
Automatic ICD-10 Classification of Diseases from Dutch Discharge
Letters
Ayoub Bagheri
1,2
, Arjan Sammani
2
, Peter G. M. Van Der Heijden
1,3
, Folkert W. Asselbergs
2,4,5
and Daniel L. Oberski
1,6
1
Department of Methodology and Statistics, Faculty of Social Sciences, Utrecht University, Utrecht, The Netherlands
2
Department of Cardiology, Division of Heart and Lungs, University Medical Center Utrecht, Utrecht, The Netherlands
3
S3RI, Faculty of Social Sciences, University of Southampton, U.K.
4
Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London, London, U.K.
5
Health Data Research UK, Institute of Health Informatics, University College London, London, U.K.
6
Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, The Netherlands
Keywords: Automated ICD Coding, Multi-label Classification, Clinical Text Mining, Dutch Discharge Letters.
Abstract: The international classification of diseases (ICD) is a widely used tool to describe patient diagnoses. At
University Medical Center Utrecht (UMCU), for example, trained medical coders translate information from
hospital discharge letters into ICD-10 codes for research and national disease epidemiology statistics, at
considerable cost. To mitigate these costs, automatic ICD coding from discharge letters would be useful.
However, this task has proven challenging in practice: it is a multi-label task with a large number of very
sparse categories, presented in a hierarchical structure. Moreover, existing ICD systems have been
benchmarked only on relatively easier versions of this task, such as single-label performance and performance
on the higher “chapter” level of the ICD hierarchy, which contains fewer categories. In this study, we
benchmark the state-of-the-art ICD classification systems and two baseline systems on a large dataset
constructed from Dutch cardiology discharge letters at UMCU hospital. Performance of all systems is
evaluated for both the easier chapter-level ICD codes and single-label version of the task found in the literature,
as well as for the lower-level ICD hierarchy and multi-label task that is needed in practice. We find that state-
of-the-art methods outperform the baseline for the single-label version of the task only. For the multi-label
task, the baselines are not defeated by any state-of-the-art system, with the exception of HA-GRU, which does
perform best in the most difficult task on accuracy. We conclude that practical performance may have been
somewhat overstated in the literature, although deep learning techniques are sufficiently good to complement,
though not replace, human ICD coding in our application.
1 INTRODUCTION
ICD-10 is the 10th edition of the International
statistical Classification of Diseases, a repository
maintained by the World Health Organization to
provide a standardized system of diagnostic codes for
classifying diseases (Atutxa et al., 2019; Baumel et
al., 2018). These classification codes are vastly used
in clinical research and are a part of the electronic
health records (EHRs) in the University Medical
Center Utrecht (UMCU), The Netherlands. Currently,
the task of assigning classification categories to the
diagnoses is carried out manually by medical staff.
Manual classification of diagnoses is a labor-
intensive process that consumes significant resources.
For this reason, a number of systems have been
proposed to automate the disease coding process with
machine learning algorithms trained on data
generated by medical experts.
The ICD coding task is challenging due to the use
of free-text, multi-label setting of diagnosis codes and
the large number of codes (Atutxa et al., 2019;
Boytcheva 2011). Several attempts have been made
to automatically assign ICD codes to medical
documents, ranging from rule-based (Baghdadi et al.,
2019; Boytcheva 2011; Koopman et al., 2015a;
Nguyen et al., 2018) to machine learning approaches
(Atutxa et al., 2019; Baumel et al., 2018; Cao et al.,
2019; Chen et al., 2017; Du et al., 2019; Duarte et al.,
2018; Karimi et al., 2017; Kemp et al., 2019;
Koopman et al., 2015b; Lin et al., 2019; Liu et al.,
2018; Miranda et al., 2018; Mujtaba et al., 2017;
Bagheri, A., Sammani, A., Van Der Heijden, P., Asselbergs, F. and Oberski, D.
Automatic ICD-10 Classification of Diseases from Dutch Discharge Letters.
DOI: 10.5220/0009372602810289
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 281-289
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
281

Mullenbach et al., 2018; Nigam et al., 2016;
Pakhomov et al., 2006; Shing et al., 2019; Xie et al.,
2019; Zweigenbaum and Lavergne, 2016). Rule-
based methods have good performance when: (1) the
terms to be categorized follow regular patterns, (2)
the number of ICD labels is quite small, and (3) the
task is limited to single-label classification (Atutxa et
al., 2019). Unfortunately, with ICD classification
these conditions seldom apply.
When a coded dataset is available and the range
of the ICDs to label is large, machine learning based
techniques have been successful (Atutxa et al., 2019;
Baumel et al., 2018; Cao et al., 2019; Duarte et al.,
2018; Miranda et al., 2018; Nigam et al., 2016). An
approach for automatic matching of ICD-10
classification of Bulgarian free text (Boytcheva,
2011) was based on support vector machines (SVM).
Zweigenbaum and Lavergne (Zweigenbaum and
Lavergne, 2016) suggested a hybrid method for ICD-
10 coding of death certificates based on a dictionary
projection method and a supervised learning
algorithm. They used the SNOMED (systemic
nomenclature of medicine) and UMLS (unified
medical language source) to set up the dictionary
projection method. Koopman et al. (Koopman et al.,
2015b) trained 86 SVM classifiers to identify cancers,
first identifying the presence of a cancer by one
classifier and later in a cascaded architecture
classifying the cancer type according to ICD-10 codes
using 85 different SVM classifiers.
