
Convolutional Neural Networks for Quantitative Prediction of Different
Organic Materials using Near-Infrared Spectrum
Dagmawi Delelegn Tegegn
1,2 a
, Italo Francesco Zoppis
1 b
, Sara Manzoni
1 c
, Cezar Sas
3 d
and Edoardo Lotti
2
1
Department of Computer Science, University of Milano-Bicocca, Milan, Italy
2
SeleTech Engineering Srl, Milan, Italy
3
Independent Researcher, Italy
Keywords:
Convolutional Neural Network (CNN), Near-Infrared (NIR), Quantitative Analysis.
Abstract:
Advances in Near-infrared (NIR) spectroscopy technology led to an increase of interest in its applications
in various industries due to its powerful non-destructive quantization tool. In this work, we used a one-
dimensional CNN to determine simultaneously quantities of organic materials in a mixture using their NIR
infrared spectra. The coefficient of determination (R
2
) and the root mean square error (RMSE) is used to test
the performance of the model. We used six materials to make pairwise combinations with distinct quantities
of each pair. We obtained 13 different pairwise mixtures, afterward, their near-infrared spectrum profiles is
extracted. The model predicted for each mixture their percentage of composition with a result of 0.9955 R
2
and
RMSE 0.0199. Furthermore, we examined the performance of our model when predicting unseen composition
percentages with unseen mixtures. To do so, two scenarios are carried out by filtering the training and testing
set: the first one where we test on unseen composition percentage (UP) of mixtures, and the second one where
we test on unseen composition percentage of unseen mixtures (UPM). The model achieved an R
2
of 0.947 and
0.627 scores respectively for UP and UPM.
1 INTRODUCTION
Near-infrared (NIR) spectroscopy has been around for
decades and a lot of research has been done in this
field (Windham et al., 1997; Berzaghi and Riovanto,
2009; Osborne, 2006;
´
Eva Szab
´
o et al., 2019; Teye
et al., 2019). In the last years, it has been mainly used
to analyze the chemical composition of organic sam-
ples, drugs, food, and other compounds. In particular,
in the food industry, it is used for the quantitative and
qualitative analysis of foods such as meat, fruit, grain,
dairy products, and beverages (Cen and He, 2007; Za-
reef et al., 2020; Huang et al., 2008).
NIR spectra of biological materials are signals
composed of peaks because of molecular vibrations
of mostly O-H, C-H, and N-H groups (Li et al., 2019;
Chen et al., 2015) caused by their interaction with in-
frared light within the NIR wavelength region (800-
a
https://orcid.org/0000-0002-5031-7589
b
https://orcid.org/0000-0001-7312-7123
c
https://orcid.org/0000-0002-6406-536X
d
https://orcid.org/0000-0002-3018-0140
2500 nm). The spectral data measured in this region
are generally composed of high noises and overlap-
ping peaks associated with the chemical composition
of the sample. The spectral information extracted
from these broad peaks for the quantitative determina-
tion of the chemical composition is often analysed us-
ing chemometry and other linear based methods (i.e.
partial least square, multivariate regression) to cap-
ture the various possible infrared spectra patterns of a
single material.
There are many applications in the field of NIR
spectroscopy and mixture analysis. However, this
work focuses on their use in the food analysis in-
dustry. Initial works in this field used Multivariate
Analysis (MVA) like Principal Component Analysis
(PCA), Partial Least Square (PLS), and Support Vec-
tor Machine (SVM). For instance, (Wu et al., 2008)
used least squares support vector machine (LS - VM)
to analyse NIR spectra of milk powder and they also
determined contents of fat, protein, and carbohydrate.
(Windham et al., 1997) and (Qingyun et al., 2007) as-
sessed the potential of NIR to determine the quality of
rice using respectively PLS and a multi-linear regres-
Tegegn, D., Zoppis, I., Manzoni, S., Sas, C. and Lotti, E.
Convolutional Neural Networks for Quantitative Prediction of Different Organic Materials using Near-Infrared Spectrum.
DOI: 10.5220/0010244101690176
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 4: BIOSIGNALS, pages 169-176
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
169

Table 1: Relevant works in the field of Near-infrared for the qualitative and quantitative analysis of organic materials.
Reference Sample Range Target Task Method Performance
(Qingyun et al., 2007) Indica Rice 540,640,970 nm Indica Rice Reg MVA R
2
= 0.71
(Wu et al., 2008) Infant Milk Powder 800-1025 nm Fat, Protein, Carbohydrates Reg, Class ICA-LS-SVM R
2
= 0.983, 0.231, 0.982 Pred = 98%
(Grossi et al., 2015) Olive Oil 569,835 nm Peroxide, Phenol Reg Undefined (Excel) R
2
= 0.883 (Peroxide), 0.895 (Phenol)
(V
´
asconez et al., 2018) Cocoa Powder 1100–2500 nm Adulterant Reg, Class PLS-DA R
2
= 0.974, Acc = 98%
(Sun et al., 2019) Tubers 1000-2500 nm Sugars Reg PLS, LS-SVM R
2
= 0.950
(Wang et al., 2019) Wheat and Potato Flour 1000-2500 nm Potato Flour Reg PLS R
2
= 0.8865
(Teye et al., 2019) Rice 740-1070 nm Rice Class KNN, SVM ACC = 91.88%
(Ng et al., 2019) Soil 350-25000 nm Sand, Clay, Organic Content Reg PLSR,Cubist-tree, CNN R
2
= 0.85-0.95, 0.91–0.97, 0.95–0.98
(Ni et al., 2019) Masson Pine Seedlings 780-2500 nm Nitrogen Reg 1D-CNN R
2
= 0.984
(Liu et al., 2020) Rice 1000-2500 nm Rice Reg PLS-DA, SVM R
2
= 0.96
(de Lima et al., 2020) Cumin, Black Pepper 1100-2500 nm Adulterant Reg MLR,PLS R
2
= 0.90
(Zhang et al., 2020) Tobacco 1000-2500 nm Tobacco Class 1D-CNN,2D-CNN Acc = 93.15%, 93.05%
sion (MLR). (V
´
asconez et al., 2018) used rapid meth-
ods, like NIR technology combined with multivariate
analysis (PCA and partial least squares discriminant
analysis (PLS-DA)), to detect fraud of cocoa powder.
