., Bakker, S. J., Jansen, D. F.,
Stolk, R. P., De Zeeuw, D., De Jong, P. E., Gan-
sevoort, R. T., and for the PREVEND study group
(2008). Gender differences in predictors of the decline
of renal function in the general population. Kidney In-
ternational, 74(4):505–512.
Li, Z.-x., Wu, S.-h., Zhou, Y., and Li, C. (2017). A com-
bined filtering search for dtw. In 2017 2nd Interna-
tional Conference on Image, Vision and Computing
(ICIVC), pages 884–888. IEEE.
Rakthanmanon, T., Campana, B., Mueen, A., Batista, G.,
Westover, B., Zhu, Q., Zakaria, J., and Keogh, E.
(2012). Searching and mining trillions of time se-
ries subsequences under dynamic time warping. In
Proceedings of the 18th ACM SIGKDD international
conference on Knowledge discovery and data mining,
pages 262–270.
Tomlinson, L. A. and Clase, C. M. (2019). Sex and the
incidence and prevalence of kidney disease.
Vikram, S., Li, L., and Russell, S. (2013). Handwriting and
gestures in the air, recognizing on the fly. In Proceed-
ings of the CHI, volume 13, pages 1179–1184.
Wang, L., Wang, X., Leckie, C., and Ramamohanarao, K.
(2008). Characteristic-based descriptors for motion
sequence recognition. In Pacific-Asia Conference on
Knowledge Discovery and Data Mining, pages 369–
380. Springer.
Wang, X., Mueen, A., Ding, H., Trajcevski, G., Scheuer-
mann, P., and Keogh, E. (2013). Experimental com-
parison of representation methods and distance mea-
sures for time series data. Data Mining and Knowl-
edge Discovery, 26(2):275–309.
Xing, W. and Bei, Y. (2019). Medical health big data clas-
sification based on knn classification algorithm. IEEE
Access, 8:28808–28819.
KDIR 2021 - 13th International Conference on Knowledge Discovery and Information Retrieval
128