Recently, deep learning methods boosted
benchmarked results in various text mining studies
(Gargiulo et al., 2018; Shickel et al., 2017;
Subramanyam and Sivanesan, 2020; Xiao, 2018),
including in automated ICD coding (Atutxa et al.,
2019; Baumel et al., 2018; Du et al., 2019; Duarte et
al., 2018; Karimi et al., 2017; Lin et al., 2019; Liu et
al., 2018; Miranda et al., 2018; Mujtaba et al., 2017;
Mullenbach et al., 2018; Nigam et al., 2016; Shing et
al., 2019). Karimi et al. (Karimi et al., 2017)
described a deep learning method for ICD coding,
reporting on tests over a dataset of radiology reports.
The authors proposed to use a convolutional neural
network (CNN) architecture, attempting to quantify
the impact of using pre-trained word embeddings for
model initialization. The best CNN model
outperformed baseline SVM, random forest, and
logistic regression models using bag-of-words
(BOW) representations. BOW is a vector
representation method, demonstrating each document
by one vector of features, i.e. words or combinations
of words (n-grams). In (Nigam et al., 2016), recurrent
1
https://www.who.int/classifications/icd/
neural networks (RNNs) have been applied to the
multi-label classification task for assigning ICD-9
labels to medical notes, finding that an RNN with
long short-term memory (LSTM) units shows an
improvement over the binary relevance logistic
regression model. Atutxa et al. (Atutxa et al., 2019)
evaluated different architectures of neural networks
for multi-class document classification as a language
modeling problem. In their experiments, the results of
ICD-10 coding using the RNN-CNN architecture
outperformed alternative approaches. Baumel et al.
(Baumel et al., 2018) investigated four models
namely SVM, continuous-BOW (CBOW), CNN and
hierarchical attention bidirectional gated recurrent
unit (HA-GRU) for attributing multiple ICD-9 codes.
The HA-GRU model achieved the best performance.
A drawback of the existing literature is that the
performance of different systems is difficult to
compare, because the ICD classification task is often
made easier by only considering the top-level
“chapters” of the ICD hierarchy, or by only
considering a single label as the output.
In the current application, we sought to implement
a system to support human ICD coding of Dutch-
language discharge letters at UMCU hospital. We
explicitly aim at multi-label classification of three-
digit ICD-10 codes, a task that is relatively difficult.
Here, we present a benchmark of five state-of-the-art
systems, all deep learning models, and two baseline
methods based on BOW and pretrained embeddings
with SVM. We aim to evaluate both the relative
performance of these systems, which were all
reported to outperform others, as well as the overall
level of performance for potential support of human
ICD coding, using a dataset of UMCU cardiology
discharge letters.
2 METHODS
2.1 Case Study
Table 1 provides the characteristics of the dataset of
discharge letters collected at the department of
Cardiology in the UMCU. A hospital discharge letter
is a medical text summary describing information
about patient’s hospital admission and treatments.
UMCU cardiology discharge letters are coded based
on the ICD-10 of cardiovascular diseases.
ICD-10 has a hierarchical structure, connecting
specific diagnostic codes through is-a relations
1
. The
hierarchy has several levels, from less specific to
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
282
more specific. ICD codes contain both diagnosis and
procedure codes. In this paper, we focus on diagnosis
codes. ICD-10 codes consist of three to seven
characters. For example, I50.0 shows the “congestive
heart failure” disease, and I50 is its rolled-up code that
shows the heart failure category in chapter IX:
“Diseases of the circulatory system”.
Table 1: UMCU dataset.
Feature Description
Taxonomy ICD-10
Language Dutch
Nb of records 5,548
Nb of unique tokens 148,726
Avg nb of tokens / records 936
Nb of full labels 1,195
Nb of rolled-up labels 608
Label cardinality 4.7
Label density 0.0039
% labels with 50+ records 8.03%
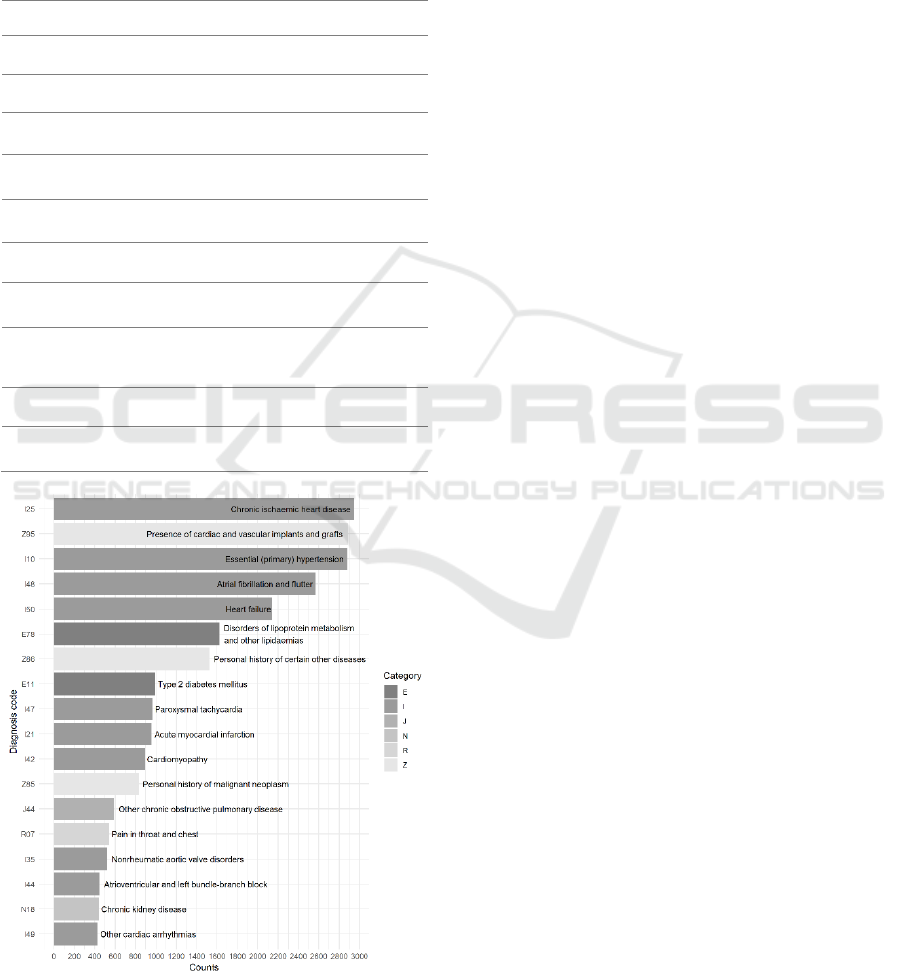
Figure 1: ICD rolled-up codes with more than 400
appearances in the UMCU dataset.