Other methods improved the multivariate analy-
sis by using kernel-based methods like Support Vec-
tor Machines (SVM), for example, (Sun et al., 2019)
investigated the feasibility of NIR spectroscopy com-
bined with kernel PLS regression algorithm for quan-
titative determination of reducing sugar content in
potato flours.
Further improvements are revealed by models that
use machine learning. In fact, machine learning ap-
proaches for spectral profiles analysis (Galli et al.,
2016; Galli et al., 2017; Zoppis et al., 2011), and
in particular Convolutional Neural Networks (CNN)
for spectroscopy signal classification have reported
promising results in the literature. (Zhang et al.,
2020) proposed a one-dimensional CNN (1D-CNN)
to classify the origin of tobacco using their NIR spec-
trum, and concluded that the performance of 1D-CNN
and 2D-CNN was better than traditional PLS mod-
els. Similarly, (Ni et al., 2019) use a 1D-CNN to per-
form a regression task, instead of classification task
as (Zhang et al., 2020), to find the amount of nitrogen
in the Masson pine seedling leaves using NIR spec-
trum. We can view a summary of works that use NIR
spectrum with their relative task and methods in Table
1.
Compared to our model, (Ni et al., 2019) per-
forms regression on a single variable (the nitrogen
content). In this work we propose a modified ver-
sion of the 1D-CNN model proposed by (Ng et al.,
2019). Their model is used on data coming from
the visible/near-infrared, mid-infrared, and a combi-
nation of both while ours uses information only on
NIR data. Moreover, they don’t perform experiments
with unseen combinations and unseen percentages.
2 CONTRIBUTIONS
Considering the promising results reached with CNNs
in the literature for spectroscopy analysis, we adopt a
1D-CNN based approach for the regression analysis
of NIR spectroscopy to predict quantities of organic
mixtures. We used six materials to make pairwise
combinations at different quantities of each, obtain-
ing 13 different mixtures. Subsequently, we extracted
their near-infrared spectrum profiles. We tested the
model performance in three different scenarios to an-
swer the following research questions:
• RQ1 - How well can we predict unseen
1
percent-
ages of mixtures?
• RQ2 - How well can we predict unseen percent-
ages of unseen mixtures?
The first scenario, Whole Prediction (WP), where
we test the model ability to predict the same com-
position percentages of mixtures seen in the training
set. In this scenario, given two materials A, B, and
their combination (A, B), the model sees the spectral
profiles with the same percentage and combination
(A=25, B=75)
2
in training and in testing set. The
WP is used as a baseline for the other scenarios.
We used the second scenario, Unseen Percentages
(UP), to test the model’s ability to predict the un-
seen composition percentage of the same mixtures.
In this scenario, the model sees the same mixtures of
materials in the training set, but with different com-
position percentages. For example, with the combi-
nation set {(A=15, B=85),(A=35, B=65), (C=85,
D=15)} in training set, the model is tested using
{(A=25, B=75),(A=50, B=50), (C=75, D=25)}.
The last scenario, Unseen Percentage and Mix-
ture (UPM), used to test the model’s ability to pre-
dict the unseen composition percentage of unseen
1
Unseen refers to the samples that are not present in the
training set.
2
The mixture of two materials A and B, where A is at 25%
of the total composition and B at 75%.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
170
mixtures. UPM is similar to UP, but here the pri-
mary goal is to train and test the model with differ-
ent composition percentage and mixtures. This means
that the model will see the following pairs and per-
centages in the training set {(A=25, B=75),(A=50,
B=50), (C=75, D=25)}, and in the testing set it will
only see {(A=15, C=85), (A=35, D=65), (B=85,
C=15)}.
This paper is structured as follows. Section 3
presents the materials and the model architecture.
Section 4 examines the experiments’ setup and the re-
sults. Section 5 presents the discussion of the results
and Section 6 closes with the conclusions.
3 MATERIALS AND METHODS
In this section, we present the data collection pro-
cess and the neural network model architecture used
to predict the quantitative measure for the mixed or-
ganic materials. The data collection is made of sev-
eral steps that includes the sample preparation proce-
dure of the six organic powders, the data acquisition
that describes the mechanism of acquiring the spectral
data. Furthermore, we describe the 1D-CNN architec-
ture and its parameters.
