
A Deep-learning based Method for the Classification of the Cellular
Images
Caleb Vununu
1
, Suk-Hwan Lee
2
and Ki-Ryong Kwon
1
1
Department of IT Convergence and Application Engineering, Pukyong National University, Busan, Republic of Korea
2
Department of Information Security, Tongmyong University, Busan, Republic of Korea
Keywords: Deep-learning, Classification of Cellular Images, Convolutional Neural Networks (CNN), HEp-2 Cell Images,
Deep Convolutional Auto-Encoders (DCAE), SNPHEp-2 Dataset.
Abstract: The present work proposes a classification method for the Human Epithelial of type 2 (HEp-2) cell images
using an unsupervised deep feature learning method. Unlike most of the state-of-the-art methods in the
literature that utilize deep learning in a strictly supervised way, we propose here the use of the deep
convolutional autoencoder (DCAE) as the principal feature extractor for classifying the different types of the
HEp-2 cellular images. The network takes the original cellular images as the inputs and learns how to
reconstruct them through an encoding-decoding process in order to capture the features related to the global
shape of the cells. A final feature vector is constructed by using the latent representations extracted from the
DCAE, giving a highly discriminative feature representation. The created features will then be fed to a
nonlinear classifier whose output will represent the final type of the cell image. We have tested the
discriminability of the proposed features on one of the most popular HEp-2 cell classification datasets, the
SNPHEp-2 dataset and the results show that the proposed features manage to capture the distinctive
characteristics of the different cell types while performing at least as well as some of the actual deep learning
based state-of-the-art methods.
1 INTRODUCTION
Computer-aided diagnostic (CAD) systems have
gained tremendous interests since the unfolding of
various machine learning techniques in the past
decades. They comprise all the systems that aim to
consolidate the automation of the disease diagnostic
procedures. One of the most challenging tasks
regarding those CAD systems is the complete
analysis and understanding of the images
representing the biological organisms. In case of the
autoimmune diseases, the automatic classification of
the different types of the Human Epithelial type 2
(HEp-2) cell patterns is one of the most important
steps of the diagnosis procedure.
Automatic feature learning methods have been
widely adopted since the unfolding of deep learning
(LeCun et al., 2015). They have shown outstanding
results in the object recognition problems (LeCun et
al., 2004; He et al., 2016) and many researchers have
adopted them as principal tool for the HEp-2 cell
classification problem (Gao et al., 2016). Unlike
conventional methods whose accuracy depends on
the subjective choice of the features, deep learning
methods, such as deep convolutional neural networks
(CNNs), have the advantage of offering an automatic
feature learning process. In fact, many works have
demonstrated the superiority of the deep learning
based features over the hand-crafted ones for the
HEp-2 cell classification task. Although the
performance obtained with the supervised learning
methodology continues to reach impressive levels,
the exigency of always having labelled datasets in
hand, knowing that deep-learning methods
necessitate huge amount of images, can represent a
relative drawback for these methods.
We propose an unsupervised deep feature learning
process that uses the deep convolutional autoencoder
(DCAE) as the principal feature extractor. The
DCAE, which learns to reproduce the original cellular
images via a deep encoding-decoding scheme, is used
for extracting the features. The DCAE takes the
original cell image as an input and will learn to
reproduce it by extracting the meaningful features
needed for the discrimination part of the method. The
latent representations trapped between the encoder
242
Vununu, C., Lee, S. and Kwon, K.
A Deep-learning based Method for the Classification of the Cellular Images.
DOI: 10.5220/0009183702420245
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 242-245
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

and the decoder of the DCAE will be extracted and
used as the final high-level features of the system.
The DCAE will help to encode the geometrical
details of the cells contained in the original pictures.
The discrimination potentiality carried by the
extracted features allows us to feed them as the inputs
of a shallow nonlinear classifier, which will certainly
find a way to discriminate them. The proposed
method was tested on the SNP HEp-2 Cell dataset
(Wiliem et al.) and the results show that the proposed
features outperform by far the conventional and
popular handcrafted features and perform at least as
well as the state-of-the-art supervised deep learning
based methods.
2 PROPOSED METHODOLOGY
Auto-encoders (Hinton et al.) are unsupervised
learning methods that are used for the purpose of
feature extraction and dimensionality reduction of
data. Neural network based auto-encoder consists of
an encoder and a decoder. The encoder takes an input
of dimension d, and maps it to a hidden
representation , of dimension r, using a
deterministic mapping function such that:
y
= f(Wx + b) (1)
where the parameters W and b are the weights and
biases associated with the encoder. The decoder then
takes the output of the encoder and uses the same
mapping function in order to provide a
reconstruction that must be of the same shape or in
the same form (which means almost equal to) as the
original input signal . Using equation (1), the output
of the decoder is also given by:
z = f(W’x + b’) (2)
where the parameters W’ and b’ are the weights and
bias associated with the decoder layer. Finally, the
network must learn the parameters W, W’, b and b’
so that z must be close or, if possible, equal to x. In
final, the network leans to minimize the differences
between the encoder’s input x and the decoder’s
output.