In Table 1, cardinality is the average number of
codes assigned to records in the dataset. Density is the
cardinality divided by the total number of codes. We
filtered out ICD codes with less than 50 observations
on their frequency. We note that there are
approximately 64 frequent labels with at least 200
records in UMCU dataset. ICD codes in this dataset
are mainly from chapters 4, 9, and 21. Figure 1
illustrates the ICD rolled-up codes with more than
400 appearance in the UMCU dataset. I25, Z95, I10,
I48 and I50 are the top frequent rolled-up codes (at
least 1000 counts) in our dataset.
In this study, we experimented with two versions
of the label set: one with the 22 ICD chapters and one
with the labels rolled up to their three-digit equivalent.
2.2 Preprocessing
Preprocessing the dataset of discharge letters
comprised the following steps: (i) we anonymize the
letters for legal and privacy reasons. We used
DEDUCE (Menger et al., 2018), a pattern matching
tool for automatic de-identification of Dutch medical
texts; (ii) we use the tm (Feinerer, 2018) and tidytext
(Silge and Robinson, 2016) packages in R to trim
whitespace, remove numbers, and convert all
characters to lower case; (iii) we tokenize all texts
using the Python scikit-learn (Pedregosa et al., 2011)
feature extractor, gensim library (Rehurek and Sojka,
2010) and the tokenizer in the keras library (Chollet
et al., 2015).
2.3 Classification Methods
To employ the classification methods, we investigate
two methods of vector representation:
• Bag-of-words (BOW; baseline)
• Word embeddings (average word vectors)
We use SVMs with each of the vector
representations. We also assess the following neural
network architecures for the automatic ICD coding of
the Dutch discharge letters.
• CNN
• LSTM and BiLSTM
• HA-GRU
With these deep learning architectures, the first
layer is the word embedding layer to represent
patients’ discharge letters. Hyperparameters of the
models are formulated on the corresponding cited
studies, while we tuned some based on the
development set using a random parameter search.
Automatic ICD-10 Classification of Diseases from Dutch Discharge Letters
283
2.3.1 Baseline: Support Vector Machines
using Bag-of-Words
We use a one-vs-all, multi-label binary SVM
classifier as the baseline learning method for ICD-10
classification. Baghdadi et al. (Baghdadi et al., 2019),
Koopman et al. (Koopman et al., 2015a), Mujtaba et
al. (Mujtaba et al., 2017) and Boytcheva (Boytcheva,
2011) applied SVM classifers for the task of ICD
coding. We calculate the BOW representations using
the preprocessed discharge letters. We also use the tf-
idf vectorizer. The baseline model fits a one-vs-all
binary SVM classifier with linear kernel for each ICD
code against the rest of the codes.
2.3.2 Word Embeddings: Support Vector
Machines using Average Word Vectors
Word embeddings (Mikolov et al., 2013a; Mikolov et
al., 2013b) are vector representations for texts,
representing words by capturing similarities between
them (for a recent review on word embeddings in
clinical natural language processing see
Subramanyam and Sivanesan, 2020). Skip-gram and
CBOW are two ways of learning word embeddings.
Both approaches use a simple neural network to
create a dense representation of words. The CBOW
tries to predict a word (target word) from the words
that appear around it (context), while skip-gram
inverts contexts and targets, and tries to predict
context from a given word. Baumel et al. (Baumel et
al. 2018) examined the word embedding
representations for ICD coding and achieved better
scores comparing to the BOW representations. In this
study, we train CBOW word embeddings in gensim.
We set the vector dimensionality to 300, the window
size to 5, and discard the words that appear only once
in the training set. We then use the average of word
embeddings to represent each discharge letter. These
embeddings are then inputs to the classification
model defined by the baseline SVM.
2.3.3 Convolutional Neural Networks
To be able to capture the order of the words as well
as multi-word expressions, the next model we
investigate is a CNN model. CNN has proven to be a
good method for text classification and is also applied
for the task of ICD coding (Baumel et al., 2018; Du
et al., 2019; Karimi et al., 2017). The CNN represents
texts at different levels of abstraction, essentially
choosing the most salient n-grams. We perform one
dimensional convolutions on the embedded
representations of the words. The architecture of this
model is very similar to the average word embeddings
model, but instead of averaging the embedded words
we apply a one dimensional convolution layer with
filter f, followed by a max pooling layer. One
dimensional convolution layers have proven effective
for deriving features from sequences data (Du et al.,
2019). In our experiments, we used the same
embedding parameters as in the average word
embeddings model. In addition, we set the number of
filters to 128, and the filter size to 5. On the output of
the max pooling layer, a fully connected neural
network (two dense layers) was applied for the
classification of the ICD-10 codes. The hidden dense
layer contains 128 units and uses the relu activation
function, and the output layer uses a softmax function
to determine if the ICD code should be assigned to the
letter. We also examine the CNN model with two
convolution layers and two max pooling layers. In
this setting, we employed a dropout layer after the
first max pooling layer with rate 0.15.