3.1 Sample Preparation
Each sample was prepared by carefully mixing a
given fraction in weight of two base materials and
placing the mixture in a container of the Petri dish
type. However, because of the unique characteristics
of the powders used, such as grain size and tendency
to form lumps, it is not possible to guarantee that the
mixture is homogeneous.
The powders used are cocoa (Cocoa), ice sugar
(IceSugar), baby milk powder (BabyMilk), potato
starch (Potato), rice starch (Rice) and baking soda
(NaHCO3). We made a total of 13 pairwise combi-
nations using the six basic powders, summarized in
Table 2. We prepared 56 samples of pairwise mixtures
with different weight proportions of the two compo-
nents, in total 62, adding the six base materials at
Table 2: The pairwise mixtures overview. Value 1 indicates
presence of powder mixture while 0 means that the powders
are not mixed. The diagonal values correspond to the base
materials at 100%.
BabyMilk IceSugar NaHCO3 Cocoa Potato Rice
BabyMilk 1
IceSugar 1 1
NaHCO3 1 1 1
Cocoa 1 1 1 1
Potato 1 1 0 1 1
Rice 1 1 1 1 0 1
100%. In particular, the percentage of each compo-
nent in a mixture is assigned from set P = {15, 25, 35,
50 , 65, 75, 85}, such that the composition percentage
of a given mixture of two materials add up to 100%,
e.g. (A=15, B=75).
3.2 Data Acquisition
We took the measurements using an automatized me-
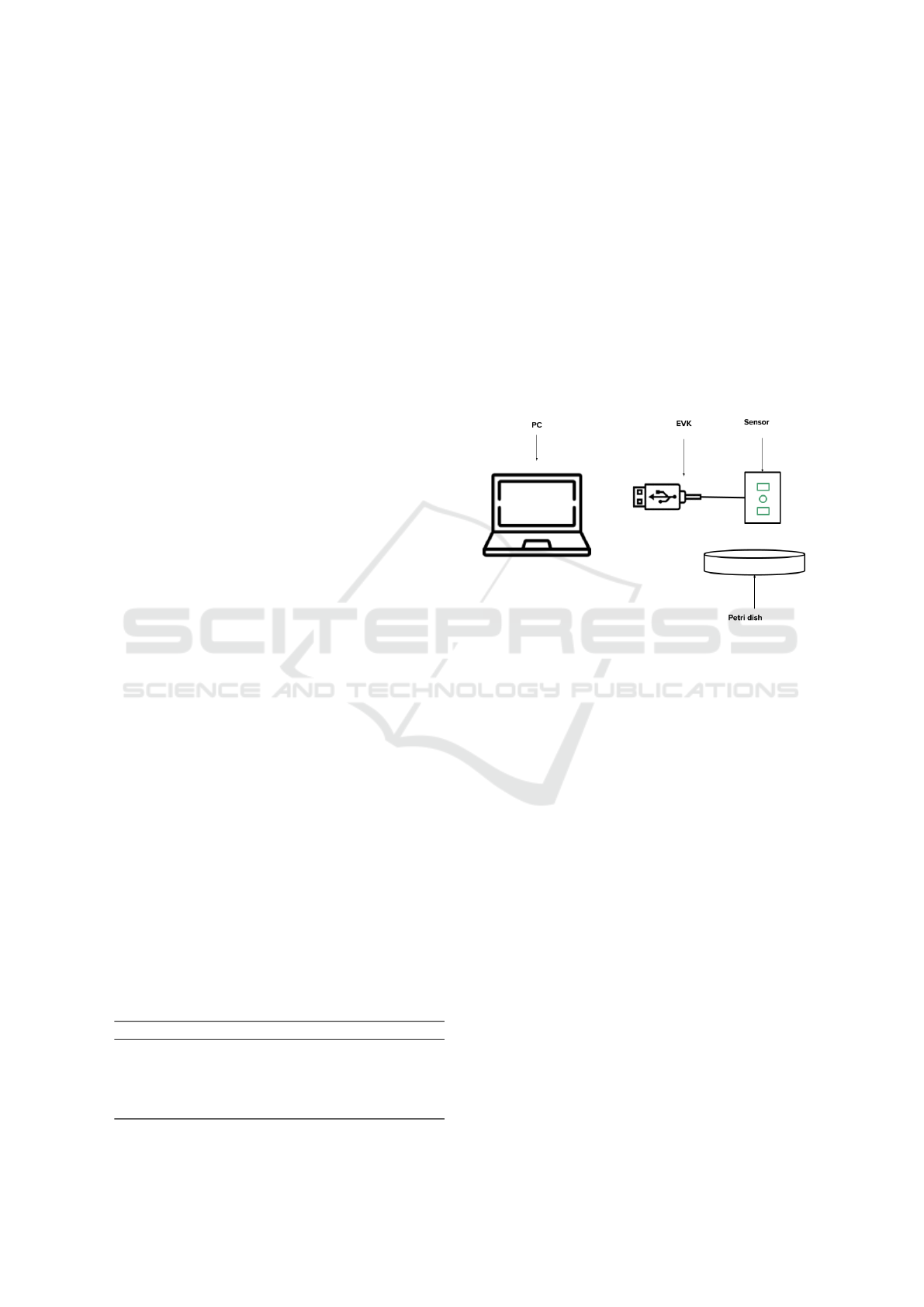
chanical setup. Figure 1 illustrates the tools used to
collect spectral profiles. We put the NIR sensor and
the Petri dish into a dark box to avoid any external
interference. The data is then viewed using a custom
made software for the sensor.