This encoding-decoding process can be done with
the use of convolutional neural networks by using
what we call the deep convolutional autoencoder
(DCAE). Unlike conventional neural networks,
where you can set the size of the output that you want
to get, the convolutional neural networks are
characterized by the process of down-sampling,
accomplished by the pooling layers, which are
incorporated in their architecture. And this sub-
sampling process has as consequence the loss of the
input’s spatial information while we go deeper inside
the network.
To tackle this problem, we can use DCAE instead
of conventional convolutional neural networks. In the
DCAE, after the down-sampling process
accomplished by the encoder, the decoder tries to up-
sample the representation until we reconstruct the
original size. This can be made by backwards
convolution often called “deconvolution” operations.
The final solution of the network can be written in the
form:
, ’, , ’
argmin
,
,,
,
(3)
where z denotes the decoder’s output and x is the
original image. The function L in equation (3)
estimates the differences between the x and z. So, the
solution of equation (3) represents the parameter
values that minimize the most the difference between
input x and the reconstruction z.
In our experiments, the feature vectors extracted
from the DCAE contain 4096 elements. The second
part of the method consists of giving this feature
vector to a shallow artificial neural network (ANN).
Finally, in order to predict the cell type, a supervised
learning process will be conducted using the extracted
features from the DCAE as the inputs and a 2 layered
ANN as the classifier.
3 RESULTS AND DISCUSSION
There are 1,884 cellular images in the dataset, all of
them extracted from the 40 different specimen
images. Different specimens were used for
constructing the training and testing image sets, and
both sets were created in such a way that they cannot
contain images from the same specimen. From the 40
specimen, 20 were used for the training sets and the
remaining 20 were used for the testing sets. In total
there are 905 and 979 cell images for the training and
testing sets, respectively. Each set (training and
testing) contains five-fold validation splits of
randomly selected images. In each set, the different
splits are used for cross validating the different
models, each split containing 450 images
approximatively. The SNPHEp-2 dataset was
presented by Wiliem et al. (2016). Figure 1 shows the
example images of the five different cell types
randomly selected from the dataset.
As previously mentioned, the created feature
vectors extracted from the DCAE contain 4096
elements. So, our network will have 4096 neurons in
A Deep-learning based Method for the Classification of the Cellular Images
243
the input layer. The best results were obtained using
a 4096-250-50-5 architecture, meaning that we have
4096 neurons in the input layer, 250 neurons in the
first hidden layer, 50 neurons in the second hidden
layer and a final layer containing 5 neurons
corresponding to the 5 cell types of our dataset. The
total accuracy reached by the network was 88.08 %.
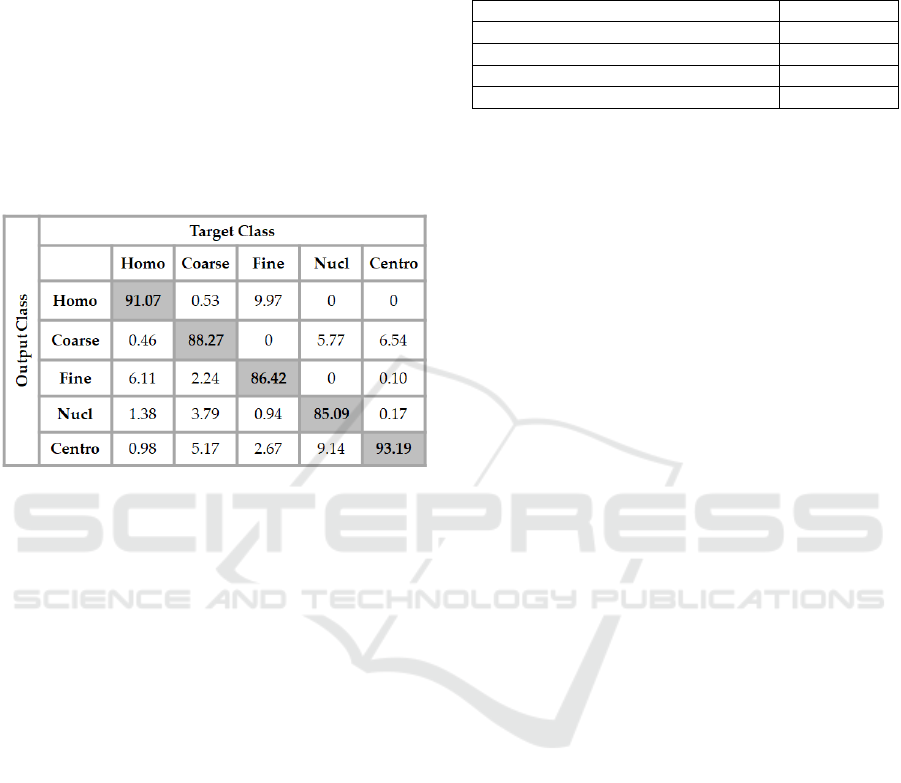
The details of the results are shown in the confusion
matrix depicted in Figure 1. In the figure, ‘Homo”,
“Coarse”, “Fine”, “Nucl” and “Centro” denote the
homogeneous, the coarse speckled, the fine speckled,
the nucleolar and the centromere cell types,
respectively.