2.3.4 Long Short-term Memory and
Bidirectional Long Short-term
Memory
Feedforward neural networks require fixed length
contexts that need to be specified ad hoc before
training (Chung et al., 2014). For automated ICD
coding, this means that neural networks see relatively
few preceding words when predicting the next one.
RNNs avoid this problem by not consuming all the
input data at once (Chung et al., 2014; Mikolov et al.,
2010; Miranda et al., 2018). An RNN is a
straightforward adaptation of the standard feed
forward neural network to allow it to model
sequential data (Hochreiter and Schmidhuber, 1997;
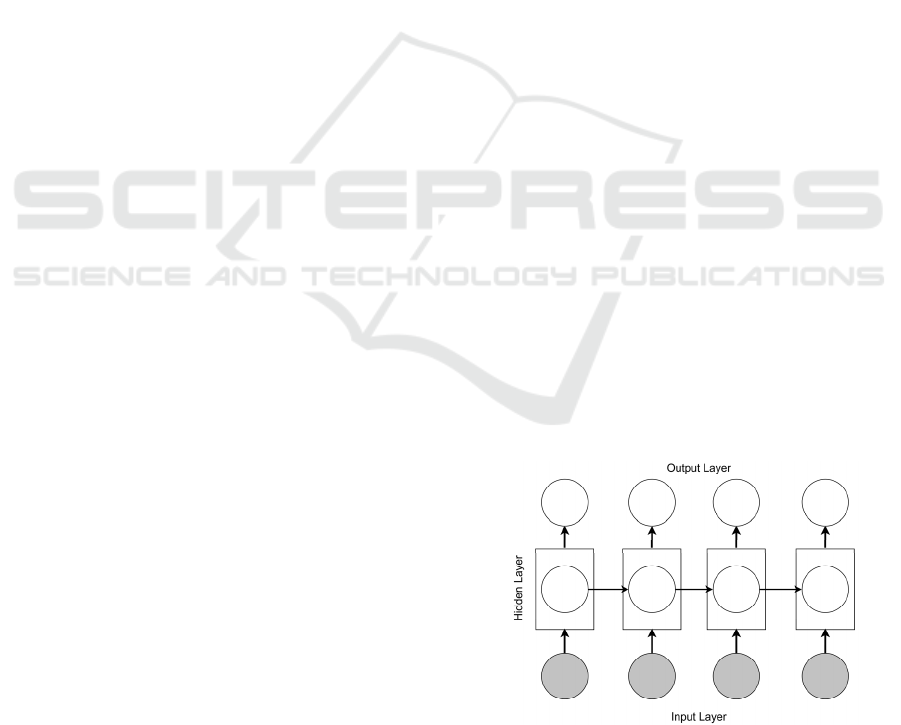
Sutskever et al., 2011). At each timestep, the RNN
receives an input, updates its hidden state, and makes
a prediction (see Figure 2).
Figure 2: RNN architecture overview.
By using recurrent connections, information can
cycle inside these networks for an arbitrarily long
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
284
time. LSTM (Hochreiter and Schmidhuber, 1997)
models are variants of RNNs with memory gates that
take a single input word at each time step and update
the models’ internal representation accordingly. RNN
is extended to use LSTM units, simply replacing the
nodes in hidden layers in Figure 2 with LSTM units.
To overcome the limitations in RNNs using all
available input information in the past and future of a
specific time frame, bidirectional LSTM (BiLSTM)
model is introduced by Schuster and Paliwal
(Schuster and Paliwal, 1997). The BiLSTM model as
shown in Figure 3 is an extension of the RNN model
using LSTM units, that combines two LSTMs with
one running forward in time and the other running
backward. Thus the context window around each
word consists of both information prior to and after
the current word.
Figure 3: BiLSTM architecture overview.
RNN models have been applied extensively on
textual data for natural language processing, as well
as in the medical domain and ICD coding (Atutxa et
al., 2019; Baumel et al., 2018; Du et al., 2019; Duarte
et al., 2018; Miranda et al., 2018; Nigam, 2016).
In this study, we used the keras library to
implement RNN models for automated ICD coding.
We implemented LSTM and BiLSTM. We keep the
same embedding parameters as in the average word
embeddings model. We experimented with RNN
models directly on the word sequence of all the
discharge letters. However, as in previous studies on
textual data, the fact that our data contains long texts
creates a challenge for preserving the gradient across
thousands of words. Therefore, we used dropout
layers to mask the network units randomly during the
training (Gal and Ghahramani, 2016). We set the
number of hidden units in the RNN layers at 100.
Dropout and recurrent dropout were added to avoid
overfitting, both at a 0.2 rate. On the output of the
recurrent layer, a fully connected neural network with
the setting in CNN was applied for classification of
the ICD-10 codes.