Figure 1: The figure shows the tools used for collecting NIR
spectrum data. The setup is composed of a Petri dish where
the powders are mixed and put inside, the sensor that col-
lects NIR spectra, an Evaluation kit (EVK) used to transfer
data from the sensor and a PC that includes a custom made
software to visualize spectrum.
3.2.1 Sensor Used
We used a device that captures two ranges of wave-
length points: [1350−1650]nm and [1750−2150]nm.
Resulting in a total of 702 wavelength points cap-
tured. We calibrated the NIR sensor with SRS-99-
020 Reflectance Standard (a white diffuse reflectance
sample) by collecting the spectra of the “white” refer-
ence at the minimum distance allowed by the scanner
and with the maximum level of light bulb ignition.
3.2.2 Capture Mode
We set the Petri dish and the NIR sensor inside a box
and inside a dark room along with the automatic ac-
quiring mechanical setup to avoid any outside inter-
ference. The NIR sensor captured the spectra of the
samples at different sensor-sample distances, mov-
ing the vertical axis of the scanner with 1mm pitch
along the entire 20mm useful range. The minimum
distance between the outer surface of the Petri dish
Convolutional Neural Networks for Quantitative Prediction of Different Organic Materials using Near-Infrared Spectrum
171

window and the detector is 5mm. For each distance,
we examined three different areas of the sample’s ex-
posed surface. We have acquired three spectra for
each zone, hence there are nine spectra acquired at the
same distance from each sample. Assuming the sam-
ple is homogeneous, the spectra of the same mixture
should be the same with each other with minor differ-
ences due to measurement noise. Instead, we found
that there are some differences, particularly between
zones. In isolated cases, we also found differences
between spectra acquired at the same location, proba-
bly because of measurement errors related to electri-
cal disturbances or mechanical vibrations. We mea-
sured all samples by varying the level of lamp inten-
sity (parameter varied between 200 and 250 with step
1).
3.2.3 Reflectance Values
The measured reflectance R of a generic sample is cal-
culated by the sensor at each wavelength as follows:
R =
I
C
− I
F
I
SRS
− I
F
R
SRS
(1)
where I
C
is the intensity of light received from the
sample, I
SRS
the intensity of light received from the
reference SRS-99-020 placed at a given distance dur-
ing calibration, I
F
the intensity of background light
(cross-talk), measured during calibration with lamp
on but without target, and R
SRS
the reflectance of the
reference SRS-99-020. In practice, since cross-talk
levels are generally small, the measured reflectance is
proportional to the ratio of the light intensity received
by the sample in question to the one measured dur-
ing calibration using the SRS-99-020 reference. For
this reason, although reflectance is an intrinsic prop-
erty of the sample, the reflectance measured using the
same sample at different distances from that used dur-
ing calibration is different. A similar effect occurs if
the measurement uses a different light bulb ignition
level than the one used when calibrating.
3.2.4 Dataset
Following the sample preparation and the acquisition
of the NIR spectra, we collected 454896 samples
3
.
Each sample has 702 features representing the cap-
tured wavelengths, and for each composition percent-
age of a mixture we have ∼ 7300 samples. The target
variable of each sample is a percentage distribution
over the six base materials describing the quantity of
that material in the spectral sample. Given that each
spectral sample represents only the mixture of two
3
The dataset is available upon request.
materials, only two elements in the target vector con-
tain the value of the individual materials represented
in the spectral sample while we set the remaining four
elements to 0. Whereas, for the mixtures containing
only one powder, we set the five remaining target vari-
ables to 0 and assigned the value 100% to the material
represented by the spectra.
3.3 Method
3.3.1 Convolutional Neural Network
Convolutional Neural Networks (CNNs) (Nebauer,
1998) are a specific type of neural networks (NN)
used to process data having a known, grid-like struc-
ture (i.e. time-series data, which can be thought of as
a 1D grid taking samples at regular time intervals, and
image data, which can be thought of as a 2D grid of
pixels).
The basic blocks of CNNs consist of convolu-
tion layers, pooling layers, and fully connected lay-
ers (Shin et al., 2016). The convolution layer uses
filters to perform convolution operations over the in-
put producing the activation map. The pooling layer
is usually after a convolution layer and performs the
down sampling of the activation map. The fully con-
nected layers are usually found at the end of the net-
work. They are used to producing an output that can
be optimized for targets like classification of the in-
put. In this work we adopted a one-dimensional CNN
for our one-dimensional spectral data. The convolu-
tional neural network consists of seven trainable lay-
ers - four convolutional layers and two fully con-
nected layers, similar to the one presented by (Ng
et al., 2019), but with different hyper-parameter val-
ues to fit our data. The output of every convolutional
layer is passed to the pooling layer. The convolution
layer contained 32 filters with a filter size of 3, stride
as one and zero padding. The number of filters on
every two convolutional layers is increased by a fac-
tor of two, while other parameters are kept the same.
The pooling size of the first layer is set to two, and
the last one to 4. The final feature maps from the
pooling layer are then flattened and used as input to
Table 3: Architecture of the 1D-CNN used in our work.