Figure 1: Confusion matrix of the results obtained with a
4096-250-50-5 neural network using the extracted features
from the DCAE as the inputs.
In the confusion matrix in Figure 1, we can see
that the most distinguishable cells for the classifier
are the centromere cells, for which the classification
accuracy reaches 93.19 %. But, in the same time, we
can notice that there is a significant confusion
between the centromere and the coarse speckled cells:
6.54 % of the coarse speckled cells were misclassified
as centromere. The confusion is confirmed by also
taking a look at the classification rate of the coarse
speckled: 5.17 % of them were misclassified as
centromere, as we can see in the second column (fifth
row) of the confusion matrix.
The homogeneous cells also are well classified in
general, over 91 % of them were correctly recognized
by the classifier. Another important confusion comes
between the homogeneous and fine speckled cells. As
we can notice in the confusion matrix, 6.11 % of the
homogeneous cells were misclassified as fine
speckled. And in the case of the fine speckled cells,
the confusion is even more noticeable. We can see in
the matrix that almost 10% (9.97) of the fine speckled
were misclassified as homogeneous. Trying to
decrease the confusion between the cells that show
strong similarities in terms of shape and intensity
level can be the direction of any consideration about
the future works. As mentioned before, the overall
classification rate of the proposed method is 88.08 %.
Table 1: Comparative results.
Method Accuracy
Texture features + SVM 80.90%
LPB descri
p
tors + SVM 85.71%
5 la
y
ers CNN 86.20%
Present work
(
DCAE features + ANN
)
88.08%
We have conducted a comparative study with the
handcrafted features and one deep learning method
using the CNN in a strictly supervised manner for the
classification of the cellular images. The results of the
comparative study are shown in Table 1. We can
clearly see that the proposed method outperforms the
handcrafted features. The proposed features from the
DCAE perform also slightly better than the
supervised deep-learning method proposed by Gao et
al. (2016) using a 5 layers’ network.
4 CONCLUSIONS
We have presented a cell classification method for the
images portraying the microscopy data, the HEp-2
cells, a method that has adopted the DCAE as the
principal feature extractor. Unlike most of the
methods in the literature that are based on the
supervised learning, we have used the DCAE in order
to construct the feature vectors in an unsupervised
way. These obtained vectors were then given to a
nonlinear classifier whose outputs determine the cell
type of the image. The results show that the proposed
feature extraction method really captures the
characteristics of each cell type. The comparative
study demonstrates that our proposed features
perform far better than the handcrafted ones and
slightly better than the supervised deep learning
method.
But, as we have discussed in the results, many cell
types exhibit strong similarities between them in
terms of shape and intensity level. These similarities
encourage a significant confusion during the
discrimination step of the proposed features. We
consider that the next step of our work is to try to find
a way of minimizing the confusion between these
cells that show strong similarities.
ACKNOWLEDGEMENTS
This research was supported by the MSIT (Ministry
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
244

of Science and ICT), Korea, under the ICT
Consilience Creative program (IITP-2019-2016-0-
00318) supervised by the IITP (Institute for
Information & communications Technology Planning
& Evaluation), and Basic Science Research Program
through the National Research Foundation of Korea
(NRF) funded by the Ministry of Science, ICT &
Future Planning (2016R1D1A3B03931003, No.
2017R1A2B2012456), and Ministry of Trade,
Industry and Energy for its financial support of the
project titled “the establishment of advanced marine
industry open laboratory and development of realistic
convergence content”.
REFERENCES
Gao, Z., Wang, L., Zhou, L., Zhang, J., 2016. Hep-2 cell
image classification with deep convolutional neural
networks. IEEE Journal of Biomedical and Health
Informatics, 21(2), 416-428.
He, K., Zhang, X., Ren, S., Sun, J., 2016. Deep residual
learning for image recognition. In CVPR’2016, 2016
IEEE Conference on Computer Vision and Pattern
Recognition (pp. 770-778).
Hinton, G.E., Sa1akhutdinov, R.R., 2006. Reducing the
dimensionality of the data with neural networks.
Nature, 313(5786), 504-507.
LeCun, Y., Bengio, Y., Hinton, G., 2015. Deep Learning.
Nature, 521, 436-444.
LeCun, Y., Huang, F.J., Bottou, L. Learning methods for
generic object recognition with invariance to pose and
lighting. In CVPR’04, 2004 IEEE Computer Society
Conference on Computer Vision and Pattern
Recognition.
Wiliem, A., Wong, Y., Sanderson, C., Hobson, P., Chen, S.,
Lovell, B.C. Classification of human epithelial type 2
cell indirect immunofluorescence images via codebook
based descriptors. In WACV’13, 2013 IEEE Workshop
on Applications of Computer Vision (pp. 95-102).
A Deep-learning based Method for the Classification of the Cellular Images
245