2.3.5 Hierarchical Attention Bidirectional
Gated Recurrent Unit
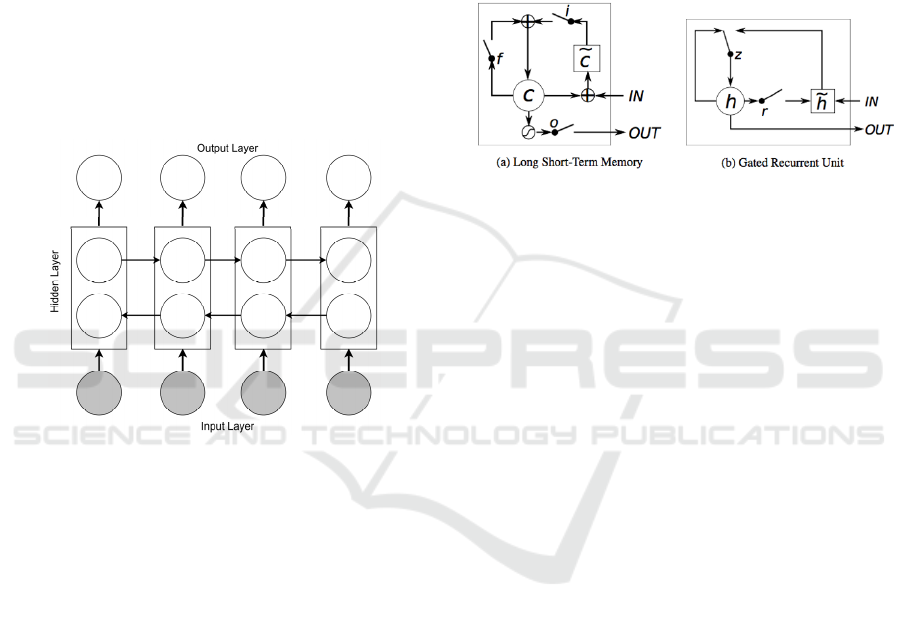
GRU can be considered as a variation on the LSTM,
that is a gating mechanism in RNN (Figure 4) aims to
solve the vanishing gradient problem (Cho et al.,
2014). Figure 4 compares the memory cell structures
of the LSTM and the GRU.
Figure 4: (a) LSTM memory cell: c is the memory cell, 𝑐̃ is
the new memory cell content. i, f and o are the input, forget
and output gates, respectively. (b) h and ℎ
are the activation
and candidate activation, respectively. r and z are the reset
and update gates.
The GRU has a slightly different architecture
where it combines both the input (gate i) and forget
(gate f) gates into a single gate called the update gate
(gate z). Also, it merges the cell state and the hidden
state. This results to a reduced number of parameters
as compared to LSTM architecture and in some cases
has resulted in faster convergence and a more
generalized model (Duarte et al., 2018).
Baumel et al. (Baumel et al., 2018) proposed a
HA-GRU model with label-dependent attention layer
to classify diseases codes. Since the GRU model is
too slow when applied to long documents as it
requires as many layers as of the document length,
they developed a HA-GRU to be able to handle multi-
label classification. In this paper, we implemented the
HA-GRU (Baumel et al., 2018) for the ICD-10
classification of cardiovascular diseases. The HA-
GRU is a hierarchical model with two levels of
bidirectional GRU encoding. The first bidirectional
GRU operates over tokens and encodes sentences.
The second bidirectional GRU encodes the entire
document, applied over the encoded sentences. In this
architecture, each GRU is applied to a much shorter
sequence compared with a single GRU.
We applied the HA-GRU model using the Dynet
deep learning library (Neubig et al., 2017) for ICD
coding. The attention mechanism in the HA-GRU has
the advantage that each label is invoked from
different parts of the text. This allows the model to
Automatic ICD-10 Classification of Diseases from Dutch Discharge Letters
285

focus on the relevant sentences for each label (Choi
et al., 2016). As for our previous deep learning
models, we kept the same embedding parameters in
the average word embeddings model. We used a
neural attention mechanism with 128 hidden units to
encode the bidirectional GRU outputs. The first GRU
layer encoded the sentences into a fixed length vector.
Then the second bidirectional GRU layer uses 128
attention layers to generate an encoding specific to
each class. Finally, we applied a fully connected layer
with softmax activation.
2.4 Evaluation Measures
Two evaluation measures are considered: accuracy,
and F1. In the single-label classification scenario,
accuracy is the fraction of correctly classified
discharge letters to the whole collection of discharge
letters. F1 is the harmonic mean of the fraction of
positively coded discharge letters and the fraction of
actual discharge letters that are positively classified.
Accuracy is a simple and intuitive measure, yet F1
takes both false positives and false negatives into
account. F1 score is a good measure for the ICD
classification task as this task has a large number of
catergories and usually contains imbalanced data. To
evaluate the multi-label classification performance,
we use the following sample-based metrics for
accuracy and F1:
𝐴𝑐𝑐𝑢𝑟𝑎𝑐𝑦 =
1
𝑛
|
𝑌
∩𝑍
|
|
𝑌
∪𝑍
|
𝐹1 =
1
𝑛
2
|
𝑌
∩𝑍
|
|
𝑌
|
+
|
𝑍
|
Where:
|
𝑌
|
= 𝑠𝑒𝑡 𝑜𝑓 𝑝𝑟𝑒𝑑𝑖𝑐𝑡𝑒𝑑 𝐼𝐶𝐷 𝑐𝑜𝑑𝑒𝑠
|
𝑍
|
= 𝑠𝑒𝑡 𝑜𝑓 𝑔𝑟𝑜𝑢𝑛𝑑 𝑡𝑟𝑢𝑡ℎ 𝐼𝐶𝐷 𝑐𝑜𝑑𝑒𝑠
𝑛 = 𝑛𝑢𝑚𝑏𝑒𝑟 𝑜𝑓 𝑠𝑎𝑚𝑝𝑙𝑒
We evaluate our experimental results in two
scenarios: (1) single-label prediction: a model assigns
one label to each patient letter; and (2) multi-label
prediction: a model assigns multiple labels per patient
letter.