Layer Output Shape # Param Kernel Filter Attributes
Conv1D (702, 32) 128 3 32
MaxPooling1D (351, 32) 0 size = 2
Conv1D (351, 32) 3104 3 32
MaxPooling1D (175, 32) 0 size = 2
Conv1D (175, 64) 6208 3 64
MaxPooling1D (87, 64) 0 size = 2
Conv1D (87, 64) 12352 3 64
MaxPooling1D (21, 64) 0 size = 4
Flatten (1344) 0
Dropout (1344) 0 rate = 0.3
Dense (512) 688640
Dense (6) 3078
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
172

a fully connected layers. The dropout layer acted as a
regularization layer to mitigate over-fitting. The final
model has 713,510 parameters, the detailed architec-
ture is summed up in Table 3. The input of the 1D-
CNN is a one dimensional spectral vector containing
values of the 702 wavelength points, and the target is
also a one dimensional vector containing the percent-
age distributions of the six materials.
3.3.2 Hyper-Parameters
Since we have to predict the percentage of composi-
tion in a mixture we use the Kullback-Leibler (KL)
divergence (Equation 2) as our loss function.
The KL is a measure of distance between two distri-
butions defined as:
D
KL
(QkZ) =
∑
x∈X
Q(x)log
Q(x)
Z(x )
(2)
Where Q and Z are two probability distributions,
that in our case correspond respectively to the true
and the predicted distribution of percentages. The KL
gives a score close to 0 when Q and Z are similar and
increases as the distributions differ.
We used Adam as the optimization algorithm
(Kingma and Ba, 2015), with a scheduled learning
rate, starting from lr = 0.001, and exponentially de-
creasing it every epoch. We also use early stopping
and limit the number of epochs to 100. However, the
training always stopped earlier.
We used the Keras as our machine learning frame-
work and we used for the entire work the Asus Vivo-
Book X580GD with an Intel(R) Core (TM) i7-8750H
CPU.
4 RESULTS
The quantitative analysis of the NIR spectra based on
the multi-modal 1D-CNN is studied in three scenarios
as detailed in Section 2. For each scenario is reported
a table that summarises the model’s overall perfor-
mance for each material, this is done by filtering the
model’s output for each of the six materials for the
corresponding scenario. In the Appendix is reported
the full output of each mixture for each scenario and
it is available here
4 5
.
4.1 Whole Prediction (WP)
The first scenario (WP) we used all the composition
percentages of all mixtures. We use this scenario as
4
https://github.com/dtegegn/CNN-NIR-Spectra
5
https://www.seletech.com
a baseline result for the next experiments, as this one
shows the ideal case where both the mixtures and the
percentages have been seen during training.
Given the 454896 samples, we split them into
train, validation, and test set, respectively with the ra-
tio of 45%, 22%, and 33% which is a good amount of
data to train and test the model’s performance.
All the three dataset have the same type and bal-
anced number of the 62 different composition per-
centage of all mixtures including the six base mate-
rials of 100%. In Table 4 we report the metrics of the
overall performance of our model for this experiment.
We can see that most of the materials have good per-
formances except for Babymilk, which has a higher
Mean Absolute Error (MAE).
Table 4: The overall performance for the WP scenario.
Material MAE MSE RMSE R
2
BabyMilk 0.0161 0.0009 0.0298 0.9910
IceSugar 0.0065 0.0002 0.0143 0.9976
NaHCO3 0.0046 0.0001 0.0114 0.9983
Cocoa 0.0089 0.0005 0.0228 0.9941
Potato 0.0060 0.0003 0.0167 0.9961
Rice 0.0073 0.0004 0.0189 0.9958
4.2 Unseen Percentage (UP)
The WP experiment settled the baseline result for the
prediction for all the mixtures and their quantities,
and we use it to see the best case scenario for our
model. Therefore, we created specific subsets from
the whole dataset used in WP, by filtering specific set
of composition percentage of the mixtures, therefore
we can test the model’s performance on predicting
unseen composition percentage of the same mixture
and compare it with the baseline scenario. Thus, in
the second experiment UP we created two subsets of
P = {15, 25,35,50,65, 75, 85}: P1 = {15,35,65, 85}
and P2 = P − P1 = {25,50,75}, then defined the set
MP1 as the mixtures of materials belonging in set P1
and used it to train the model. We also defined the set
MP2 that comprises of the mixtures of materials be-
longing to set P2 and the six base materials at 100%,
and used it for testing. We use 33% of the training set
MP1 for the validation set, totaling 118380 samples
for the training, 58308 for the validation and 278208
for the testing sets.
With the UP experiment, the number of mixtures
in the training (13 mixtures) set outnumbered the ones
in the test set (6 mixtures), therefore, the set of mix-
tures in the training set is a subset of the mixtures in
the test set but in different quantities. This setup al-
lows us to test the model’s predicting ability only on
Convolutional Neural Networks for Quantitative Prediction of Different Organic Materials using Near-Infrared Spectrum
173

Table 5: The overall performance for the UP scenario.
Material MAE MSE RMSE R
2
BabyMilk 0.166 0.013 0.115 0.865
IceSugar 0.060 0.003 0.052 0.969
NaHCO3 0.046 0.001 0.031 0.989
Cocoa 0.069 0.003 0.058 0.950
Potato 0.041 0.002 0.046 0.969
Rice 0.062 0.004 0.063 0.941
the composition percentage. We can see from Table 5
the model’s overall performance for each material.