3 RESULTS
We used the train-test split function from the model
selection module implemented in the scikit-learn
library to randomly split the dataset into train and test
sets. We separate 25% of the data as the test set and
the rest as for training. To evaluate the proposed
models on the dataset of cardiovascular discharge
letters, we conducted the following experiments. In
the first setting, we trained the models on the training
set separately using chapters as the labels. All models
were evaluated on the test set according to the
evaluation measures. In the second setting, we only
considered the rolled-up ICD-10 codes to their three-
digit codes.
3.1 Single-label Prediction
Performance
Table 2 presents the obtained results for each
model for both experimental settings (ICD chapters
and rolled-up ICD codes) on the single-label scenario.
In this case, a single code is predicted for every
testing patient’s letter. Bolded values in Table 2
indicate the best-performing model for each category.
Table 2: Single-label performance: accuracy and F1 score
on two settings (ICD chapters and rolled-up ICDs) for the
models when trained on the UMCU discharge letters.
ICD chapters
Rolled-up ICD
codes
Accurac
y
F1 Accurac
y
F1
BOW SVM
(
baseline
)
54.8 54.8 14.1 14.1
Average
word
embeddings
(
SVM
)
54.9 54.9 18.2 18.2
CNN(1conv) 57.3 49.2 22.1 17.4
CNN(2conv) 59.2 54.0 22.5 18.1
LSTM 73.0 38.1 19.1 14.1
BiLSTM 73.9 41.3 23.2 21.8
HA-GRU 72.5 43.5 23.7 19.8
BiLSTM gives the best accuracy in the ICD-10
chapters i.e. 73.9%, while the SVM classifier using
the average word embedding has the highest F1 score
of 54.9%. HA-GRU gives the best accuracy results in
the rolled-up ICD-10 setting i.e. 23.7%, while the
BiLSTM model has the highest value in F1 score with
21.8%.
Table 2 shows that the difference between the
results of the rolled-up ICDs and the ones for the
chapters is considerable. This is expected given the
large number of the rolled-up ICD codes comparing
to the number of the ICD chapters. We note that the
SVM classifier is still competitive with the deep
learning architectures in our application.
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
286

3.2 Multi-label Prediction Performance
Table 3 presents the results for the multi-label task. In
this scenario, corresponding to the prediction made
by the classification models, every ICD label that
presents a probability above a defined threshold is
considered as a predicted output code. We assign the
threshold in such a way that the label cardinality for
the test set is in the same order as the label cardinality
in the training set. Bolded values in Table 3 indicate
the best-performing model for each category.
Table 3: Multi-label performance: accuracy and F1 score
on two settings for the models when trained on the UMCU
discharge letters.
ICD chapters
Rolled-up ICD
codes
Accuracy F1 Accuracy F1
BOW SVM
(baseline)
62.3 74.3 11.6 20.2
Average
word
embeddings
(SVM)
60.4 72.6 12.5 25.8
CNN(1conv) 38.1 46.3 09.0 16.1
CNN(2conv) 42.2 49.0 12.4 19.1
LSTM 53.4 59.6 11.7 18.8
BiLSTM 55.0 70.1 13.7 23.2
HA-GRU 56.8 71.3 15.9 24.3
For the multi-label scenario, the SVM classifier
gives the best results in F1 score for the chapter labels
and for the rolled-up codes with values equal to
74.3% and 25.8%, respectively. The former is the F1
score for the BOW representation and the latter is the
one for the word embeddings. In terms of accuracy,
when the number of ICDs to be coded are large the
HA-GRU has the best results with 15.9%.
By comparing Table 2 and Table 3, it is notable
that the difference between the results on chapters and
the results on the rolled-up codes is more consistent
when we applied the CNN models using our case
study. With regard to the single-label task, CNNs
have the highest values of F1 of about 54% and
18.1%, respectively, for the ICD chapters and the
rolled-up codes. For the multi-label task these values
are equal to 49% and 19.1%.
4 DISCUSSION
Automated ICD-10 classification can potentially save
valuable time and resources in a clinical setting. In
this study, we compared several state-of-the-art ICD
coding systems on a dataset of Dutch-language
discharge letters.
Classification performance of the 22 higher-level
codes is very promising, especially when only a
single label is considered. For this version of the task,
RNNs (LSTM, BiLSTM, and HA-GRU) showed
good performance, as reported in the literature.
However, in many practical applications, including
our own, a lower level of classification is required,
and each letter receives multiple ICD codes. For this
version of the task, performance was somewhat
disappointing, and state-of-the-art systems failed to
outperform the baseline BOW SVM with linear
kernel. An exception is the HA-GRU system, which
had the best accuracy, and showed an F1 performance
close to that of the baseline.
While none of the systems were able to achieve a
level of classification accuracy on the most difficult
versions of the ICD classification task that would
allow them to completely replace a human coder,
they do show performance that is good enough to
suggest codes in an interaction with the human.
Future work could investigate the performance of
human-in-the-loop systems, for example by
employing active learning.
A question that may arise is whether machine
learning could be supplanted with a rule-based
system. This is possible for the higher-level codes
using information retrieval and natural language
processing methods (Pakhomov et al., 2006).
However, developing rule-based systems with
manually coded rules is tremendously difficult for the
lower levels of ICDs. There are a large number of
ICD codes in lower levels of the ICD hierarchy, and
a small number of observations per ICD code. Deep
learning-based models are useful here because they
obviate the need for manual feature engineering
(Atutxa et al., 2019). For this reason, we believe
machine learning remains an attractive alternative to
rule-based systems.
A second consideration is the question of model
interpretability. Here, the deep learning models that
form the current state of the art are especially
challenging in this regard, and this may be a point in
favor of “simpler” methods such as BOW: the more
opaque the model, the less willing clinicians may be
to accept artificial intelligence recommendations.
Although it is not clear whether this is a problem for
ICD-10 coding specifically, future work could focus
Automatic ICD-10 Classification of Diseases from Dutch Discharge Letters
287

on developing more interpretable systems or generic
prediction explanation methods that mitigate this
problem. Moreover, such systems could be very
powerful when combined with a human-in-the-loop
approach, by allowing the human to learn how text
can be written to teach the correct code to the system.
REFERENCES
Atutxa, A., de Ilarraza, A.D., Gojenola, K., Oronoz, M.,
Perez-de-Viñaspre, O., 2019. Interpretable deep
learning to map diagnostic texts to ICD-10
codes. International Journal of Medical
Informatics, 129, pp.49-59.
Baghdadi, Y., Bourrée, A., Robert, A., Rey, G., Gallay, A.,
Zweigenbaum, P., Grouin, C., Fouillet, A., 2019.
Automatic classification of free-text medical causes
from death certificates for reactive mortality
surveillance in France. International journal of medical
informatics, 131, p.103915.
Baumel, T., Nassour-Kassis, J., Cohen, R., Elhadad, M.,
Elhadad, N., 2018, June. Multi-label classification of
patient notes: case study on ICD code assignment.
In Workshops at the Thirty-Second AAAI Conference
on Artificial Intelligence.
Boytcheva, S., 2011, September. Automatic matching of
ICD-10 codes to diagnoses in discharge letters.
In Proceedings of the Second Workshop on Biomedical
Natural Language Processing (pp. 11-18).
Cao, L., Gu, D., Ni, Y., Xie, G., 2019. Automatic ICD Code
Assignment based on ICD’s Hierarchy Structure for
Chinese Electronic Medical Records. AMIA Summits
on Translational Science Proceedings, 2019, p.417.
Chen, Y., Lu, H., Li, L., 2017. Automatic ICD-10 coding
algorithm using an improved longest common
subsequence based on semantic similarity. PloS
one, 12(3), p.e0173410.
Cho, K., Van Merriënboer, B., Gulcehre, C., Bahdanau, D.,
Bougares, F., Schwenk, H. and Bengio, Y., 2014.
Learning phrase representations using RNN encoder-
decoder for statistical machine translation. arXiv
preprint arXiv:1406.1078.
Choi, E., Bahadori, M.T., Schuetz, A., Stewart, W.F. and
Sun, J., 2016, December. Doctor ai: Predicting clinical
events via recurrent neural networks. In Machine
Learning for Healthcare Conference (pp. 301-318).
Chollet, F., and others, 2015. Keras, https://keras.io.
Chung, J., Gulcehre, C., Cho, K., Bengio, Y., 2014.
Empirical evaluation of gated recurrent neural networks
on sequence modeling. arXiv preprint arXiv:1412.
3555.
Du, J., Chen, Q., Peng, Y., Xiang, Y., Tao, C., Lu, Z., 2019.
ML-Net: multi-label classification of biomedical texts
with deep neural networks. Journal of the American
Medical Informatics Association, 26(11), pp.1279-
1285.
Duarte, F., Martins, B., Pinto, C.S., Silva, M.J., 2018. Deep
neural models for ICD-10 coding of death certificates
and autopsy reports in free-text. Journal of biomedical
informatics, 80, pp.64-77.
Feinerer, I., 2018. Introduction to the tm Package Text
Mining in R. Retrieved, March 1, p.2019.
Gal, Y. and Ghahramani, Z., 2016. A theoretically
grounded application of dropout in recurrent neural
networks. In Advances in neural information
processing systems (pp. 1019-1027).
Gargiulo, F., Silvestri, S., Ciampi, M., 2018. Deep
Convolution Neural Network for Extreme Multi-label
Text Classification. In HEALTHINF (pp. 641-650).
Hochreiter, S. and Schmidhuber, J., 1997. Long short-term
memory. Neural computation, 9(8), pp.1735-1780.
Karimi, S., Dai, X., Hassanzadeh, H., Nguyen, A., 2017,
August. Automatic diagnosis coding of radiology
reports: a comparison of deep learning and
conventional classification methods. In BioNLP 2017
(pp. 328-332).
Kemp, J., Rajkomar, A., Dai, A.M., 2019. Improved Patient
Classification with Language Model Pretraining Over
Clinical Notes. arXiv preprint arXiv:1909.03039.
Koh, P.W. and Liang, P., 2017, August. Understanding
black-box predictions via influence functions.
In Proceedings of the 34th International Conference on
Machine Learning-Volume 70 (pp. 1885-1894). JMLR.
org.
Koopman, B., Karimi, S., Nguyen, A., McGuire, R.,
Muscatello, D., Kemp, M., Truran, D., Zhang, M.,
Thackway, S., 2015. Automatic classification of
diseases from free-text death certificates for real-time
surveillance. BMC medical informatics and decision
making, 15(1), p.53.
Koopman, B., Zuccon, G., Nguyen, A., Bergheim, A.,
Grayson, N., 2015. Automatic ICD-10 classification of
cancers from free-text death certificates. International
journal of medical informatics, 84(11), pp.956-965.
Lin, C., Lou, Y.S., Tsai, D.J., Lee, C.C., Hsu, C.J., Wu,
D.C., Wang, M.C., Fang, W.H., 2019. Projection Word
Embedding Model with Hybrid Sampling Training for
Classifying ICD-10-CM Codes: Longitudinal
Observational Study. JMIR medical informatics, 7(3),
p.e14499.
Liu, J., Zhang, Z., Razavian, N., 2018. Deep ehr: Chronic
disease prediction using medical notes. arXiv preprint
arXiv:1808.04928.