4.3 Unseen Percentage and Mixture
(UPM)
The goal of the UPM experiment is to evaluate the
model performance on predicting the unseen com-
position percentage of the unseen mixtures, thus the
model’s ability to extract from the mixture spectral
data the single component’s features and its ability to
use these to generalize on the unseen percentage with
unseen mixtures.
The experiment UP had already two sets for the
training and testing, MP1 and MP2 respectively. In
the UP experiment, the set MP1 contained a greater
number of different mixtures than those found in
MP2. While in the UPM experiment, we used the
MP2 set as the training set and the MP1 set as the test-
ing set. Consequently, this procedure had the num-
ber of mixtures for the test set outnumbering the ones
found in the training set, unlike for the UP experi-
ment. This allowed us to test the different portions
of the test set, as we created subsets of the testing
set. For the first testing subset (MP1
S1
), we used mix-
tures that are found also in the training set, while for
the second testing subset (MP1
S2
), we used the mix-
tures that are not found in the training set. Reminding
that the composition percentage of the mixtures in the
training and test sets are totally different.
Finally, we obtained three results, the whole test
set for this scenarios (MP1), the first subset (MP1
S1
),
and the second subset (MP1
S2
). The subset MP1
S1
contained all mixtures that are also included in the
training set except the base materials at 100%. The
MP1
S2
subset contained all mixtures that not found in
the training set, which is the experiment for the UPM
scores. The total test set MP1 contained MP1
S1
and
MP1
S2
. Our focus here is to see the model prediction
on the subset MP1
S2
for the composition percentage
of unseen mixtures, and we can see the model’s per-
formance for each material in Table 8. In Table 6 and
7 we can see the overall performance of the model for
each material respectively using the whole testing set
MP1 and the subset MP1
S1
.
Table 6: The overall performance for the UPM scenario.
The results are for the whole test set (MP1) of the UPM
scenario.
MAE MSE RMSE R
2
BabyMilk 0.123 0.0426 0.2063 0.5135
IceSugar 0.0566 0.0136 0.1165 0.8361
NaHCO3 0.0434 0.0146 0.1209 0.791
Cocoa 0.0703 0.0201 0.1419 0.7933
Potato 0.0425 0.0168 0.1297 0.7765
Rice 0.0593 0.0175 0.1322 0.813
Table 7: The overall performance for the UPM scenario.
The table shows the results for the MP1
S1
subset.
MAE MSE RMSE R
2
BabyMilk 0.0489 0.0048 0.0696 0.9535
IceSugar 0.0115 0.0007 0.0257 0.991
NaHCO3 0.014 0.0011 0.0336 0.9883
Cocoa 0.0135 0.0017 0.0411 0.97
Potato 0.0078 0.0006 0.0236 0.9901
Rice 0.0144 0.0016 0.0396 0.9723
Table 8: The overall performance for the UPM scenario.
The table shows the main results for the UPM experiment
using MP1
S2
testing subset.
MAE MSE RMSE R
2
IceSugar 0.0972 0.0252 0.1587 0.718
NaHCO3 0.0699 0.0267 0.1635 0.3653
Cocoa 0.1213 0.0367 0.1916 0.6769
Potato 0.0737 0.0314 0.1773 0.6486
Rice 0.0996 0.0318 0.1783 0.7266
5 DISCUSSION
The results achieved from the WP experiment are en-
couraging. The 1D-CNN predicted all the composi-
tion percentages of the mixtures with a very low error
as seen from Table 4, with an average of R
2
= 0.99.
This result is promising since the model is able to ex-
tract the features of the specific composition percent-
ages of mixtures. The outcome of the WP experiment
encouraged us for the much harder tasks that are the
UP an the UPM experiments.
The UP experiment is created to see how well we
can predict the unseen composition percentages by
training the model with the same mixtures as in the
testing set but different composition percentage of the
same mixtures. Training the model with MP1 and
testing it with MP2 gave good results in terms of the
determination coefficient, R
2
= 0.9471, with a 5% de-
crease in respect to the WP average R
2
score.
The UPM experiment have fewer variation of mix-
tures and quantities in the training set than in the test
set and scored an average of R
2
= 0.7539 using all the
test set MP1, with 25% decrease in respect to WP av-
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
174

erage R
2
score. Using the MP1
S1
subset led to better
result since the model had to predict only the unseen
composition percentage but same mixture, just as the
UP experiment, the average determination coefficient
for this testing subset is R
2
= 0.9775 with less that 2%
decrease in respect to the WP experiment. The results
for the MP1
S1
set can be compared also to the UP ex-
periment: the MP1
S1
results showed a 3% improve-
ment in respect to UP experiment. This is because
the UPM training set contained uniformly distributed
composition percentage of each material that are in
{25,50,75} and the six base materials at 100%, while
the UPM missed the mixtures that contains 65% of
one material and 35% of the other leading to unevenly
distributed composition percentage for one mixture.
While using the MP1
S2
subset as the test set, we
have an average determination coefficient of R
2
=
0.627, with a significant 36% loss in respect the aver-
age R
2
of the WP experiment. This experiment is set
up to to see if the model learned the representation of
the single materials in different compositions so that it
can predict unseen composition percentage of unseen
mixtures. We must take into account also the fact that
the materials quantities are prepared by weight rather
than volume, this mean that we can have powders
like BabyMilk that have a greater volume for a small
amount. This characteristic can affect the spectral ac-
quisition since the material with higher volumes tend
to occupy most of the Petri dish causing little signal
for the other materials mixed with them. In this spe-
cific test case scenario of UPM the model is trained
with mixtures containing only BabyMilk mixed with
the other 5 materials in different compositions and
one other mixture of IceSugar and NaHCO3, in par-
ticular the subset MP1
S2
of the test set for this exper-
iment doesn’t contain any combination of BabyMilk,
but it is trained with mixtures that contain BabyMilk.
The worst results in terms of the R
2
score NaHCO3
gave the worst results because we have only two com-
bination of this materials in the test set, with Cocoa
and Rice, while in the training set there is no such
mixture, adding also the fact that the NaHCO3 have
higher density in terms of g/cm
3
in respect to Cocoa
and Rice. Therefore, the spectra of of NaHCO3 mixed
with Cocoa and then with Rice can be very difficult to
interpolate without being trained.
In WP experiment the model was able to over-
come the errors during mixtures and the weight-
volume ratio and gave good result this is thanks to the
huge amount of sample it is trained on. In the UP and
especially the UPM experiments the errors due to the
preparation of the materials and the the models ability
to overcome them became very clear.
The UP and UPM experiments hold the answers
for the questions RQ1 and RQ2:
• A1 - The UP experiment let us predict the un-
seen new percentages of the same mixtures with
a good approximation, and also the WP experi-
ment model, that have all the percentages, can pre-
dict every composition percentage in the range of
[0 − 100] with higher accuracy.
• A2 - The model’s prediction on unseen com-
position percentage of unseen mixtures showed
promising results. We think that if we take into
account the weight-volume ratio, we can improve
the spectra acquired and therefore the final results.
6 CONCLUSION
In this work, we analysed the problem of predicting
composition percentage of organic material mixtures.
The NIR spectra of organic materials holds intrinsic
information on the analyte, including its quantity. To
uncover these intrinsic characteristics of the 1D-CNN
showed great performance, in the WP experiment, by
extracting directly relevant wavelength (feature) from
the NIR spectrum that described the quantity of the
analyte. This led to better performance of the model
avoiding the accumulation of errors caused by manual
wavelength selection.
The NIR spectra of the mixtures are most probably
affected by the density, in terms of g/cm
3
, of each
material in the mixture. Thus, affecting the result of
each experiment especially in the UPM experiment.
The research question RQ2 leads to future devel-
opments of this work. The results of the UPM using
the MP1
S2
testing subset can be improved by taking
into account the weight-volume ratio and by model-
ing new 1D-CNN architecture for the unseen compo-
sition of unseen mixtures. It also interesting to extend
the WP experiment’s model by testing it on different
composition percentage of more than two mixtures to
see if the model is acquire a good generalizing ability.
REFERENCES
Berzaghi, P. and Riovanto, R. (2009). Near infrared spec-
troscopy in animal science production: principles
and applications. Italian Journal of Animal Science,
8(sup3):39–62.
Cen, H. and He, Y. (2007). Theory and application of near
infrared reflectance spectroscopy in determination of
food quality. Trends in Food Science and Technology,
18(2):72 – 83.
Convolutional Neural Networks for Quantitative Prediction of Different Organic Materials using Near-Infrared Spectrum
175

Chen, Q., Zhang, D., Pan, W., Ouyang, Q., Li, H., Ur-
mila, K., and Zhao, J. (2015). Recent developments of
green analytical techniques in analysis of tea’s quality
and nutrition. Trends in Food Science & Technology,
43(1):63 – 82.
de Lima, A. B. S., Batista, A. S., de Jesus, J. C., de Je-
sus Silva, J., de Ara
´
ujo, A. C. M., and Santos, L. S.
(2020). Fast quantitative detection of black pepper and
cumin adulterations by near-infrared spectroscopy and
multivariate modeling. Food Control, 107:106802.
Galli, M., Pagni, F., De Sio, G., Smith, A., Chinello, C.,
Stella, M., L’Imperio, V., Manzoni, M., Garancini, M.,
Massimini, D., et al. (2017). Proteomic profiles of thy-
roid tumors by mass spectrometry-imaging on tissue
microarrays. Biochimica et Biophysica Acta (BBA)-
Proteins and Proteomics, 1865(7):817–827.
Galli, M., Zoppis, I., De Sio, G., Chinello, C., Pagni, F.,
Magni, F., and Mauri, G. (2016). A support vector ma-
chine classification of thyroid bioptic specimens using
maldi-msi data. Advances in bioinformatics, 2016.
Grossi, M., Di Lecce, G., Arru, M., Gallina Toschi, T., and
Ricc
`
o, B. (2015). An opto-electronic system for in-
situ determination of peroxide value and total phenol
content in olive oil. J. of Food Engineering, 146:1 – 7.
Huang, H., Yu, H., Xu, H., and Ying, Y. (2008). Near in-
frared spectroscopy for on/in-line monitoring of qual-
ity in foods and beverages: A review. J. of Food Engi-
neering, 87(3):303 – 313.