Menger, V., Scheepers, F., van Wijk, L.M., Spruit, M.,
2018. DEDUCE: A pattern matching method for
automatic de-identification of Dutch medical
text. Telematics and Informatics, 35(4), pp.727-736.
Mikolov, T., Chen, K., Corrado, G. and Dean, J., 2013.
Efficient estimation of word representations in vector
space. arXiv preprint arXiv:1301.3781.
Mikolov, T., Karafiát, M., Burget, L., Černocký, J.,
Khudanpur, S., 2010. Recurrent neural network based
language model. In Eleventh annual conference of the
international speech communication association.
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S. and
Dean, J., 2013. Distributed representations of words
and phrases and their compositionality. In Advances in
C2C 2020 - Workshop on COMP2CLINIC: Biomedical Researchers Clinicians Closing The Gap Between Translational Research And
Healthcare Practice
288

neural information processing systems (pp. 3111-
3119).
Miranda, R., Martins, B., Silva, M., Silva, N., Leite, F.,
2018. Deep Learning for Multi-Label ICD-9
Classification of Hospital Discharge Summaries, Thesis
report, University of Lisbon, Lisbon, Portugal.
Molnar, C., 2019. Interpretable machine learning. Lulu.
com.
Mujtaba, G., Shuib, L., Raj, R.G., Rajandram, R., Shaikh,
K., Al-Garadi, M.A., 2017. Automatic ICD-10 multi-
class classification of cause of death from plaintext
autopsy reports through expert-driven feature selection.
PloS one, 12(2), p.e0170242.
Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., Eisenstein,
J., 2018. Explainable prediction of medical codes from
clinical text. arXiv preprint arXiv:1802.05695.
Neubig, G., Dyer, C., Goldberg, Y., Matthews, A., Ammar,
W., Anastasopoulos, A., Ballesteros, M., Chiang, D.,
Clothiaux, D., Cohn, T. and Duh, K., 2017. Dynet: The
dynamic neural network toolkit. arXiv preprint
arXiv:1701.03980.
Nguyen, A.N., Truran, D., Kemp, M., Koopman, B.,
Conlan, D., O’Dwyer, J., Zhang, M., Karimi, S.,
Hassanzadeh, H., Lawley, M.J., Green, D., 2018.
Computer-Assisted Diagnostic Coding: Effectiveness
of an NLP-based approach using SNOMED CT to ICD-
10 mappings. In AMIA Annual Symposium
Proceedings (Vol. 2018, p. 807). American Medical
Informatics Association.
Nigam, P., 2016. Applying deep learning to ICD-9 multi-
label classification from medical records. Technical
report, Stanford University.
Pakhomov, S.V., Buntrock, J.D., Chute, C.G., 2006.
Automating the assignment of diagnosis codes to
patient encounters using example-based and machine
learning techniques. Journal of the American Medical
Informatics Association, 13(5), pp.516-525.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V. and Vanderplas, J., 2011.
Scikit-learn: Machine learning in Python. Journal of
machine learning research, 12(Oct), pp.2825-2830.
Rehurek, R. and Sojka, P., 2010. Software framework for
topic modelling with large corpora. In Proceedings of
the LREC 2010 Workshop on New Challenges for NLP
Frameworks, pp. 45-50.
Schuster, M. and Paliwal, K.K., 1997. Bidirectional
recurrent neural networks. IEEE Transactions on
Signal Processing, 45(11), pp.2673-2681.
Shickel, B., Tighe, P.J., Bihorac, A., Rashidi, P., 2017.
Deep EHR: a survey of recent advances in deep
learning techniques for electronic health record (EHR)
analysis. IEEE journal of biomedical and health
informatics, 22(5), pp.1589-1604.
Shing, H.C., Wang, G., Resnik, P., 2019. Assigning
Medical Codes at the Encounter Level by Paying
Attention to Documents.
arXiv preprint
arXiv:1911.06848.
Silge, J. and Robinson, D., 2016. tidytext: Text Mining and
Analysis Using Tidy Data Principles in R. J. Open
Source Software, 1(3), p.37.
Subramanyam, K.K., Sivanesan, S., 2020. SECNLP: A
Survey of Embeddings in Clinical Natural Language
Processing. Journal of biomedical informatics,
p.103323.
Sutskever, I., Martens, J. and Hinton, G.E., 2011.
Generating text with recurrent neural networks.
In Proceedings of the 28th International Conference on
Machine Learning (ICML-11) (pp. 1017-1024).
Xiao, C., Choi, E. and Sun, J., 2018. Opportunities and
challenges in developing deep learning models using
electronic health records data: a systematic
review. Journal of the American Medical Informatics
Association, 25(10), pp.1419-1428.
Xie, X., Xiong, Y., Yu, P.S., Zhu, Y., 2019, November.
EHR Coding with Multi-scale Feature Attention and
Structured Knowledge Graph Propagation.
In Proceedings of the 28th ACM International
Conference on Information and Knowledge
Management (pp. 649-658). ACM.
Yang, Z., Yang, D., Dyer, C., He, X., Smola, A., Hovy, E.,
2016, June. Hierarchical attention networks for
document classification. In Proceedings of the 2016
conference of the North American chapter of the
association for computational linguistics: human
language technologies (pp. 1480-1489).
Zweigenbaum, P., Lavergne, T., 2016, November. Hybrid
methods for ICD-10 coding of death certificates.
In Proceedings of the Seventh International Workshop
on Health Text Mining and Information Analysis (pp.
96-105).
Automatic ICD-10 Classification of Diseases from Dutch Discharge Letters
289