Kingma, D. P. and Ba, J. (2015). Adam: A method for
stochastic optimization. In Bengio, Y. and LeCun,
Y., editors, 3rd International Conference on Learn-
ing Representations, ICLR 2015, San Diego, CA, USA,
May 7-9, 2015, Conference Track Proceedings.
Li, Z., Tang, X., Shen, Z., Yang, K., Zhao, L., and Li,
Y. (2019). Comprehensive comparison of multiple
quantitative near-infrared spectroscopy models for as-
pergillus flavus contamination detection in peanut. J.
of the Science of Food and Agriculture, 99(13):5671–
5679.
Liu, Y., Li, Y., Peng, Y., Yang, Y., and Wang, Q. (2020). De-
tection of fraud in high-quality rice by near-infrared
spectroscopy. Journal of Food Science.
Nebauer, C. (1998). Evaluation of convolutional neural net-
works for visual recognition. IEEE Transactions on
Neural Networks, 9(4):685–696.
Ng, W., Minasny, B., Montazerolghaem, M., Padarian, J.,
Ferguson, R., Bailey, S., and McBratney, A. B. (2019).
Convolutional neural network for simultaneous pre-
diction of several soil properties using visible/near-
infrared, mid-infrared, and their combined spectra.
Geoderma, 352:251 – 267.
Ni, C., Wang, D., and Tao, Y. (2019). Variable weighted
convolutional neural network for the nitrogen content
quantization of masson pine seedling leaves with near-
infrared spectroscopy. Spectrochimica Acta Part A:
Molecular and Biomolecular Spectroscopy, 209:32 –
39.
Osborne, B. G. (2006). Near-infrared spectroscopy in food
analysis. Enc. of analytical chemistry: applications,
theory and instrumentation.
Qingyun, L., Yeming, C., Mikami, T., Kawano, M., and
Zaigui, L. (2007). Adaptability of four-samples sen-
sory tests and prediction of visual and near-infrared
reflectance spectroscopy for chinese indica rice. J. of
Food Engineering, 79(4):1445 – 1451.
Shin, H., Roth, H. R., Gao, M., Lu, L., Xu, Z., Nogues,
I., Yao, J., Mollura, D., and Summers, R. M. (2016).
Deep convolutional neural networks for computer-
aided detection: Cnn architectures, dataset charac-
teristics and transfer learning. IEEE Trans. on Med.
Imaging, 35(5):1285–1298.
Sun, X., Zhu, K., and Liu, J. (2019). Nondestructive detec-
tion of reducing sugar of potato flours by near infrared
spectroscopy and kernel partial least square algorithm.
Journal of Food Measurement and Characterization,
13(1):231–237.
Teye, E., Amuah, C. L., McGrath, T., and Elliott, C.
(2019). Innovative and rapid analysis for rice authen-
ticity using hand-held nir spectrometry and chemo-
metrics. Spectrochimica Acta Part A: Molecular and
Biomolecular Spectroscopy, 217:147 – 154.
´
Eva Szab
´
o, Gergely, S., Spaits, T., Simon, T., and Salg
´
o,
A. (2019). Near-infrared spectroscopy-based methods
for quantitative determination of active pharmaceuti-
cal ingredient in transdermal gel formulations. Spec-
troscopy Letters, 52(10):599–611.
V
´
asconez, M.,
´
Edgar P
´
erez-Esteve, Arnau-Bonachera, A.,
Barat, J., and Talens, P. (2018). Rapid fraud detection
of cocoa powder with carob flour using near infrared
spectroscopy. Food Control, 92:183 – 189.
Wang, H., Lv, D., Dong, N., Wang, S., and Liu, J. (2019).
Application of near-infrared spectroscopy for screen-
ing the potato flour content in chinese steamed bread.
Food science and biotechnology, 28(4):955–963.
Windham, W. R., Lyon, B. G., Champagne, E. T., Bar-
ton, F. E., Webb, B. D., McClung, A. M., Molden-
hauer, K. A., Linscombe, S., and McKenzie, K. S.
(1997). Prediction of cooked rice texture quality us-
ing near-infrared reflectance analysis of whole-grain
milled samples. Cereal Chemistry, 74(5):626–632.
Wu, D., Feng, S., and He, Y. (2008). Short-wave near-
infrared spectroscopy of milk powder for brand iden-
tification and component analysis. Journal of Dairy
Science, 91(3):939 – 949.
Zareef, M., Chen, Q., Hassan, M. M., Arslan, M., Hashim,
M. M., Ahmad, W., Kutsanedzie, F. Y., and Agyekum,
A. A. (2020). An overview on the applications of
typical non-linear algorithms coupled with nir spec-
troscopy in food analysis. Food Engineering Reviews,
pages 1–18.
Zhang, L., Ding, X., and Hou, R. (2020). Classification
modeling method for near-infrared spectroscopy of to-
bacco based on multimodal convolution neural net-
works. Journal of Analytical Methods in Chemistry,
2020.
Zoppis, I., Gianazza, E., Borsani, M., Chinello, C., Mainini,
V., Galbusera, C., Ferrarese, C., Galimberti, G., Sorbi,
S., Borroni, B., et al. (2011). Mutual information opti-
mization for mass spectra data alignment. IEEE/ACM
transactions on computational biology and bioinfor-
matics, 9(3):934–939.
BIOSIGNALS 2021 - 14th International Conference on Bio-inspired Systems and Signal Processing
176
